




















中国农业科技导报 ›› 2022, Vol. 24 ›› Issue (4): 63-74.DOI: 10.13304/j.nykjdb.2021.0926
李美丽1( ), 宿俊吉1, 杨永林2, 秦江鸿2, 李鲜鲜1, 杨德龙1(
), 宿俊吉1, 杨永林2, 秦江鸿2, 李鲜鲜1, 杨德龙1( ), 马麒3(
), 马麒3( ), 王彩香1(
), 王彩香1( )
)
收稿日期:2021-11-02
接受日期:2021-12-09
出版日期:2022-04-15
发布日期:2022-04-19
通讯作者:
杨德龙,马麒,王彩香
作者简介:李美丽 E-mail: 2538268277@qq.com
基金资助:
Meili LI1( ), Junji SU1, Yonglin YANG2, Jianghong QIN2, Xianxian LI1, Delong YANG1(
), Junji SU1, Yonglin YANG2, Jianghong QIN2, Xianxian LI1, Delong YANG1( ), Qi MA3(
), Qi MA3( ), Caixiang WANG1(
), Caixiang WANG1( )
)
Received:2021-11-02
Accepted:2021-12-09
Online:2022-04-15
Published:2022-04-19
Contact:
Delong YANG,Qi MA,Caixiang WANG
摘要:
COI1(COR-insensitive 1)与Skp1-Cullin-F-box蛋白组成茉莉素信号转导的核心组分SCFCOI1复合物,在植物生长发育和响应逆境胁迫中发挥重要作用。从全基因组水平上鉴定出48个陆地棉COI家族基因,其不均匀地分布在21条染色体上,主要定位于细胞核和细胞质中。根据系统发育关系将其分为3个亚族(Ⅰ~Ⅲ),共线性分析发现COI基因以片段复制方式在陆地棉中进行扩张。陆地棉COI基因含有1~9个外显子,其启动子区域共存在6种逆境胁迫响应元件。转录组数据显示,分别有66.7%、56.25%、68.8%和27.1%的基因在NaCl、PEG、热和冷胁迫下表达量升高,且部分基因同时响应多种非生物胁迫;qRT-PCR验证结果显示,GhCOI5-A06强烈响应NaCl和PEG胁迫,是潜在的候选基因,表明GhCOI参与非生物胁迫应答。上述结果为后续深入解析该家族基因的功能奠定基础。
中图分类号:
李美丽, 宿俊吉, 杨永林, 秦江鸿, 李鲜鲜, 杨德龙, 马麒, 王彩香. 陆地棉COI家族基因鉴定及在干旱和盐胁迫下的表达分析[J]. 中国农业科技导报, 2022, 24(4): 63-74.
Meili LI, Junji SU, Yonglin YANG, Jianghong QIN, Xianxian LI, Delong YANG, Qi MA, Caixiang WANG. Identification of COI Family Genes and Their Expression in Gossypium hirsutum L. Under Drought and Salt Stress[J]. Journal of Agricultural Science and Technology, 2022, 24(4): 63-74.
引物名称 Primer name | 正向引物 Forward primer(5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| GhCOI1⁃D01 | GTGACTTGACCGATGCAGGA | GAAAGCGCAAGAACCACAGG |
| GhCOI1⁃D06 | CTCGTTTCGATGCTGTCACG | CGAGTCAAATTCCGGCAACG |
| GhCOI3⁃D04 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI3⁃A05 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI5⁃A05 | CAGCTCTGCGTTCTGGTTTG | AGCAATCTCCTGGCAACCAT |
| GhCOI5⁃A06 | TGGCGGAGCAAATGTCTGAT | ACTCGGAATGACCCGAAACC |
| GhCOI7⁃A07 | CCGGCGACAGTAACAAAGGA | GGACATCCAAAGGGATCGCA |
| GhCOI9⁃D08 | AGCTCGTAAGCCAGTTTCCC | CCTCAACGGCCAACAATTCG |
| GhCOI10⁃A09 | TCAAGTTCCTCGCTCAACGG | TCATCTCTGCGACGCCTTTT |
| GhCOI10⁃D09 | TTTGGCAACTGAGTGTGGGA | GCCTCCAGTGATTTGTCGGT |
| GhActin | ATCCTCCGTCTTGACCTTG | TGTCCGTCAGGCAACTCAT |
表1 实时荧光定量PCR引物
Table 1 Primer sequence for qRT?PCR
引物名称 Primer name | 正向引物 Forward primer(5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| GhCOI1⁃D01 | GTGACTTGACCGATGCAGGA | GAAAGCGCAAGAACCACAGG |
| GhCOI1⁃D06 | CTCGTTTCGATGCTGTCACG | CGAGTCAAATTCCGGCAACG |
| GhCOI3⁃D04 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI3⁃A05 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI5⁃A05 | CAGCTCTGCGTTCTGGTTTG | AGCAATCTCCTGGCAACCAT |
| GhCOI5⁃A06 | TGGCGGAGCAAATGTCTGAT | ACTCGGAATGACCCGAAACC |
| GhCOI7⁃A07 | CCGGCGACAGTAACAAAGGA | GGACATCCAAAGGGATCGCA |
| GhCOI9⁃D08 | AGCTCGTAAGCCAGTTTCCC | CCTCAACGGCCAACAATTCG |
| GhCOI10⁃A09 | TCAAGTTCCTCGCTCAACGG | TCATCTCTGCGACGCCTTTT |
| GhCOI10⁃D09 | TTTGGCAACTGAGTGTGGGA | GCCTCCAGTGATTTGTCGGT |
| GhActin | ATCCTCCGTCTTGACCTTG | TGTCCGTCAGGCAACTCAT |
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI1⁃A01 | GH_A01G1190 | 534 | 57.36 | 8.7 | 叶绿体、细胞核Chloroplast, nucleus |
| GhCOI1⁃D01 | GH_D01G1258 | 534 | 57.54 | 8.82 | 细胞核Nucleus |
| GhCOI1⁃A05 | GH_A05G2065 | 534 | 57.38 | 8.86 | 细胞核Nucleus |
| GhCOI1⁃D05 | GH_D05G2095 | 535 | 57.44 | 8.83 | 细胞核Nucleus |
| GhCOI1⁃A06 | GH_A06G0210 | 528 | 56.82 | 8.74 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI1⁃D06 | GH_D06G0195 | 528 | 56.7 | 8.86 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI2⁃A02 | GH_A02G1803 | 544 | 58.97 | 8.49 | 细胞核Nucleus |
| GhCOI2⁃D03 | GH_D03G0256 | 514 | 55.87 | 7.87 | 细胞质Cytoplasm |
| GhCOI2⁃A10 | GH_A10G0913 | 545 | 59.31 | 8.6 | 细胞质Cytoplasm |
| GhCOI2⁃D10 | GH_D10G1048 | 545 | 59.21 | 8.59 | 细胞质Cytoplasm |
| GhCOI3⁃D04 | GH_D04G0696 | 591 | 66.99 | 5.98 | 细胞质Cytoplasm |
| GhCOI3⁃A05 | GH_A05G3439 | 589 | 66.74 | 5.81 | 细胞质Cytoplasm |
| GhCOI4⁃A05 | GH_A05G0481 | 362 | 39.99 | 6.32 | 细胞质Cytoplasm |
| GhCOI4⁃D05 | GH_D05G0480 | 362 | 40.08 | 6.96 | 细胞质Cytoplasm |
| GhCOI5⁃A05 | GH_A05G2217 | 611 | 68.38 | 5.97 | 细胞核Nucleus |
| GhCOI5⁃D05 | GH_D05G2242 | 611 | 68.32 | 5.88 | 叶绿体Chloroplast |
| GhCOI5⁃A06 | GH_A06G0343 | 615 | 69.03 | 5.67 | 细胞核Nucleus |
| GhCOI5⁃D06 | GH_D06G0328 | 615 | 69.11 | 5.54 | 细胞核Nucleus |
| GhCOI6⁃A06 | GH_A06G1509 | 701 | 78.94 | 6.57 | 叶绿体 Chloroplast |
| GhCOI6⁃D06 | GH_D06G1549 | 428 | 47.46 | 8.17 | 细胞质Cytoplasm |
| GhCOI6⁃A10 | GH_A10G0382 | 650 | 72.69 | 6.75 | 叶绿体Chloroplast |
| GhCOI6⁃D10 | GH_D10G0398 | 650 | 72.59 | 6.75 | 细胞核Nucleus |
| GhCOI6⁃A12 | GH_A12G1052 | 702 | 79.13 | 5.83 | 细胞核Nucleus |
| GhCOI6⁃D12 | GH_D12G1027 | 702 | 79.06 | 5.95 | 细胞核Nucleus |
| GhCOI7⁃A07 | GH_A07G2611 | 579 | 65.23 | 7.93 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI7⁃D07 | GH_D07G2552 | 572 | 64.35 | 8.23 | 细胞核Nucleus |
表2 GhCOI基因理化性质及亚细胞定位预测
Table 2 Characteristics and subcellular localization prediction of GhCOI genes
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI1⁃A01 | GH_A01G1190 | 534 | 57.36 | 8.7 | 叶绿体、细胞核Chloroplast, nucleus |
| GhCOI1⁃D01 | GH_D01G1258 | 534 | 57.54 | 8.82 | 细胞核Nucleus |
| GhCOI1⁃A05 | GH_A05G2065 | 534 | 57.38 | 8.86 | 细胞核Nucleus |
| GhCOI1⁃D05 | GH_D05G2095 | 535 | 57.44 | 8.83 | 细胞核Nucleus |
| GhCOI1⁃A06 | GH_A06G0210 | 528 | 56.82 | 8.74 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI1⁃D06 | GH_D06G0195 | 528 | 56.7 | 8.86 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI2⁃A02 | GH_A02G1803 | 544 | 58.97 | 8.49 | 细胞核Nucleus |
| GhCOI2⁃D03 | GH_D03G0256 | 514 | 55.87 | 7.87 | 细胞质Cytoplasm |
| GhCOI2⁃A10 | GH_A10G0913 | 545 | 59.31 | 8.6 | 细胞质Cytoplasm |
| GhCOI2⁃D10 | GH_D10G1048 | 545 | 59.21 | 8.59 | 细胞质Cytoplasm |
| GhCOI3⁃D04 | GH_D04G0696 | 591 | 66.99 | 5.98 | 细胞质Cytoplasm |
| GhCOI3⁃A05 | GH_A05G3439 | 589 | 66.74 | 5.81 | 细胞质Cytoplasm |
| GhCOI4⁃A05 | GH_A05G0481 | 362 | 39.99 | 6.32 | 细胞质Cytoplasm |
| GhCOI4⁃D05 | GH_D05G0480 | 362 | 40.08 | 6.96 | 细胞质Cytoplasm |
| GhCOI5⁃A05 | GH_A05G2217 | 611 | 68.38 | 5.97 | 细胞核Nucleus |
| GhCOI5⁃D05 | GH_D05G2242 | 611 | 68.32 | 5.88 | 叶绿体Chloroplast |
| GhCOI5⁃A06 | GH_A06G0343 | 615 | 69.03 | 5.67 | 细胞核Nucleus |
| GhCOI5⁃D06 | GH_D06G0328 | 615 | 69.11 | 5.54 | 细胞核Nucleus |
| GhCOI6⁃A06 | GH_A06G1509 | 701 | 78.94 | 6.57 | 叶绿体 Chloroplast |
| GhCOI6⁃D06 | GH_D06G1549 | 428 | 47.46 | 8.17 | 细胞质Cytoplasm |
| GhCOI6⁃A10 | GH_A10G0382 | 650 | 72.69 | 6.75 | 叶绿体Chloroplast |
| GhCOI6⁃D10 | GH_D10G0398 | 650 | 72.59 | 6.75 | 细胞核Nucleus |
| GhCOI6⁃A12 | GH_A12G1052 | 702 | 79.13 | 5.83 | 细胞核Nucleus |
| GhCOI6⁃D12 | GH_D12G1027 | 702 | 79.06 | 5.95 | 细胞核Nucleus |
| GhCOI7⁃A07 | GH_A07G2611 | 579 | 65.23 | 7.93 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI7⁃D07 | GH_D07G2552 | 572 | 64.35 | 8.23 | 细胞核Nucleus |
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI7⁃A11 | GH_A11G0655 | 572 | 64.46 | 7.42 | 细胞核Nucleus |
| GhCOI7⁃D11 | GH_D11G0684 | 572 | 64.28 | 8.2 | 细胞核Nucleus |
| GhCOI8A⁃A08 | GH_A08G0483 | 587 | 65.9 | 5.8 | 细胞核Nucleus |
| GhCOI8B⁃A08 | GH_A08G0792 | 586 | 65.92 | 6.13 | 细胞核Nucleus |
| GhCOI8C⁃A08 | GH_A08G1389 | 586 | 65.62 | 5.5 | 叶绿体、细胞质Chloroplast, cytoplasm |
| GhCOI8A⁃D08 | GH_D08G0497 | 587 | 65.67 | 5.61 | 细胞核Nucleus |
| GhCOI8B⁃D08 | GH_D08G0784 | 586 | 65.91 | 5.97 | 细胞质Cytoplasm |
| GhCOI8C⁃D08 | GH_D08G1411 | 586 | 65.57 | 5.51 | 叶绿体Chloroplast |
| GhCOI8⁃A11 | GH_A11G1227 | 585 | 65.64 | 6.54 | 细胞质Cytoplasm |
| GhCOI8⁃D11 | GH_D11G1257 | 585 | 65.49 | 5.76 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI9⁃A08 | GH_A08G2517 | 467 | 52.62 | 7.74 | 细胞核Nucleus |
| GhCOI9⁃D08 | GH_D08G2524 | 467 | 52.6 | 6.91 | 细胞质Cytoplasm |
| GhCOI9A⁃A11 | GH_A11G1519 | 395 | 43.46 | 6.63 | 细胞质Cytoplasm |
| GhCOI9B⁃A11 | GH_A11G3019 | 654 | 72.04 | 8.25 | 叶绿体、细胞核、细胞质Chloroplast, nucleus, cytoplasm |
| GhCOI9A⁃D11 | GH_D11G1550 | 395 | 43.43 | 6.98 | 细胞质Cytoplasm |
| GhCOI9B⁃D11 | GH_D11G3049 | 654 | 72.27 | 8.17 | 细胞核Nucleus |
| GhCOI10⁃A09 | GH_A09G2525 | 592 | 64.35 | 7.91 | 细胞质Cytoplasm |
| GhCOI10⁃D09 | GH_D09G2461 | 607 | 65.93 | 8.11 | 细胞质Cytoplasm |
| GhCOI10⁃A12 | GH_A12G1492 | 411 | 44.68 | 5.8 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI10⁃D12 | GH_D12G1505 | 411 | 44.61 | 5.95 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI11⁃A10 | GH_A10G0227 | 586 | 65.71 | 5.97 | 细胞核Nucleus |
| GhCOI11⁃D10 | GH_D10G0239 | 586 | 65.87 | 5.92 | 细胞核Nucleus |
表2 GhCOI基因理化性质及亚细胞定位预测 (续表Continuted)
Table 2 Characteristics and subcellular localization prediction of GhCOI genes
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI7⁃A11 | GH_A11G0655 | 572 | 64.46 | 7.42 | 细胞核Nucleus |
| GhCOI7⁃D11 | GH_D11G0684 | 572 | 64.28 | 8.2 | 细胞核Nucleus |
| GhCOI8A⁃A08 | GH_A08G0483 | 587 | 65.9 | 5.8 | 细胞核Nucleus |
| GhCOI8B⁃A08 | GH_A08G0792 | 586 | 65.92 | 6.13 | 细胞核Nucleus |
| GhCOI8C⁃A08 | GH_A08G1389 | 586 | 65.62 | 5.5 | 叶绿体、细胞质Chloroplast, cytoplasm |
| GhCOI8A⁃D08 | GH_D08G0497 | 587 | 65.67 | 5.61 | 细胞核Nucleus |
| GhCOI8B⁃D08 | GH_D08G0784 | 586 | 65.91 | 5.97 | 细胞质Cytoplasm |
| GhCOI8C⁃D08 | GH_D08G1411 | 586 | 65.57 | 5.51 | 叶绿体Chloroplast |
| GhCOI8⁃A11 | GH_A11G1227 | 585 | 65.64 | 6.54 | 细胞质Cytoplasm |
| GhCOI8⁃D11 | GH_D11G1257 | 585 | 65.49 | 5.76 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI9⁃A08 | GH_A08G2517 | 467 | 52.62 | 7.74 | 细胞核Nucleus |
| GhCOI9⁃D08 | GH_D08G2524 | 467 | 52.6 | 6.91 | 细胞质Cytoplasm |
| GhCOI9A⁃A11 | GH_A11G1519 | 395 | 43.46 | 6.63 | 细胞质Cytoplasm |
| GhCOI9B⁃A11 | GH_A11G3019 | 654 | 72.04 | 8.25 | 叶绿体、细胞核、细胞质Chloroplast, nucleus, cytoplasm |
| GhCOI9A⁃D11 | GH_D11G1550 | 395 | 43.43 | 6.98 | 细胞质Cytoplasm |
| GhCOI9B⁃D11 | GH_D11G3049 | 654 | 72.27 | 8.17 | 细胞核Nucleus |
| GhCOI10⁃A09 | GH_A09G2525 | 592 | 64.35 | 7.91 | 细胞质Cytoplasm |
| GhCOI10⁃D09 | GH_D09G2461 | 607 | 65.93 | 8.11 | 细胞质Cytoplasm |
| GhCOI10⁃A12 | GH_A12G1492 | 411 | 44.68 | 5.8 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI10⁃D12 | GH_D12G1505 | 411 | 44.61 | 5.95 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI11⁃A10 | GH_A10G0227 | 586 | 65.71 | 5.97 | 细胞核Nucleus |
| GhCOI11⁃D10 | GH_D10G0239 | 586 | 65.87 | 5.92 | 细胞核Nucleus |
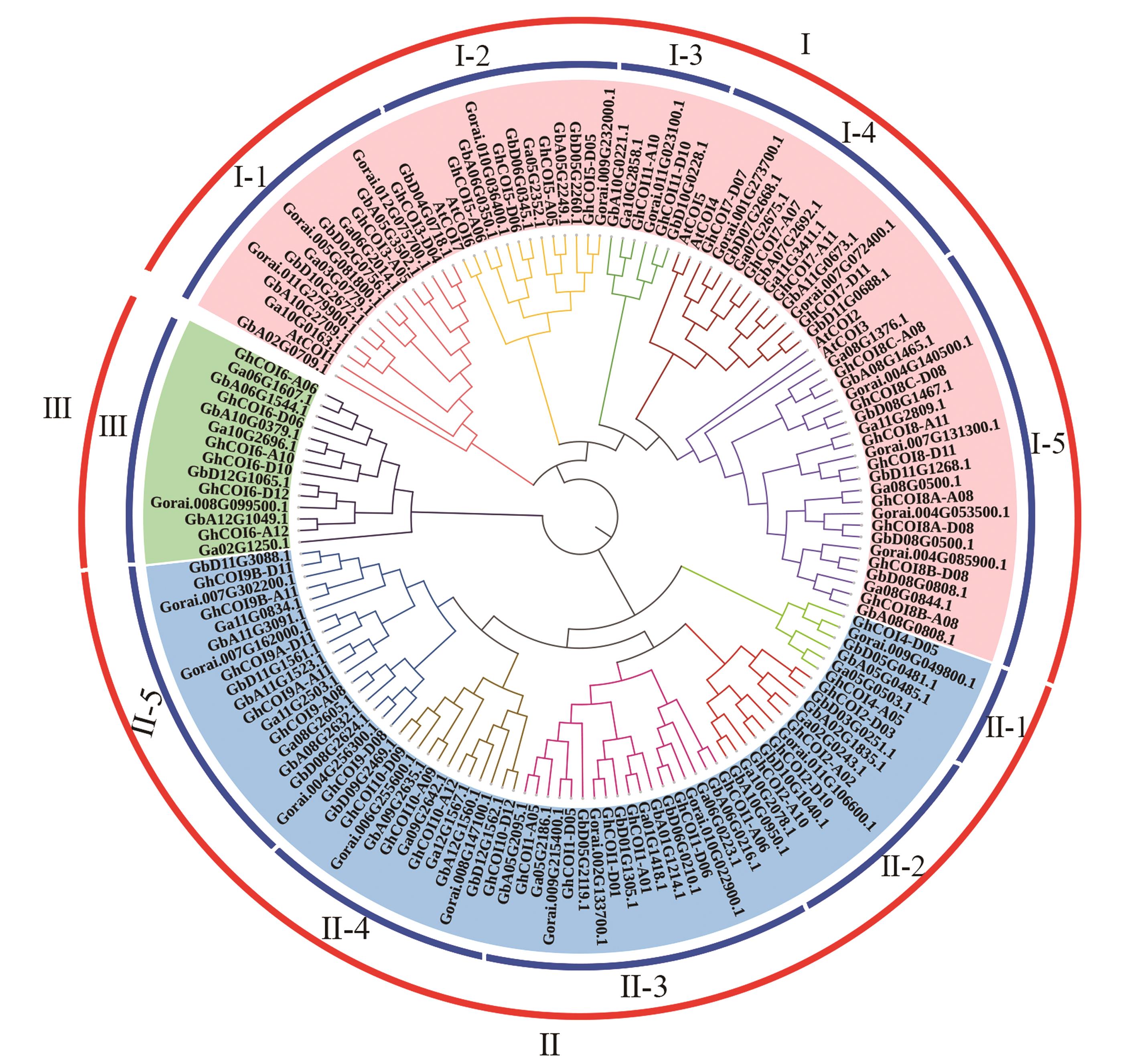
图2 COI基因系统发育分析注:Gb代表海岛棉,Gh代表陆地棉,Ga代表亚洲棉,Gorai代表雷蒙德氏棉,At代表拟南芥。
Fig.2 Phylogeny analysis of COI genesNote: Gb represents Gossypium barbadense, Gh represents Gossypium hirsutum, Ga represents Gossypium arboreum, Gorai represents Gossypium raimondii, and At represents Arabidopsis thaliana.
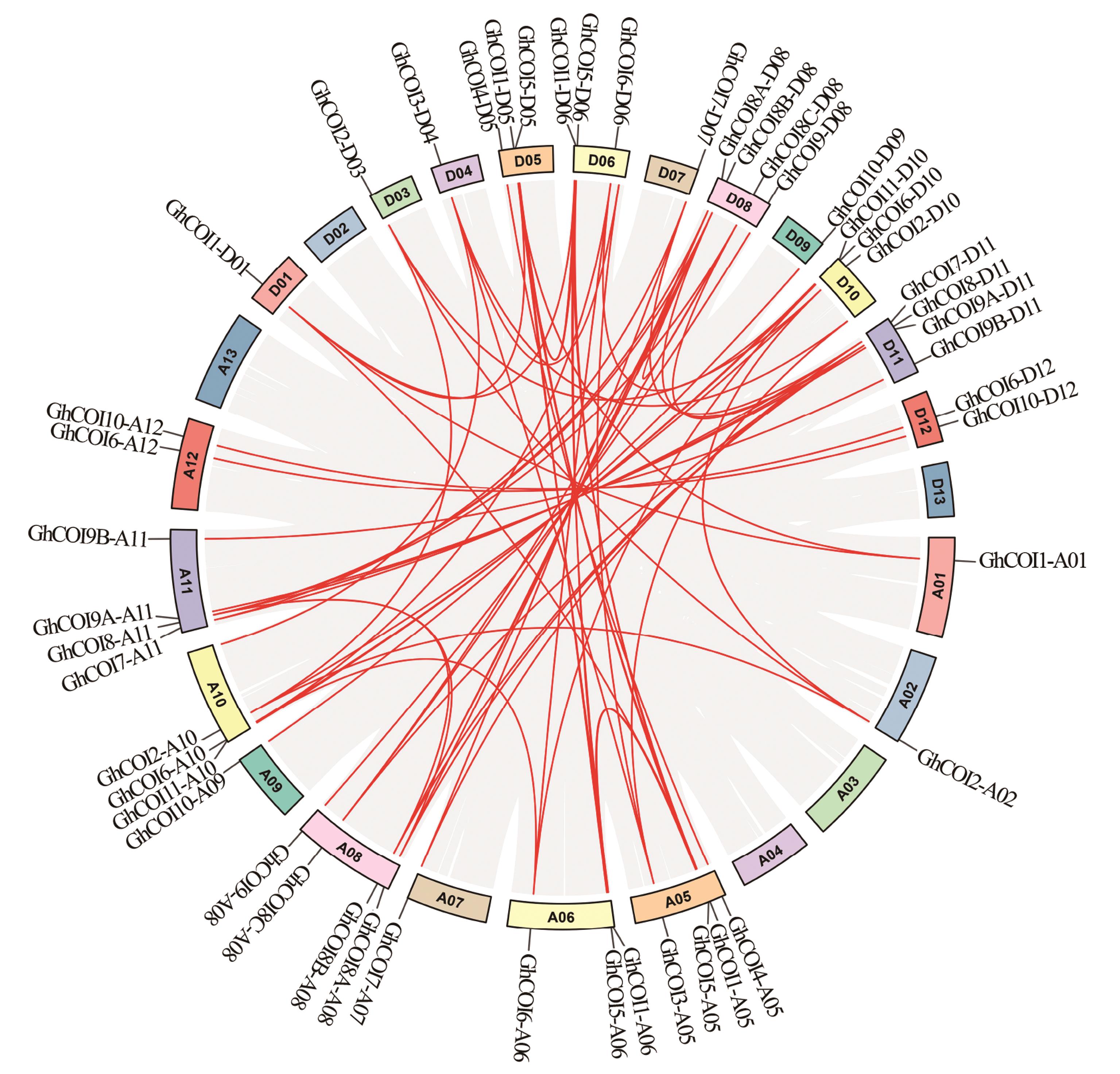
图3 COI基因系统共线性分析注:Gh代表陆地棉,红线代表片段复制基因段。
Fig. 3 Collinearity analysis of COI genesNote: Gh represents Gossypium hirsutum, and the red line indicates segmental duplication.
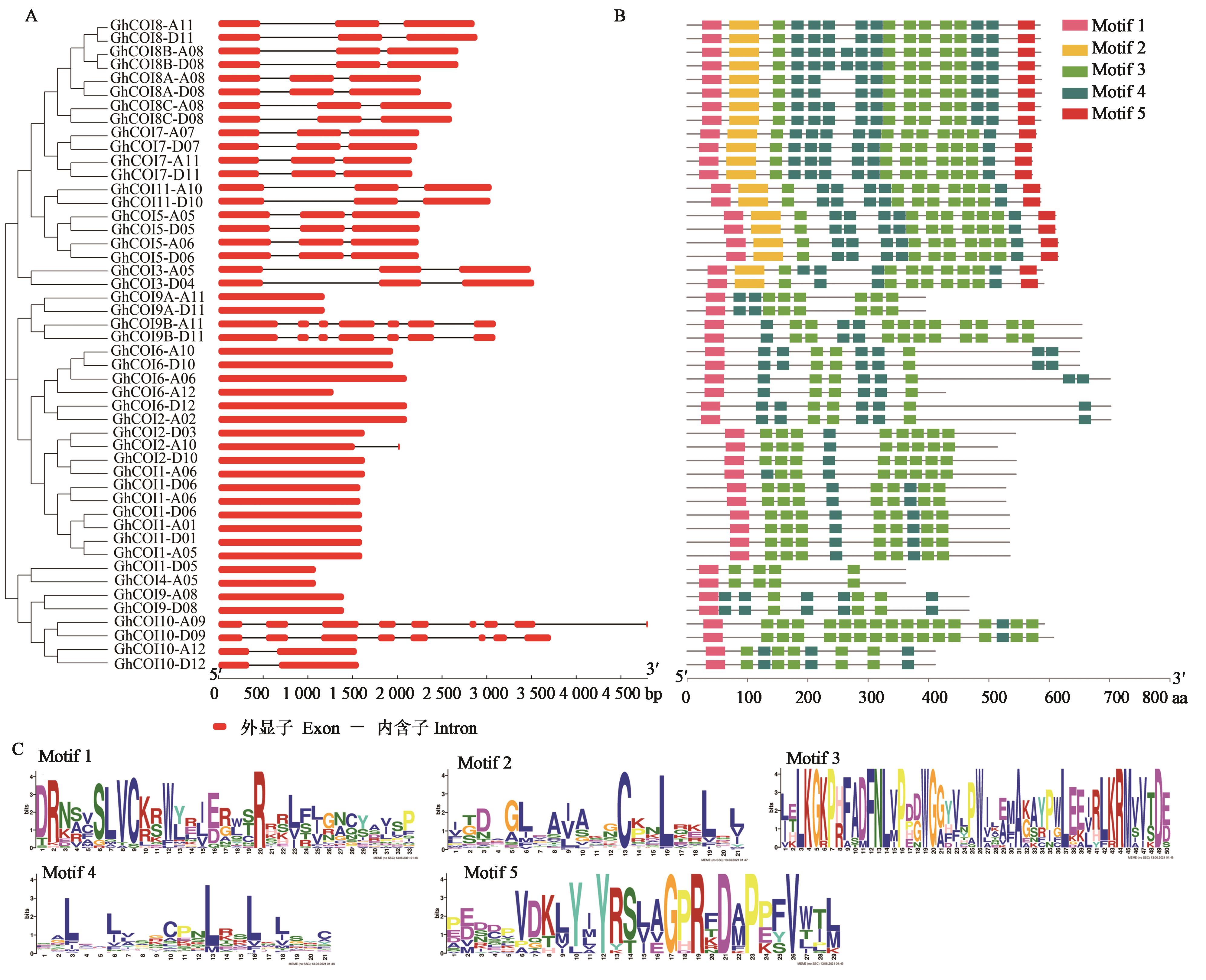
图4 GhCOI家族成员结构特征和保守基序分析A:基因结构;B:保守基序;C:保守基序序列
Fig. 4 Structure and conservative motif analysis of the GhCOIsA: Gene strcture; B: Conserved motif; C: Conserved motif sequences

图5 GhCOI基因顺式作用元件预测和在非生物胁迫下的表达A:GhCOI启动子顺式作用元件; B:GhCOI基因在NaCl和PEG胁迫下的表达; C:GhCOI基因在热和冷胁迫下的表达
Fig.5 cis?acting elements and expression analysis of GhCOI genesA: ,cis-acting elements of GhCOIs promoter; B: Expression analysis of GhCOIs under NaCl and PEG stress; C: Expression analysis of GhCOIs under cold and high-temperature stress

图6 9个GhCOI基因在NaCl和PEG胁迫下的表达分析A:GhCOI基因在NaCl胁迫下的表达分析;B:GhCOI基因在PEG胁迫下的表达分析
Fig. 6 Expression analysis of 9 GhCOIs under NaCl and PEG stressA: Expression analysis of 9 GhCOIs under NaCl stress; B: Expression analysis of 9 GhCOIs under PEG stress
| 1 | CREELMAN R A, MULLET J E. Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress [J]. Proc. Natl. Acad. Sci. USA, 1995, 92(10): 4114-4119. |
| 2 | YAN J, ZHANG C, GU M, et al.. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor [J]. Plant Cell, 2009, 21(8): 2220-2236. |
| 3 | XIE D X, FEYS B F, JAMES S, et al.. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility [J]. Science, 1998, 280(5366): 1091-1094. |
| 4 | SKOWYRA D, CRAIG K L, TYERS M, et al.. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex [J]. Cell, 1997, 91(2): 209-219. |
| 5 | CASTILLO M C, LEÓN J. Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis [J]. J. Exp. Bot., 2008, 59(8): 2171-2179. |
| 6 | SHAN X, WANG J, CHUA L, et al.. The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence [J]. Plant Physiol., 2011, 155(2): 751-764. |
| 7 | REINBOTHE C, SPRINGER A, SAMOL I, et al.. Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence [J]. FEBS J., 2009, 276(17): 4666-4681. |
| 8 | UEDA J, KATO J. Isolation and identification of a senescence-promoting substance from Wormwood (Artemisia absinthium L.) [J]. Plant Physiol., 1980, 66(2): 246-249. |
| 9 | KIM J, DOTSON B, REY C, et al.. New clothes for the jasmonic acid receptor COI1: delayed abscission, meristem arrest and apical dominance [J/OL]. PLoS One, 2013, 8(4): e60505 [ 2021-11-17 ]. . |
| 10 | 李娟,阳文龙,刘冬成,等.小麦K-型细胞质雄性不育系COI1基因的克隆与表达分析[J].分子植物育种,2015,13(3):518-530. |
| LI J, YANG W L, LIU D C, et al.. Cloning and expression analysis of COI1 gene from a wheat K-type cytoplasmic male sterility line [J]. Mol. Plant Breed., 2015, 13(3): 518-530. | |
| 11 | YE M, LUO S M, XIE J F, et al.. Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense [J/OL]. PLoS One, 2012, 7(4): e36214 [2021-11-17 ]. . |
| 12 | 王文静,杨小川,丁永强,等.甘蓝型油菜COI1的调控功能分析[J].中国农业科学,2015,48(10):1882-1891. |
| WANG W J, YANG X C, DING Y Q, et al.. Functional analysis of COI1 genes in Oilseed Rape(Brassica napus L.)[J]. Sci. Agric. Sin., 2015, 48(10): 1882-1891. | |
| 13 | 段龙飞.茉莉酸信号途径上关键基因家族COI/JAZ/MYC分子进化分析[D].陕西杨凌:西北农林科技大学,2013. |
| DUAN L F. Molecular evolutionary analysis of the key gene families COI/JAZ/MYC in jasmonic acid signaling pathway [D]. Shaanxi Yangling: Northwest A&F University, 2013. | |
| 14 | HU Y, CHEN J, FANG L, et al.. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton [J]. Nat. Genet., 2019, 51(4): 739-748. |
| 15 | ZHU T, LIANG C, MENG Z, et al.. CottonFGD: an integrated functional genomics database for cotton [J]. BMC Plant Biol., 2017, 17(1): 101-109. |
| 16 | BERARDINI T Z, REISER L, LI D, et al.. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome [J]. Genesis, 2015, 53(8): 474-485. |
| 17 | EL-GEBALI S, MISTRY J, BATEMAN A, et al.. The Pfam protein families database in 2019 [J]. Nucleic Acids Res., 2019, 47(D1): D427-D432. |
| 18 | LETUNIC I, BORK P. 20 years of the SMART protein domain annotation resource [J]. Nucleic Acids Res., 2018, 46(D1): D493-D496. |
| 19 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002, 30(1): 325-327. |
| 20 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2-ΔΔ CT method [J]. Methods, 2001, 25(4): 402-408. |
| 21 | 宋云,李林宣,卓凤萍,等.茉莉酸信号传导在植物抗逆性方面研究进展[J].中国农业科技导报, 2015,17(2): 17-24. |
| SONG Y, LI L X, ZHUO F P, et al.. Progress on jasmonic acid signaling in plant sress resistant [J]. J. Agric. Sci. Technol., 2015, 17(2): 17-24. | |
| 22 | MOORE R C, PURUGGANAN M D. The early stages of duplicate gene evolution [J]. Proc. Natl. Acad. Sci. USA, 2003, 100(26): 15682-15687. |
| 23 | CANNON S B, MITRA A, BAUMGARTEN A, et al.. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J]. BMC Plant Biol., 2004, 4(1): 10-30. |
| 24 | BAI J F, WANG Y K, WANG P, et al.. Genome-wide identification and analysis of the COI gene family in wheat (Triticum aestivum L.) [J]. BMC Genomics, 2018, 19(1): 754-770. |
| 25 | 杨秀,许艳超,杨芳芳,等.棉花CML基因家族成员鉴定与功能分析[J].棉花学报,2019, 31(4):307-318. |
| YANG X, XU Y C, YANG F F, et al.. Identification and functional analysis of CML gene family in cotton [J]. Cotton Sci., 2019, 31(4): 307-318. | |
| 26 | ZHU Y, WU N, SONG W, et al.. Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies [J]. BMC Plant Biol., 2014, 14(1): 93-111. |
| 27 | 段龙飞,李文燕,慕小倩,等.茉莉酸信号受体COI蛋白家族的分子进化与基因表达分析[J].生物信息学,2016,14(3):146-155. |
| DUAN L F, LI W Y, MU X Y, et al.. Molecular evolution and gene expression of jasmonic acid receptor COI protein family [J]. Chin. J. Bioinform., 2016, 14(3): 146-155. |
| [1] | 默韶京, 王志城, 王省芬, 刘正文, 吴立强, 张桂寅, 马峙英, 张艳, 段会军. 陆地棉GELP家族基因鉴定及其响应胁迫的表达分析[J]. 中国农业科技导报, 2022, 24(2): 93-103. |
| [2] | 王琴琴, 陈修贵, 陆许可, 王帅, 张悦新, 范亚朋, 陈全家, 叶武威. 陆地棉GhPKE1的生物信息学分析及功能验证[J]. 中国农业科技导报, 2022, 24(1): 38-45. |
| [3] | 李生梅, 杨涛, 黄雅婕, 任丹, 耿世伟, 李典鹏, 芮存, 高文伟. 海陆回交群体主要农艺性状与纤维品质关系的探讨[J]. 中国农业科技导报, 2021, 23(8): 16-24. |
| [4] | 崔江慧§,杨溥原§,常金华*. 高粱GRF基因家族鉴定及在非生物胁迫下的表达分析[J]. 中国农业科技导报, 2021, 23(4): 37-46. |
| [5] | 陈斌, 石荣康, 王志城, 刘松, 李青, 刘正文, 孙正文, 王国宁, 吴金华, 马峙英, 张艳, 王省芬. 陆地棉核心种质抗黄萎病鉴定与优异种质筛选[J]. 中国农业科技导报, 2021, 23(10): 45-51. |
| [6] | 李子玮1,陈思蒙1,王发展1,张豪洋1,张莉2*,许自成1*. 硫化氢在植物中抵御非生物胁迫机制的研究进展[J]. 中国农业科技导报, 2020, 22(4): 24-32. |
| [7] | 陈思蒙,李子玮,张璐翔,魏艳秋,朱智威,张环纬,许自成,黄五星,邵惠芳*. 硒在植物抵御胁迫中作用的研究进展[J]. 中国农业科技导报, 2020, 22(3): 6-13. |
| [8] | 周秒依,任雯,赵冰兵,李韩帅,刘亚*. 植物MAPK级联途径应答的非生物胁迫研究进展[J]. 中国农业科技导报, 2020, 22(2): 22-29. |
| [9] | 梁桂红,华营鹏,周婷,宋海星,张振华*. 植物液泡膜H+-ATPase和H+-PPase研究进展[J]. 中国农业科技导报, 2020, 22(1): 19-27. |
| [10] | 韩佳慧1,刘盈盈1,江世杰2,陈云1,耿秀秀1,2,平淑珍1,王劲1,2*. 非生物胁迫下戈壁异常球菌LEA3蛋白Dgl3抗逆功能研究[J]. 中国农业科技导报, 2019, 21(9): 69-76. |
| [11] | 王俊铎1,曾辉2,龚照龙1,梁亚军1,艾先涛1,郭江平1,莫明1,李雪源1,郑巨云1*. 陆地棉品种资源耐复合盐碱性综合评价分析[J]. 中国农业科技导报, 2019, 21(10): 1-11. |
| [12] | 杨笑敏,芮存,张悦新,王德龙,王俊娟,陆许可,陈修贵,郭丽雪,王帅,陈超,叶武威*. 棉花DNA甲基转移酶GhDMT9抗逆性分析[J]. 中国农业科技导报, 2019, 21(10): 12-19. |
| [13] | 王昭玉1,吕婷婷1,李爱芹2,王頔1,张子玉1,闫瑞娴1,. 陆地棉钾吸收相关基因GhHAK1的克隆与功能分析[J]. 中国农业科技导报, 2017, 19(10): 21-27. |
| [14] | 高丽华1,刘博欣1,2,李金博3,吴燕民1,唐益雄1*. 陆地棉bHLH转录因子GhMYC4基因的克隆及功能分析[J]. 中国农业科技导报, 2016, 18(5): 33-41. |
| [15] | 谢政文1,王连军2,陈锦洋1,王娇1,苏一钧1,杨新笋2,曹清河1*. 植物WRKY转录因子及其生物学功能研究进展[J]. 中国农业科技导报, 2016, 18(3): 46-54. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号