




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (12): 67-84.DOI: 10.13304/j.nykjdb.2023.0132
唐靓婷1,2( ), 黄世会1, 牛熙1, 李升1, 王嘉福1(
), 黄世会1, 牛熙1, 李升1, 王嘉福1( ), 冉雪琴1(
), 冉雪琴1( )
)
出版日期:2023-12-15
发布日期:2023-12-12
通讯作者:
王嘉福,冉雪琴
Liangting TANG1,2( ), Shihui HUANG1, Xi NIU1, Sheng LI1, Jiafu WANG1(
), Shihui HUANG1, Xi NIU1, Sheng LI1, Jiafu WANG1( ), Xueqin RAN1(
), Xueqin RAN1( )
)
Online:2023-12-15
Published:2023-12-12
Contact:
Jiafu WANG,Xueqin RAN
About author:TANG Liangting E-mail:714941286@qq.com
Supported by:摘要:
生物钟在生物体(如猪)的生殖系统中起着重要作用。为研究发情周期对香猪卵巢生物钟相关基因表达的影响,本研究分析了香猪卵巢在发情期和未发情期生物钟相关基因的表达和可变剪接。结果在香猪卵巢中共检测到90个生物钟相关基因的表达,其中有33个生物钟相关基因在发情期和未发情期差异表达。从34个生物钟相关基因的转录本中鉴定出44个差异可变剪接事件。此外,还发现包括核心时钟成分 arntl和 cry1在内的20个基因仅在可变剪接水平上被差异调节,包括 per1和 clock在内的14个基因同时在基因表达和可变剪接水平被差异调节。通过RT-qPCR 实验,证实了核心时钟基因 per1、cry1、 clock和 arntl以及时钟相关基因 ppp1cb和 ntrk1在香猪卵巢中的表达具有节律性。结果表明,香猪卵巢生物钟可能通过转录和转录后调控水平在调节卵巢生理功能中发挥着重要作用。
中图分类号:
唐靓婷, 黄世会, 牛熙, 李升, 王嘉福, 冉雪琴. 发情周期对香猪卵巢生物钟相关基因表达的影响[J]. 中国农业科技导报, 2023, 25(12): 67-84.
Liangting TANG, Shihui HUANG, Xi NIU, Sheng LI, Jiafu WANG, Xueqin RAN. Effects of Estrus Cycle on Expression of Ovarian Biological Clock-related Genes of Xiang Pig[J]. Journal of Agricultural Science and Technology, 2023, 25(12): 67-84.
| qPCR detection | RT-PCR detection | |||||
|---|---|---|---|---|---|---|
| Primer name | Primer sequence (5’-3’) | Size/bp | Primer name | Primer sequence (5’-3’) | Size/bp | |
| ppp1cb-F | AAGAGGAAACCATGAGTGTGCTAG | 112 | ppp1cb-MXE-1F | GGAAGACCTTCACTGATTGT | 131 | |
| ppp1cb-R | GCAGTTGAAACAATCAGTGAAGGTC | ppp1cb-MXE-1R | CCAACCTTGCACATCCTTA | |||
| atf4-F | TTGGTCAGTGCCTCAGACAACAG | 95 | ppp1cb- MXE-2F | GCAGTCTATGGAGCAGATT | 156 | |
| atf4-R | GCATCGAAGTCGAACTCCTTCAGAT | ppp1cb-MXE-2R | TCATTCCACCAGCATTATCA | |||
| csnk1d-F | GCGGGTCCTTCGGAGACATCTAT | 102 | thrap3- MXE- 1F | CCTGACTGTAAATTGGAGAAG | 112 | |
| csnk1d-F | GCGGGTCCTTCGGAGACATCTAT | thrap3- MXE- 1R | GAACGAGACCTGGAACTTAT | |||
| per1-F | CATCGCTTACAGCCTCCTGAGTG | 105 | thrap3- MXE- 2F | AGTCTGGATCTCGCTCTT | 143 | |
| per1-R | ATGAGTTCTTTCTGGGTCCTTGCC | thrap3- MXE- 2R | ATTCTCTGTCTTCTGCTCAT | |||
| klf9-F | CCATTACAGAGTGCATACAGGTGAA | 139 | top2a- MXE- 1F | TCTGCTAGTCCTCGATACATCT | 112 | |
| klf9-R | ACACAGCGGACAGCGGAACT | top2a- MXE- 1R | GCCACTTCACCACTAATCACA | |||
| nampt-F | AGATCCAAGAAGCCAAAGAGGTGTA | 141 | top2a- MXE- 2F | CCCAACTTTGATGTTCGTGAAG | 125 | |
| namptR | GAATGACAGAGCCCTCAGGAACAG | top2a- MXE- 2R | GGTGTCTTCTCAGTGCCATTC | |||
| bhlhe40-F | ACTGGAGCCTGGAGACCTACCA | 91 | clock- SE-F | GGTGGTCAGTTTCAGTTCT | 111 | |
| bhlhe40-R | GCTGTCCTCGCTCCGCTTTATTC | clock- SE-R | TTCTTCTTGTTGCCGATGA | |||
Table 1 Primers sequence
| qPCR detection | RT-PCR detection | |||||
|---|---|---|---|---|---|---|
| Primer name | Primer sequence (5’-3’) | Size/bp | Primer name | Primer sequence (5’-3’) | Size/bp | |
| ppp1cb-F | AAGAGGAAACCATGAGTGTGCTAG | 112 | ppp1cb-MXE-1F | GGAAGACCTTCACTGATTGT | 131 | |
| ppp1cb-R | GCAGTTGAAACAATCAGTGAAGGTC | ppp1cb-MXE-1R | CCAACCTTGCACATCCTTA | |||
| atf4-F | TTGGTCAGTGCCTCAGACAACAG | 95 | ppp1cb- MXE-2F | GCAGTCTATGGAGCAGATT | 156 | |
| atf4-R | GCATCGAAGTCGAACTCCTTCAGAT | ppp1cb-MXE-2R | TCATTCCACCAGCATTATCA | |||
| csnk1d-F | GCGGGTCCTTCGGAGACATCTAT | 102 | thrap3- MXE- 1F | CCTGACTGTAAATTGGAGAAG | 112 | |
| csnk1d-F | GCGGGTCCTTCGGAGACATCTAT | thrap3- MXE- 1R | GAACGAGACCTGGAACTTAT | |||
| per1-F | CATCGCTTACAGCCTCCTGAGTG | 105 | thrap3- MXE- 2F | AGTCTGGATCTCGCTCTT | 143 | |
| per1-R | ATGAGTTCTTTCTGGGTCCTTGCC | thrap3- MXE- 2R | ATTCTCTGTCTTCTGCTCAT | |||
| klf9-F | CCATTACAGAGTGCATACAGGTGAA | 139 | top2a- MXE- 1F | TCTGCTAGTCCTCGATACATCT | 112 | |
| klf9-R | ACACAGCGGACAGCGGAACT | top2a- MXE- 1R | GCCACTTCACCACTAATCACA | |||
| nampt-F | AGATCCAAGAAGCCAAAGAGGTGTA | 141 | top2a- MXE- 2F | CCCAACTTTGATGTTCGTGAAG | 125 | |
| namptR | GAATGACAGAGCCCTCAGGAACAG | top2a- MXE- 2R | GGTGTCTTCTCAGTGCCATTC | |||
| bhlhe40-F | ACTGGAGCCTGGAGACCTACCA | 91 | clock- SE-F | GGTGGTCAGTTTCAGTTCT | 111 | |
| bhlhe40-R | GCTGTCCTCGCTCCGCTTTATTC | clock- SE-R | TTCTTCTTGTTGCCGATGA | |||
| qPCR detection | RT-PCR detection | |||||
|---|---|---|---|---|---|---|
| Primer name | Primer sequence (5’-3’) | Size/bp | Primer name | Primer sequence (5’-3’) | Size/bp | |
| id4-F | CAGTGCGATATGAACGACTGCTA | 89 | cipc- SE-F | GCAGAGCCAATAACCCTAA | 236 | |
| id4-R | AGGATCTCCACTTTGCTGACTTTCT | cipc- SE-R | CGAGTTGACGAGACACTT | |||
| ass1-F | F:CTGCATCCTCGTGTGGCTGAA | 95 | mapk9- SE-F | CGTTACCAGCAGTTGAAACCA | 157 | |
| ass1-R | R:TCTTCCTGGCTTCCTCAAAGTCTTC | mapk9- SE-R | AGAGGACAAGTTCACGATAAGC | |||
| ntrk1-F | TACATCGAGAACCAGCAGCACCT | 146 | cry- RI-F | AGCAAGGTAAGAATGTAGCA | 147 | |
| ntrk1-R | AGATGACTGAGCCGAGGAGTGAA | cry- RI-R | CGTGTCCTCTTCCTGACT | |||
| per1- RI-F | CTGCTGTAGTCTCCTGTCT | 189 | ||||
| per1- RI-R | GGAAGGTCACCTGTGGAT | |||||
| ppp1cb- RI-F | GCGTGACTTGTAGGTGAG | 105 | ||||
| ppp1cb- RI-R | CCATCTTGTCGGCACTAG | |||||
Table 1 Primers sequence (Continued)
| qPCR detection | RT-PCR detection | |||||
|---|---|---|---|---|---|---|
| Primer name | Primer sequence (5’-3’) | Size/bp | Primer name | Primer sequence (5’-3’) | Size/bp | |
| id4-F | CAGTGCGATATGAACGACTGCTA | 89 | cipc- SE-F | GCAGAGCCAATAACCCTAA | 236 | |
| id4-R | AGGATCTCCACTTTGCTGACTTTCT | cipc- SE-R | CGAGTTGACGAGACACTT | |||
| ass1-F | F:CTGCATCCTCGTGTGGCTGAA | 95 | mapk9- SE-F | CGTTACCAGCAGTTGAAACCA | 157 | |
| ass1-R | R:TCTTCCTGGCTTCCTCAAAGTCTTC | mapk9- SE-R | AGAGGACAAGTTCACGATAAGC | |||
| ntrk1-F | TACATCGAGAACCAGCAGCACCT | 146 | cry- RI-F | AGCAAGGTAAGAATGTAGCA | 147 | |
| ntrk1-R | AGATGACTGAGCCGAGGAGTGAA | cry- RI-R | CGTGTCCTCTTCCTGACT | |||
| per1- RI-F | CTGCTGTAGTCTCCTGTCT | 189 | ||||
| per1- RI-R | GGAAGGTCACCTGTGGAT | |||||
| ppp1cb- RI-F | GCGTGACTTGTAGGTGAG | 105 | ||||
| ppp1cb- RI-R | CCATCTTGTCGGCACTAG | |||||
| Period | Sample | Input read pairs | Both surviving | Forward only surviving | Reverse only surviving | Dropped | Percentage of mapped reads/% |
|---|---|---|---|---|---|---|---|
| Diestrus | D1 | 77 089 626 | 74 382 355 (96.49%) | 2 471 254 (3.21%) | 164 408 (0.21%) | 69 380 (0.09%) | 96.01 |
| D2 | 77 040 960 | 74 260 107 (96.39%) | 2 550 481 (3.31%) | 160 145 (0.21%) | 70 227 (0.09%) | 95.88 | |
| D3 | 76 170 080 | 73 520 243 (96.52%) | 2 408 511 (3.16%) | 167 409 (0.22%) | 73 917 (0.10%) | 95.93 | |
| D4 | 77 100 634 | 74 147 772 (96.17%) | 2 713 372 (3.52%) | 165 857 (0.22%) | 73 633 (0.10%) | 96.26 | |
| D5 | 76 860 177 | 74 164 234 (96.49%) | 2 449 821 (3.19%) | 177 911 (0.23%) | 68 211 (0.09%) | 96.06 | |
| D6 | 76 903 234 | 74 486 606 (96.86%) | 2 179 432 (2.83%) | 173 980 (0.23%) | 63 216 (0.08%) | 96.11 | |
| D7 | 77 054 008 | 74 411 266 (96.57%) | 2 408 339 (3.13%) | 163 511 (0.21%) | 70 892 (0.09%) | 95.95 | |
| Estrus | E1 | 75 005 968 | 74 960 964 (99.94%) | 45 004 (0.06%) | 0 (0.00%) | 0 (0.00%) | 95.98 |
| E2 | 74 990 854 | 74 960 858 (99.96%) | 29 996 (0.04%) | 0 (0.00%) | 0 (0.00%) | 95.90 | |
| E3 | 75 206 715 | 75 176 632 (99.96%) | 30 083 (0.04%) | 0 (0.00%) | 0 (0.00%) | 95.93 | |
| E4 | 75 187 643 | 75 135 012 (99.93%) | 52 631 (0.07%) | 31 (0.00%) | 0 (0.00%) | 96.24 | |
| E5 | 75 184 086 | 75 154 012 (99.96%) | 30 074 (0.04%) | 31 (0.00%) | 1 (0.00%) | 96.15 | |
| E6 | 76 109 159 | 74 099 877 (97.36%) | 1 682 012 (2.21%) | 243 549 (0.32%) | 83 720 (0.11%) | 94.13 | |
| E7 | 75 002 160 | 73 149 607 (97.53%) | 1 545 044 (2.06%) | 225 006 (0.30%) | 82 502 (0.11%) | 93.57 |
Table 2 RNA-Seq data description
| Period | Sample | Input read pairs | Both surviving | Forward only surviving | Reverse only surviving | Dropped | Percentage of mapped reads/% |
|---|---|---|---|---|---|---|---|
| Diestrus | D1 | 77 089 626 | 74 382 355 (96.49%) | 2 471 254 (3.21%) | 164 408 (0.21%) | 69 380 (0.09%) | 96.01 |
| D2 | 77 040 960 | 74 260 107 (96.39%) | 2 550 481 (3.31%) | 160 145 (0.21%) | 70 227 (0.09%) | 95.88 | |
| D3 | 76 170 080 | 73 520 243 (96.52%) | 2 408 511 (3.16%) | 167 409 (0.22%) | 73 917 (0.10%) | 95.93 | |
| D4 | 77 100 634 | 74 147 772 (96.17%) | 2 713 372 (3.52%) | 165 857 (0.22%) | 73 633 (0.10%) | 96.26 | |
| D5 | 76 860 177 | 74 164 234 (96.49%) | 2 449 821 (3.19%) | 177 911 (0.23%) | 68 211 (0.09%) | 96.06 | |
| D6 | 76 903 234 | 74 486 606 (96.86%) | 2 179 432 (2.83%) | 173 980 (0.23%) | 63 216 (0.08%) | 96.11 | |
| D7 | 77 054 008 | 74 411 266 (96.57%) | 2 408 339 (3.13%) | 163 511 (0.21%) | 70 892 (0.09%) | 95.95 | |
| Estrus | E1 | 75 005 968 | 74 960 964 (99.94%) | 45 004 (0.06%) | 0 (0.00%) | 0 (0.00%) | 95.98 |
| E2 | 74 990 854 | 74 960 858 (99.96%) | 29 996 (0.04%) | 0 (0.00%) | 0 (0.00%) | 95.90 | |
| E3 | 75 206 715 | 75 176 632 (99.96%) | 30 083 (0.04%) | 0 (0.00%) | 0 (0.00%) | 95.93 | |
| E4 | 75 187 643 | 75 135 012 (99.93%) | 52 631 (0.07%) | 31 (0.00%) | 0 (0.00%) | 96.24 | |
| E5 | 75 184 086 | 75 154 012 (99.96%) | 30 074 (0.04%) | 31 (0.00%) | 1 (0.00%) | 96.15 | |
| E6 | 76 109 159 | 74 099 877 (97.36%) | 1 682 012 (2.21%) | 243 549 (0.32%) | 83 720 (0.11%) | 94.13 | |
| E7 | 75 002 160 | 73 149 607 (97.53%) | 1 545 044 (2.06%) | 225 006 (0.30%) | 82 502 (0.11%) | 93.57 |
| Gene_ID | Gene name | D1 | D2 | D3 | D4 | D5 | D6 | D7 | Ave | E1 | E2 | E3 | E4 | E5 | E6 | E7 | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSSSCG00000035690 | sfpq | 458.35 | 542.25 | 498.24 | 522.37 | 564.03 | 562.76 | 580.45 | 532.63 | 397.46 | 626.01 | 441.45 | 605.76 | 370.15 | 343.85 | 338.28 | 446.14 |
| ENSSSCG00000017473 | top2a | 345.41 | 554.28 | 288.81 | 867.20 | 484.25 | 463.57 | 581.44 | 512.14 | 111.89 | 81.99 | 80.71 | 52.11 | 80.55 | 121.22 | 94.22 | 88.95 |
| ENSSSCG00000023245 | nono | 405.64 | 435.83 | 402.61 | 448.48 | 422.10 | 507.09 | 428.38 | 435.73 | 401.72 | 596.11 | 462.85 | 455.95 | 462.21 | 432.43 | 378.01 | 455.61 |
| ENSSSCG00000029954 | thrap3 | 266.35 | 288.70 | 268.72 | 305.83 | 280.16 | 278.34 | 268.36 | 279.50 | 164.10 | 213.17 | 210.03 | 237.74 | 199.46 | 205.14 | 173.68 | 200.48 |
| ENSSSCG00000012318 | maged1 | 233.41 | 253.54 | 208.48 | 381.77 | 258.82 | 339.07 | 241.52 | 273.80 | 222.71 | 306.74 | 232.40 | 214.95 | 257.00 | 196.98 | 179.35 | 230.02 |
| ENSSSCG00000000068 | ep300 | 288.00 | 226.71 | 264.90 | 274.01 | 214.29 | 246.97 | 226.61 | 248.78 | 133.20 | 146.62 | 144.88 | 197.58 | 162.06 | 141.04 | 139.62 | 152.14 |
| ENSSSCG00000021038 | nrip1 | 239.06 | 224.86 | 208.48 | 317.12 | 211.51 | 280.37 | 257.42 | 248.40 | 72.46 | 73.31 | 109.88 | 140.04 | 85.35 | 66.44 | 91.95 | 91.35 |
| ENSSSCG00000008540 | ppp1cb | 197.64 | 269.27 | 254.38 | 218.60 | 252.33 | 257.09 | 283.27 | 247.51 | 462.46 | 394.51 | 416.17 | 605.76 | 378.78 | 405.62 | 348.49 | 430.26 |
| ENSSSCG00000013335 | lgr4 | 282.35 | 183.22 | 174.05 | 228.86 | 145.65 | 256.08 | 293.21 | 223.34 | 318.61 | 102.25 | 132.24 | 108.56 | 125.62 | 169.01 | 256.54 | 173.26 |
| ENSSSCG00000006274 | prkdc | 178.82 | 184.14 | 185.52 | 226.81 | 180.90 | 228.75 | 182.88 | 195.40 | 56.48 | 63.66 | 74.87 | 85.76 | 73.84 | 50.12 | 63.57 | 66.90 |
| ENSSSCG00000004832 | ube3a | 147.76 | 173.96 | 192.22 | 145.73 | 172.55 | 171.06 | 205.74 | 172.72 | 94.84 | 109.00 | 101.13 | 118.33 | 96.85 | 101.41 | 107.84 | 104.20 |
| ENSSSCG00000005724 | setx | 144.94 | 234.11 | 187.44 | 135.47 | 161.42 | 158.91 | 160.02 | 168.90 | 60.74 | 62.70 | 112.79 | 70.56 | 74.80 | 65.27 | 71.51 | 74.05 |
| ENSSSCG00000000084 | atf4 | 167.53 | 171.19 | 157.79 | 158.05 | 156.78 | 174.09 | 150.08 | 162.21 | 350.58 | 422.49 | 337.41 | 509.14 | 331.79 | 362.50 | 233.84 | 363.96 |
| ENSSSCG00000030480 | dyrk1a | 154.35 | 161.93 | 164.49 | 153.94 | 155.85 | 162.96 | 169.96 | 160.50 | 77.79 | 86.81 | 79.73 | 105.30 | 91.10 | 80.43 | 69.24 | 84.34 |
| ENSSSCG00000001880 | sin3a | 145.88 | 159.16 | 154.92 | 163.18 | 144.72 | 159.92 | 145.11 | 153.27 | 91.64 | 100.32 | 94.32 | 93.36 | 92.06 | 85.09 | 77.19 | 90.57 |
Table 3 Top 15 circadian rhythm-related genes with the highest expression levels in the ovaries of Xiang pig
| Gene_ID | Gene name | D1 | D2 | D3 | D4 | D5 | D6 | D7 | Ave | E1 | E2 | E3 | E4 | E5 | E6 | E7 | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSSSCG00000035690 | sfpq | 458.35 | 542.25 | 498.24 | 522.37 | 564.03 | 562.76 | 580.45 | 532.63 | 397.46 | 626.01 | 441.45 | 605.76 | 370.15 | 343.85 | 338.28 | 446.14 |
| ENSSSCG00000017473 | top2a | 345.41 | 554.28 | 288.81 | 867.20 | 484.25 | 463.57 | 581.44 | 512.14 | 111.89 | 81.99 | 80.71 | 52.11 | 80.55 | 121.22 | 94.22 | 88.95 |
| ENSSSCG00000023245 | nono | 405.64 | 435.83 | 402.61 | 448.48 | 422.10 | 507.09 | 428.38 | 435.73 | 401.72 | 596.11 | 462.85 | 455.95 | 462.21 | 432.43 | 378.01 | 455.61 |
| ENSSSCG00000029954 | thrap3 | 266.35 | 288.70 | 268.72 | 305.83 | 280.16 | 278.34 | 268.36 | 279.50 | 164.10 | 213.17 | 210.03 | 237.74 | 199.46 | 205.14 | 173.68 | 200.48 |
| ENSSSCG00000012318 | maged1 | 233.41 | 253.54 | 208.48 | 381.77 | 258.82 | 339.07 | 241.52 | 273.80 | 222.71 | 306.74 | 232.40 | 214.95 | 257.00 | 196.98 | 179.35 | 230.02 |
| ENSSSCG00000000068 | ep300 | 288.00 | 226.71 | 264.90 | 274.01 | 214.29 | 246.97 | 226.61 | 248.78 | 133.20 | 146.62 | 144.88 | 197.58 | 162.06 | 141.04 | 139.62 | 152.14 |
| ENSSSCG00000021038 | nrip1 | 239.06 | 224.86 | 208.48 | 317.12 | 211.51 | 280.37 | 257.42 | 248.40 | 72.46 | 73.31 | 109.88 | 140.04 | 85.35 | 66.44 | 91.95 | 91.35 |
| ENSSSCG00000008540 | ppp1cb | 197.64 | 269.27 | 254.38 | 218.60 | 252.33 | 257.09 | 283.27 | 247.51 | 462.46 | 394.51 | 416.17 | 605.76 | 378.78 | 405.62 | 348.49 | 430.26 |
| ENSSSCG00000013335 | lgr4 | 282.35 | 183.22 | 174.05 | 228.86 | 145.65 | 256.08 | 293.21 | 223.34 | 318.61 | 102.25 | 132.24 | 108.56 | 125.62 | 169.01 | 256.54 | 173.26 |
| ENSSSCG00000006274 | prkdc | 178.82 | 184.14 | 185.52 | 226.81 | 180.90 | 228.75 | 182.88 | 195.40 | 56.48 | 63.66 | 74.87 | 85.76 | 73.84 | 50.12 | 63.57 | 66.90 |
| ENSSSCG00000004832 | ube3a | 147.76 | 173.96 | 192.22 | 145.73 | 172.55 | 171.06 | 205.74 | 172.72 | 94.84 | 109.00 | 101.13 | 118.33 | 96.85 | 101.41 | 107.84 | 104.20 |
| ENSSSCG00000005724 | setx | 144.94 | 234.11 | 187.44 | 135.47 | 161.42 | 158.91 | 160.02 | 168.90 | 60.74 | 62.70 | 112.79 | 70.56 | 74.80 | 65.27 | 71.51 | 74.05 |
| ENSSSCG00000000084 | atf4 | 167.53 | 171.19 | 157.79 | 158.05 | 156.78 | 174.09 | 150.08 | 162.21 | 350.58 | 422.49 | 337.41 | 509.14 | 331.79 | 362.50 | 233.84 | 363.96 |
| ENSSSCG00000030480 | dyrk1a | 154.35 | 161.93 | 164.49 | 153.94 | 155.85 | 162.96 | 169.96 | 160.50 | 77.79 | 86.81 | 79.73 | 105.30 | 91.10 | 80.43 | 69.24 | 84.34 |
| ENSSSCG00000001880 | sin3a | 145.88 | 159.16 | 154.92 | 163.18 | 144.72 | 159.92 | 145.11 | 153.27 | 91.64 | 100.32 | 94.32 | 93.36 | 92.06 | 85.09 | 77.19 | 90.57 |
| Gene_ID | Gene name | Gene description | DESeq2 | Diestrus group mean | Estrus group mean | Limma | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Base mean | log2FC | Padj | logFC | Ave Expr | Padj | B | |||||
| ENSSSCG00000017473 | top2a | DNA topoisomerase II alpha | 302.438 7 | -2.521 9 | 0.000 0 | 512.135 0 | 88.954 1 | -2.525 4 | 300.500 0 | 0.000 2 | 2.339 0 |
| ENSSSCG00000000068 | ep300 | E1A binding protein p300 | 201.785 4 | -0.705 9 | 0.041 8 | 248.784 1 | 152.141 8 | -0.709 5 | 200.571 4 | 0.000 0 | 4.160 8 |
| ENSSSCG00000021038 | nrip1 | Nuclear receptor interacting protein 1 | 170.763 4 | -1.443 9 | 0.000 0 | 248.401 2 | 91.345 2 | -1.443 3 | 169.714 3 | 0.000 0 | 5.939 3 |
| ENSSSCG00000008540 | ppp1cb | Protein phosphatase 1 catalytic subunit beta | 341.087 9 | 0.801 7 | 0.036 8 | 247.510 7 | 430.257 0 | 0.797 7 | 338.857 1 | 0.000 5 | 1.557 7 |
| ENSSSCG00000006274 | prkdc | Protein kinase, DNA-activated, catalytic subunit | 132.072 6 | -1.543 9 | 0.000 0 | 195.402 7 | 66.899 7 | -1.546 4 | 131.285 7 | 0.000 0 | 9.550 5 |
| ENSSSCG00000004832 | ube3a | Ubiquitin protein ligase E3A | 139.305 3 | -0.727 6 | 0.026 3 | 172.716 9 | 104.198 1 | -0.729 1 | 138.500 0 | 0.000 0 | 4.959 5 |
| ENSSSCG00000005724 | setx | Senataxin | 122.150 9 | -1.180 9 | 0.000 2 | 168.899 9 | 74.053 9 | -1.189 5 | 121.500 0 | 0.000 1 | 3.434 5 |
| ENSSSCG00000000084 | atf4 | Activating transcription factor 4 | 264.791 5 | 1.167 4 | 0.000 9 | 162.214 5 | 363.963 2 | 1.165 9 | 263.071 4 | 0.000 1 | 2.859 5 |
| ENSSSCG00000030480 | dyrk1a | Dual specificity tyrosine phosphorylation regulated kinase 1A | 123.069 5 | -0.925 8 | 0.003 5 | 160.496 8 | 84.343 4 | -0.928 2 | 122.357 1 | 0.000 0 | 10.657 4 |
| ENSSSCG00000001880 | sin3a | SIN3 transcription regulator family member A | 122.580 3 | -0.759 2 | 0.018 6 | 153.269 7 | 90.567 4 | -0.759 0 | 121.857 1 | 0.000 0 | 10.630 5 |
| ENSSSCG00000008909 | clock | Clock circadian regulator | 108.607 9 | -0.726 2 | 0.023 6 | 134.616 2 | 81.396 9 | -0.725 8 | 108.000 0 | 0.000 0 | 5.107 3 |
| ENSSSCG00000006194 | ncoa2 | Nuclear receptor coactivator 2 | 91.956 8 | -1.140 0 | 0.000 2 | 125.890 0 | 57.162 7 | -1.139 0 | 91.428 6 | 0.000 0 | 9.636 0 |
| ENSSSCG00000011579 | pparg | Peroxisome proliferator activated receptor gamma | 74.673 9 | -1.724 5 | 0.000 0 | 114.062 0 | 34.382 4 | -1.730 1 | 74.214 3 | 0.000 2 | 2.561 6 |
| ENSSSCG00000034395 | creb1 | CAMP responsive element binding protein 1 | 89.508 5 | -0.780 0 | 0.013 8 | 112.686 0 | 65.439 1 | -0.784 1 | 89.000 0 | 0.000 0 | 6.077 5 |
| ENSSSCG00000009569 | pspc1 | Paraspeckle component 1 | 73.279 2 | -1.478 6 | 0.000 0 | 107.345 8 | 38.455 7 | -1.481 0 | 72.857 1 | 0.000 0 | 12.522 2 |
| ENSSSCG00000035852 | csnk1d | Casein kinase 1 delta | 148.010 1 | 1.086 4 | 0.000 7 | 94.216 4 | 199.661 1 | 1.083 5 | 147.000 0 | 0.000 0 | 7.773 9 |
Table 4 Differential expression of circadian rhythm genes in ovary of Xiang pig
| Gene_ID | Gene name | Gene description | DESeq2 | Diestrus group mean | Estrus group mean | Limma | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Base mean | log2FC | Padj | logFC | Ave Expr | Padj | B | |||||
| ENSSSCG00000017473 | top2a | DNA topoisomerase II alpha | 302.438 7 | -2.521 9 | 0.000 0 | 512.135 0 | 88.954 1 | -2.525 4 | 300.500 0 | 0.000 2 | 2.339 0 |
| ENSSSCG00000000068 | ep300 | E1A binding protein p300 | 201.785 4 | -0.705 9 | 0.041 8 | 248.784 1 | 152.141 8 | -0.709 5 | 200.571 4 | 0.000 0 | 4.160 8 |
| ENSSSCG00000021038 | nrip1 | Nuclear receptor interacting protein 1 | 170.763 4 | -1.443 9 | 0.000 0 | 248.401 2 | 91.345 2 | -1.443 3 | 169.714 3 | 0.000 0 | 5.939 3 |
| ENSSSCG00000008540 | ppp1cb | Protein phosphatase 1 catalytic subunit beta | 341.087 9 | 0.801 7 | 0.036 8 | 247.510 7 | 430.257 0 | 0.797 7 | 338.857 1 | 0.000 5 | 1.557 7 |
| ENSSSCG00000006274 | prkdc | Protein kinase, DNA-activated, catalytic subunit | 132.072 6 | -1.543 9 | 0.000 0 | 195.402 7 | 66.899 7 | -1.546 4 | 131.285 7 | 0.000 0 | 9.550 5 |
| ENSSSCG00000004832 | ube3a | Ubiquitin protein ligase E3A | 139.305 3 | -0.727 6 | 0.026 3 | 172.716 9 | 104.198 1 | -0.729 1 | 138.500 0 | 0.000 0 | 4.959 5 |
| ENSSSCG00000005724 | setx | Senataxin | 122.150 9 | -1.180 9 | 0.000 2 | 168.899 9 | 74.053 9 | -1.189 5 | 121.500 0 | 0.000 1 | 3.434 5 |
| ENSSSCG00000000084 | atf4 | Activating transcription factor 4 | 264.791 5 | 1.167 4 | 0.000 9 | 162.214 5 | 363.963 2 | 1.165 9 | 263.071 4 | 0.000 1 | 2.859 5 |
| ENSSSCG00000030480 | dyrk1a | Dual specificity tyrosine phosphorylation regulated kinase 1A | 123.069 5 | -0.925 8 | 0.003 5 | 160.496 8 | 84.343 4 | -0.928 2 | 122.357 1 | 0.000 0 | 10.657 4 |
| ENSSSCG00000001880 | sin3a | SIN3 transcription regulator family member A | 122.580 3 | -0.759 2 | 0.018 6 | 153.269 7 | 90.567 4 | -0.759 0 | 121.857 1 | 0.000 0 | 10.630 5 |
| ENSSSCG00000008909 | clock | Clock circadian regulator | 108.607 9 | -0.726 2 | 0.023 6 | 134.616 2 | 81.396 9 | -0.725 8 | 108.000 0 | 0.000 0 | 5.107 3 |
| ENSSSCG00000006194 | ncoa2 | Nuclear receptor coactivator 2 | 91.956 8 | -1.140 0 | 0.000 2 | 125.890 0 | 57.162 7 | -1.139 0 | 91.428 6 | 0.000 0 | 9.636 0 |
| ENSSSCG00000011579 | pparg | Peroxisome proliferator activated receptor gamma | 74.673 9 | -1.724 5 | 0.000 0 | 114.062 0 | 34.382 4 | -1.730 1 | 74.214 3 | 0.000 2 | 2.561 6 |
| ENSSSCG00000034395 | creb1 | CAMP responsive element binding protein 1 | 89.508 5 | -0.780 0 | 0.013 8 | 112.686 0 | 65.439 1 | -0.784 1 | 89.000 0 | 0.000 0 | 6.077 5 |
| ENSSSCG00000009569 | pspc1 | Paraspeckle component 1 | 73.279 2 | -1.478 6 | 0.000 0 | 107.345 8 | 38.455 7 | -1.481 0 | 72.857 1 | 0.000 0 | 12.522 2 |
| ENSSSCG00000035852 | csnk1d | Casein kinase 1 delta | 148.010 1 | 1.086 4 | 0.000 7 | 94.216 4 | 199.661 1 | 1.083 5 | 147.000 0 | 0.000 0 | 7.773 9 |
| Gene ID | Gene symbol | Chromosome | Strand | Long exon start | Long exon end | Short exon start | Short exon end | Flanking exon start | Flanking exon end | ID | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A5SS.MATS. | ENSSSCG00000000107 | casein kinase | 5 | + | 9 613 116 | 9 613 949 | 9 613 116 | 9 613 265 | 9 614 046 | 9 615 759 | 302 |
| ENSSSCG00000005701 | ass1 | 1 | + | 270 581 146 | 270 582 896 | 270 581 146 | 270 581 303 | 270 585 083 | 270 585 149 | 575 | |
| ENSSSCG00000023245 | nono | X | + | 57 293 430 | 57 298 994 | 57 293 430 | 57 293 509 | 57 305 745 | 57 305 905 | 490 | |
| RI.MATS. | ENSSSCG00000000164 | cry1 | 5 | + | 13 362 925 | 13 364 594 | 13 362 925 | 13 363 183 | 13 363 958 | 13 364 594 | 1 188 |
| ENSSSCG00000017983 | per1 | 12 | - | 53 367 594 | 53 368 034 | 53 367 594 | 53 367 700 | 53 367 902 | 53 368 034 | 2 377 | |
| ENSSSCG00000008645 | id2 | 3 | - | 127 499 409 | 127 502 384 | 127 499 409 | 127 501 531 | 12 7502 320 | 127 502 384 | 911 | |
| ENSSSCG00000012913 | ppp1ca | 2 | + | 5 122 681 | 5 126 953 | 5 122 681 | 5 122 816 | 5 122 899 | 5 126 953 | 622 | |
| ENSSSCG00000036933 | nr1d1 | 12 | + | 22 270 244 | 22 275 515 | 22 270 244 | 22 270 455 | 22 271 146 | 22 275 515 | 717 | |
| ENSSSCG00000008540 | ppp1cb | 3 | - | 110 474 324 | 110 474 733 | 110 474 324 | 110 474 464 | 110 474 677 | 110 474 733 | 182 | |
| ENSSSCG00000015120 | usp2 | 9 | - | 46 565 719 | 46 566 122 | 46 565 719 | 46 565 827 | 46 566 043 | 46 566 122 | 45 | |
| ENSSSCG00000009828 | ppp1cc | 14 | - | 32 069 627 | 32 071 873 | 32 069 627 | 32 070 938 | 32 071 812 | 32 071 873 | 2 314 | |
| ENSSSCG00000011372 | impdh2 | 13 | - | 31 655 808 | 31 658 900 | 31 655 808 | 31 658 713 | 31 658 816 | 3 1658 900 | 327 | |
| ENSSSCG00000000107 | casein kinase | 5 | + | 9 613 116 | 9 613 949 | 9 613 116 | 9 613 265 | 9 613 845 | 9 613 949 | 910 | |
| ENSSSCG00000023245 | nono | X | + | 57293430 | 57 298 994 | 57 293 430 | 57 293 509 | 57 298 717 | 57 298 994 | 14 38 | |
| ENSSSCG00000001880 | sin3a | 7 | + | 58236964 | 58 238 004 | 58 236 964 | 58 237 057 | 58 237 909 | 58 238 004 | 1 997 | |
| SE.MATS. | ENSSSCG00000017983 | per1 | 12 | - | 53376206 | 53 376 343 | 53 370 625 | 53 371 042 | 53 376 572 | 53 376 723 | 47 170 |
| ENSSSCG00000008909 | clock | 8 | - | 54809139 | 54 809 229 | 54 806 213 | 54 806 366 | 54 809 707 | 54 809 808 | 42 410 | |
| ENSSSCG00000029760 | cipc | 7 | + | 100183106 | 100 183 214 | 100 181 042 | 10 0181 090 | 100 186 329 | 100 186 517 | 42 027 | |
| ENSSSCG00000021038 | nrip1 | 13 | - | 179842517 | 179 842 709 | 179 826 140 | 179 831 189 | 179 896 139 | 179 896 221 | 2 799 | |
| ENSSSCG00000008540 | ppp1cb | 3 | - | 110 442 414 | 110 442 549 | 110 438 245 | 110 439 529 | 110 451 383 | 110 451 455 | 3 297 | |
| ENSSSCG00000009249 | hnrnpd | 8 | + | 135 846 884 | 135 846 941 | 135 844 544 | 135 845 076 | 135 856 791 | 135 856 960 | 42 053 | |
| ENSSSCG00000012318 | maged1 | X | + | 45 327 174 | 45 327 217 | 45 327 003 | 45 327 083 | 45 327 488 | 45 327 551 | 49 608 | |
| ENSSSCG00000004832 | ube3a | 1 | + | 141 923 693 | 141 923 813 | 141 887 176 | 141 887 772 | 141 926 999 | 141 927 044 | 16 039 | |
| ENSSSCG00000014010 | mapk9 | 2 | + | 78 352 935 | 78 353 390 | 78 352 352 | 78 352 523 | 78 364 932 | 78 365 062 | 26 439 | |
| ENSSSCG00000023245 | nono | X | + | 57 293 430 | 57 298 994 | 57 290 456 | 57 290 524 | 57 305 745 | 57 305 905 | 29 912 | |
| ENSSSCG00000023307 | fbxw11 | 16 | + | 52 251 834 | 52 251 936 | 52 212 805 | 52 212 850 | 52 295 470 | 52 295 696 | 38 446 | |
| ENSSSCG00000024598 | prkaa1 | 16 | - | 25 782 025 | 25 782 151 | 25 764 765 | 25 764 907 | 25 783 381 | 25 783 524 | 28 263 | |
| ENSSSCG00000028840 | ezh2 | 9 | + | 109 421 603 | 109 421 782 | 109 412 680 | 109 412 809 | 109 425 114 | 109 425 231 | 15 128 | |
| ENSSSCG00000011579 | pparg | 13 | + | 68 326 644 | 68 326 718 | 68 301 565 | 68 301 682 | 68 383 627 | 68 383 855 | 50 750 | |
| MXE.MATS. | ENSSSCG00000013397 | arntl | 2 | - | 45 926 959 | 45 927 077 | 45 923 697 | 45 923 847 | 45 936 353 | 45 936 511 | 3 757 |
| 45 934 501 | 45 934 581 |
Table 5 Differential alternative splicing of circadian rhythm genes in ovary of Xiang pig
| Gene ID | Gene symbol | Chromosome | Strand | Long exon start | Long exon end | Short exon start | Short exon end | Flanking exon start | Flanking exon end | ID | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A5SS.MATS. | ENSSSCG00000000107 | casein kinase | 5 | + | 9 613 116 | 9 613 949 | 9 613 116 | 9 613 265 | 9 614 046 | 9 615 759 | 302 |
| ENSSSCG00000005701 | ass1 | 1 | + | 270 581 146 | 270 582 896 | 270 581 146 | 270 581 303 | 270 585 083 | 270 585 149 | 575 | |
| ENSSSCG00000023245 | nono | X | + | 57 293 430 | 57 298 994 | 57 293 430 | 57 293 509 | 57 305 745 | 57 305 905 | 490 | |
| RI.MATS. | ENSSSCG00000000164 | cry1 | 5 | + | 13 362 925 | 13 364 594 | 13 362 925 | 13 363 183 | 13 363 958 | 13 364 594 | 1 188 |
| ENSSSCG00000017983 | per1 | 12 | - | 53 367 594 | 53 368 034 | 53 367 594 | 53 367 700 | 53 367 902 | 53 368 034 | 2 377 | |
| ENSSSCG00000008645 | id2 | 3 | - | 127 499 409 | 127 502 384 | 127 499 409 | 127 501 531 | 12 7502 320 | 127 502 384 | 911 | |
| ENSSSCG00000012913 | ppp1ca | 2 | + | 5 122 681 | 5 126 953 | 5 122 681 | 5 122 816 | 5 122 899 | 5 126 953 | 622 | |
| ENSSSCG00000036933 | nr1d1 | 12 | + | 22 270 244 | 22 275 515 | 22 270 244 | 22 270 455 | 22 271 146 | 22 275 515 | 717 | |
| ENSSSCG00000008540 | ppp1cb | 3 | - | 110 474 324 | 110 474 733 | 110 474 324 | 110 474 464 | 110 474 677 | 110 474 733 | 182 | |
| ENSSSCG00000015120 | usp2 | 9 | - | 46 565 719 | 46 566 122 | 46 565 719 | 46 565 827 | 46 566 043 | 46 566 122 | 45 | |
| ENSSSCG00000009828 | ppp1cc | 14 | - | 32 069 627 | 32 071 873 | 32 069 627 | 32 070 938 | 32 071 812 | 32 071 873 | 2 314 | |
| ENSSSCG00000011372 | impdh2 | 13 | - | 31 655 808 | 31 658 900 | 31 655 808 | 31 658 713 | 31 658 816 | 3 1658 900 | 327 | |
| ENSSSCG00000000107 | casein kinase | 5 | + | 9 613 116 | 9 613 949 | 9 613 116 | 9 613 265 | 9 613 845 | 9 613 949 | 910 | |
| ENSSSCG00000023245 | nono | X | + | 57293430 | 57 298 994 | 57 293 430 | 57 293 509 | 57 298 717 | 57 298 994 | 14 38 | |
| ENSSSCG00000001880 | sin3a | 7 | + | 58236964 | 58 238 004 | 58 236 964 | 58 237 057 | 58 237 909 | 58 238 004 | 1 997 | |
| SE.MATS. | ENSSSCG00000017983 | per1 | 12 | - | 53376206 | 53 376 343 | 53 370 625 | 53 371 042 | 53 376 572 | 53 376 723 | 47 170 |
| ENSSSCG00000008909 | clock | 8 | - | 54809139 | 54 809 229 | 54 806 213 | 54 806 366 | 54 809 707 | 54 809 808 | 42 410 | |
| ENSSSCG00000029760 | cipc | 7 | + | 100183106 | 100 183 214 | 100 181 042 | 10 0181 090 | 100 186 329 | 100 186 517 | 42 027 | |
| ENSSSCG00000021038 | nrip1 | 13 | - | 179842517 | 179 842 709 | 179 826 140 | 179 831 189 | 179 896 139 | 179 896 221 | 2 799 | |
| ENSSSCG00000008540 | ppp1cb | 3 | - | 110 442 414 | 110 442 549 | 110 438 245 | 110 439 529 | 110 451 383 | 110 451 455 | 3 297 | |
| ENSSSCG00000009249 | hnrnpd | 8 | + | 135 846 884 | 135 846 941 | 135 844 544 | 135 845 076 | 135 856 791 | 135 856 960 | 42 053 | |
| ENSSSCG00000012318 | maged1 | X | + | 45 327 174 | 45 327 217 | 45 327 003 | 45 327 083 | 45 327 488 | 45 327 551 | 49 608 | |
| ENSSSCG00000004832 | ube3a | 1 | + | 141 923 693 | 141 923 813 | 141 887 176 | 141 887 772 | 141 926 999 | 141 927 044 | 16 039 | |
| ENSSSCG00000014010 | mapk9 | 2 | + | 78 352 935 | 78 353 390 | 78 352 352 | 78 352 523 | 78 364 932 | 78 365 062 | 26 439 | |
| ENSSSCG00000023245 | nono | X | + | 57 293 430 | 57 298 994 | 57 290 456 | 57 290 524 | 57 305 745 | 57 305 905 | 29 912 | |
| ENSSSCG00000023307 | fbxw11 | 16 | + | 52 251 834 | 52 251 936 | 52 212 805 | 52 212 850 | 52 295 470 | 52 295 696 | 38 446 | |
| ENSSSCG00000024598 | prkaa1 | 16 | - | 25 782 025 | 25 782 151 | 25 764 765 | 25 764 907 | 25 783 381 | 25 783 524 | 28 263 | |
| ENSSSCG00000028840 | ezh2 | 9 | + | 109 421 603 | 109 421 782 | 109 412 680 | 109 412 809 | 109 425 114 | 109 425 231 | 15 128 | |
| ENSSSCG00000011579 | pparg | 13 | + | 68 326 644 | 68 326 718 | 68 301 565 | 68 301 682 | 68 383 627 | 68 383 855 | 50 750 | |
| MXE.MATS. | ENSSSCG00000013397 | arntl | 2 | - | 45 926 959 | 45 927 077 | 45 923 697 | 45 923 847 | 45 936 353 | 45 936 511 | 3 757 |
| 45 934 501 | 45 934 581 |
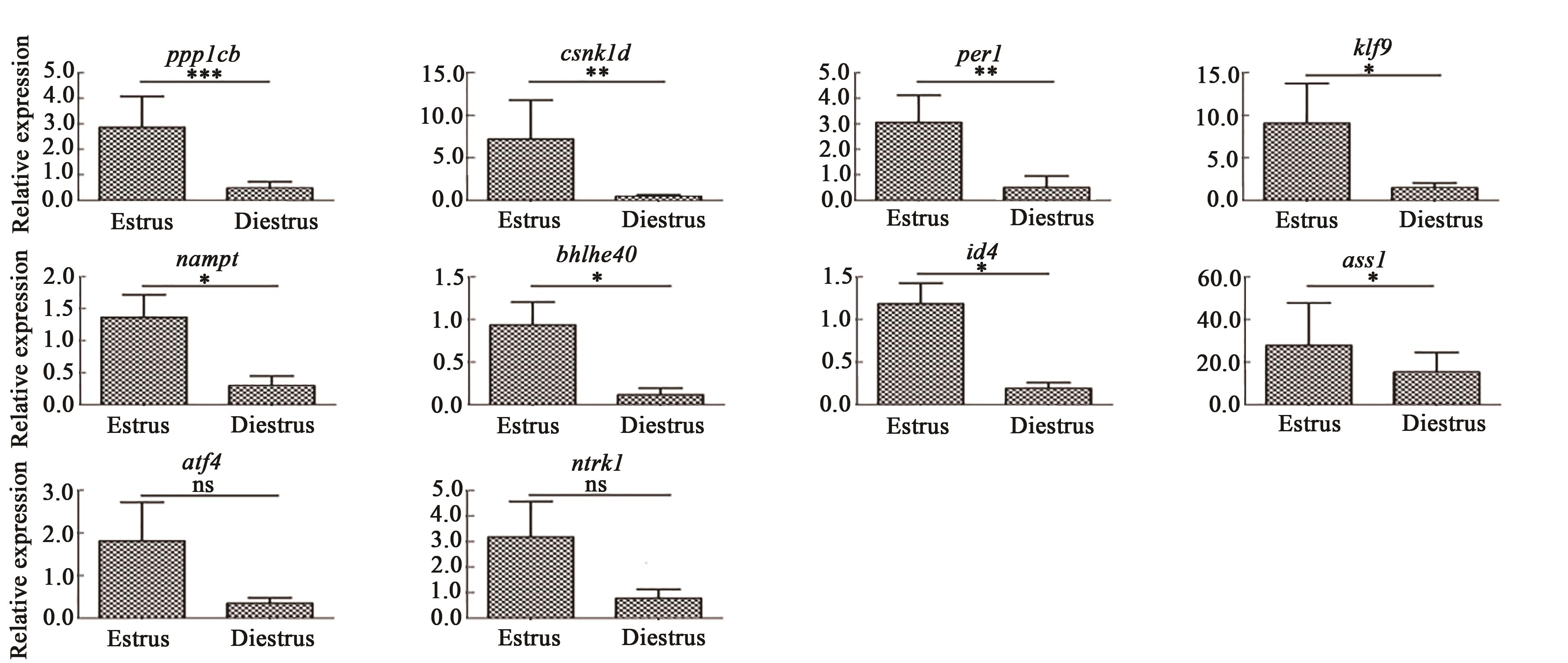
Fig. 2 Verification of differential expression by RT-qPCRNote: *, ** and *** indicate significant differences between estrus and diestrus at P<0.05, P<0.01 and P<0.001 levels, respectively; ns indicates no significant difference.

Fig. 3 RT-PCR verification of alternative splicing eventsA: Mutually exclusive exon event of thrap3 gene; B: Exon skipping event of clock gene; C: Intron retain event of cry1 gene; D: Intron retain event of per1 gene; E: Exon skipping event of cipc gene; F: Mutually exclusive exon event of top2a gene; G: Mutually exclusive exon event of ppp1cb gene; H: Exon skipping event of mapk9 gene. The figures on the left represent the alternative splicing pattern made by rMATS, and the figures on the right represent the electropherogram that proved the existence of the alternative splicing event
| 1 | SCHIBLER U, RIPPERGER J, BROWN S A. Peripheral circadian oscillators in mammals: time and food [J]. J. Biol. Rhythms, 2003, 18( 3): 250- 260. |
| 2 | VIEYRA E, RAMÍREZ D A, LAGUNAS N, et al.. Unilaterally blocking the muscarinic receptors in the suprachiasmatic nucleus in proestrus rats prevents pre-ovulatory LH secretion and ovulation [J]. Reprod. Biol. Endocrinol, 2016, 14( 1): 34- 34. |
| 3 | MANSANO N D S, PARADELA R S, BOHLEN T M, et al.. Vasoactive intestinal peptide exerts an excitatory effect on hypothalamic kisspeptin neurons during estrogen negative feedback [J/OL]. Mol. Cell. Endocrinol., 2022, 542: 111532 [ 2023-01-20]. . |
| 4 | TAKAHASHI J S. Transcriptional architecture of the mammalian circadian clock [J]. Nat. Rev. Genet., 2017, 18( 3): 164- 179. |
| 5 | REPPERT S M, WEAVER D R. Coordination of circadian timing in mammals [J]. Nature, 2002, 418( 6901): 935- 941. |
| 6 | SEN A, SELLIX M T. The circadian timing system and environmental circadian disruption: from follicles to fertility [J]. Endocrinology, 2016, 157( 9): 3366- 3373. |
| 7 | GRAY G D, SÖDERSTEIN P, TALLENTIRE D, et al.. Effects of lesions in various structures of the suprachiasmatic-preoptic region on LH regulation and sexual behavior in female rats [J]. Neuroendocrinology, 1978, 25( 3): 174- 191. |
| 8 | CHRISTIAN C A, MOBLEY J L, MOENTER S M. Diurnal and estradiol-dependent changes in gonadotropin-releasing hormone neuron firing activity [J]. Proc. Natl. Acad Sci. USA, 2005, 102( 43): 15682- 15687. |
| 9 | MIAO X, LUO Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing [J]. Reproduction, 2013, 145( 6): 587- 596. |
| 10 | TERENINA E, FABRE S, BONNET A, et al.. Differentially expressed genes and gene networks involved in pig ovarian follicular atresia [J]. Physiol. Genomics, 2017, 49( 2): 67- 80. |
| 11 | CHACKO E, RANGANATHAN S. Genome-wide analysis of alternative splicing in cow: implications in bovine as a model for human diseases [J/OL]. BMC Genomics, 2009, 10( S3):S 11 [ 2023-01-20]. . |
| 12 | KIANIANMOMENI A, ONG C S, RÄTSCH G, et al.. Genome-wide analysis of alternative splicing in Volvox carteri [J/OL]. BMC. Genomics, 2014, 15: 1117 [ 2023-01-20]. . |
| 13 | KARMAN B N, TISCHKAU S A. Circadian clock gene expression in the ovary: effects of luteinizing hormone [J]. Biol. Reprod., 2006, 75( 4): 624- 632. |
| 14 | FAHRENKRUG J, GEORG B, HANNIBAL J, et al.. Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary [J]. Endocrinology, 2006, 147( 8): 3769- 3776. |
| 15 | HE P J, HIRATA M, YAMAUCHI N, et al.. Gonadotropic regulation of circadian clockwork in rat granulosa cells [J]. Mol. Cell Biochem., 2007, 302( 1-2): 111- 118. |
| 16 | YOSHIKAWA T, SELLIX M, PEZUK P, et al.. Timing of the ovarian circadian clock is regulated by gonadotropins [J]. Endocrinology, 2009, 150( 9): 4338- 4347. |
| 17 | HE P J, HIRATA M, YAMAUCHI N, et al.. The disruption of circadian clockwork in differentiating cells from rat reproductive tissues as identified by in vitro real-time monitoring system [J]. J. Endocrinol., 2007, 193( 3): 413- 420. |
| 18 | JOHNSON M H, LIM A, FERNANDO D, et al.. Circadian clockwork genes are expressed in the reproductive tract and conceptus of the early pregnant mouse [J]. Reprod. Biomed. Online, 2002, 4( 2): 140- 145. |
| 19 | CUSHMAN R A, ALLAN M F, JONES S A, et al.. Localization of Period 1 mRNA in the ruminant oocyte and investigations of its role in ovarian function [J]. Anim. Reprod. Sci., 2007, 99( 1-2): 93- 105. |
| 20 | SELLIX M T. Circadian clock function in the mammalian ovary [J]. J. Biol. Rhythms, 2015, 30( 1): 7- 19. |
| 21 | ZHENG B, ALBRECHT U, KAASIK K, et al.. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock [J]. Cell, 2001, 105( 5): 683- 694. |
| 22 | PILORZ V, STEINLECHNER S. Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? [J] Reproduction, 2008, 135( 4): 559- 568. |
| 23 | BODEN M J, VARCOE T J, VOULTSIOS A, et al.. Reproductive biology of female Bmal1 null mice [J]. Reproduction, 2010, 139( 6): 1077- 1090. |
| 24 | ALVAREZ J D, HANSEN A, ORD T, , et al.. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice [J]. J. Biol. Rhythms, 2008, 23( 1): 26- 36. |
| 25 | LIU Y, JOHNSON B P, SHEN A L, et al.. Loss of BMAL1 in ovarian steroidogenic cells results in implantation failure in female mice [J]. Proc. Natl. Acad. Sci. USA, 2014, 111( 39): 14295- 14300. |
| 26 | MILLER B H, OLSON S L, TUREK F W, et al.. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy [J]. Curr. Biol., 2004, 14( 15): 1367- 1373. |
| 27 | CHEN H T, CHU G Y, ZHAO L J, et al.. Rev-erbα regulates circadian rhythms and StAR expression in rat granulosa cells as identified by the agonist GSK4112 [J]. Biochem. Biophys. Res. Commun., 2012, 420( 2): 374- 379. |
| 28 | CHU G Y, MISAWA I, CHEN H T, et al.. Contribution of FSH and triiodothyronine to the development of circadian clocks during granulosa cell maturation [J]. Am. J. Physiol-Endoc. M, 2012, 302( 6): 645- 653. |
| 29 | CHEN H T, ZHAO L J, CHU G Y, et al.. FSH induces the development of circadian clockwork in rat granulosa cells via a gap junction protein Cx43-dependent pathway [J]. Am. J. Physiol-endoc. M, 2013, 304( 6): E566- 575. |
| 30 | BROWN-GRANT K, RAISMAN G. Abnormalities in reproductive function associated with the destruction of the suprachiasmatic nuclei in female rats [J]. P. Roy. Soc. B-biol. Sci., 1977, 198( 1132): 279- 296. |
| 31 | LUO Z Y, DAI X L, RAN X Q, et al.. Identification and profile of microRNAs in Xiang pig testes in four different ages detected by Solexa sequencing [J]. Theriogenology, 2018, 117: 61- 71. |
| 32 | SHEN S, PARK J W, LU Z X, et al.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data [J]. Proc. Natl. Acad. Sci. USA, 2014, 111( 51): 5593- 5601. |
| 33 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method [J]. Methods, 2001, 25( 4): 402- 408. |
| 34 | ROSENWASSER A M, TUREK F W. Neurobiology of circadian rhythm regulation [J]. Sleep Med. Clin., 2015, 10( 4): 403- 412. |
| 35 | LI R W, CHENG S T, WANG Z R. Circadian clock gene plays a key role on ovarian cycle and spontaneous abortion [J]. Cell Physiol. Biochem., 2015, 37( 3): 911- 920. |
| 36 | NAKAMURA T J, SELLIX M T, KUDO T, et al.. Influence of the estrous cycle on clock gene expression in reproductive tissues: effects of fluctuating ovarian steroid hormone levels [J]. Steroids, 2010, 75( 3): 203- 212. |
| 37 | MANSUY V, RISOLD P Y, GLAUSER M, et al.. Expression of the GABAA receptor associated protein Gec1 is circadian and dependent upon the cellular clock machinery in GnRH secreting GnV-3 cells [J]. Mol. Cell Endocrinol., 2009, 307( 1-2): 68- 76. |
| 38 | MENDOZA J, LOPEZ-LOPEZ C, REVEL F G, et al.. Dimorphic effects of leptin on the circadian and hypocretinergic systems of mice [J]. J. Neuroendocrinol., 2011, 23( 1): 28- 38. |
| 39 | SMARR B L, GILE J J, DE LA IGLESIA H O. Oestrogen-independent circadian clock gene expression in the anteroventral periventricular nucleus in female rats: possible role as an integrator for circadian and ovarian signals timing the luteinising hormone surge [J]. J. Neuroendocrinol., 2013, 25( 12): 1273- 1279. |
| 40 | ZHANG Y, MENG N, BAO H, et al.. Circadian gene PER1 senses progesterone signal during human endometrial decidualization [J/OL]. J. Endocrinol., 2019: 0284 [ 2023-01-20]. . |
| 41 | LÓPEZ-RODRÍGUEZ D, FRANSSEN D, SEVRIN E, et al.. Persistent vs transient alteration of folliculogenesis and estrous cycle after neonatal vs adult exposure to bisphenol A [J]. Endocrinology, 2019, 160( 11): 2558- 2572. |
| 42 | RAVINDER R, KAIPA O, BADDELA V S, et al.. Saliva ferning, an unorthodox estrus detection method in water buffaloes ( Bubalus bubalis) [J]. Theriogenology, 2016, 86( 5): 1147- 1155. |
| 43 | NODA M, IWAMOTO I, TABATA H, et al.. Role of Per3, a circadian clock gene, in embryonic development of mouse cerebral cortex [J/OL]. Sci. Rep., 2019, 9( 1): 5874 [ 2023-01-20]. . |
| 44 | CHU Q, ZHOU B, XU F, et al. Genome-wide differential mRNA expression profiles in follicles of two breeds and at two stages of estrus cycle of gilts [J/OL]. Sci. Rep., 2017, 7( 1): 5052 [ 2023-01-20]. . |
| 45 | GOSSAN N C, ZHANG F, GUO B, et al.. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor [J]. Nucl. Acids Res., 2014, 42( 9): 5765- 5775. |
| 46 | KENNAWAY D J, VARCOE T J, VOULTSIOS A, et al.. Acute inhibition of casein kinase 1delta/epsilon rapidly delays peripheral clock gene rhythms [J]. Mol. Cell. Biochem., 2015, 398( 1-2): 195- 206. |
| 47 | SCHMUTZ I, WENDT S, SCHNELL A, et al.. Protein phosphatase 1 (PP1) is a post-translational regulator of the mammalian circadian clock [J/OL]. PLoS. One, 2011, 6( 6):e 21325 [ 2023-01-20]. . |
| 48 | KURABAYASHI N, HIROTA T, SAKAI M, et al.. DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping [J]. Mol. Cell Biol., 2010, 30( 7): 1757- 1768. |
| 49 | DEVOS J W S V, WEVRICK R. Magel2, a Prader-Willi syndrome candidate gene, modulates the activities of circadian rhythm proteins in cultured cells [J/OL]. J. Circadian Rhythms, 2011, 9: 12 [ 2023-01-20]. . |
| 50 | WANG R H, ZHAO T, CUI K, et al.. Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging [J/OL]. Sci. Rep., 2016, 6: 28633 [ 2023-01-20]. . |
| 51 | KOIKE N, YOO S H, HUANG H C, et al.. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals [J]. Science, 2012, 338( 6105): 349- 354. |
| 52 | OGAWA Y, KAWANO Y, YAMAZAKI Y, et al.. Shikonin shortens the circadian period: possible involvement of Top2 inhibition [J]. Biochem. Biophys. Res. Commun., 2014, 443( 1): 339- 343. |
| 53 | HOSODA H, KATO K, ASANO H, et al.. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription [J/OL]. Mol. Brain, 2009, 2: 34 [ 2023-01-20]. . |
| 54 | HOU T Y, WARD S M, MURAD J M, et al.. ID2 (inhibitor of DNA binding 2) is a rhythmically expressed transcriptional repressor required for circadian clock output in mouse liver [J]. J. Biol. Chem., 2009, 284( 46): 31735- 31745. |
| 55 | KNOEDLER J R, AVILA-MENDOZA J, SUBRAMANI A, et al.. The paralogous kruppel-like factors 9 and 13 regulate the mammalian cellular circadian clock output gene Dbp [J]. J. Biol. Rhythms, 2020, 35( 3): 257- 274. |
| 56 | NAKAO N, YASUO S, NISHIMURA A, et al.. Circadian clock gene regulation of steroidogenic acute regulatory protein gene expression in preovulatory ovarian follicles [J]. Endocrinology, 2007, 148( 7): 3031- 3038. |
| 57 | WU Y G, BENNETT J, TALLA D, et al.. Testosterone, not 5alpha-dihydrotestosterone, stimulates LRH-1 leading to FSH-independent expression of Cyp19 and P450scc in granulosa cells [J]. Mol. Endocrinol., 2011, 25( 4): 656- 668. |
| 58 | ISAYAMA K, CHEN H, YAMAUCHI N, et al.. REV-ERBalpha inhibits the PTGS2 expression in bovine uterus endometrium stromal and epithelial cells exposed to ovarian steroids [J]. J. Reprod. Dev., 2014, 60( 5): 362- 370. |
| 59 | MOHAWK J A, GREEN C B, TAKAHASHI J S. Central and peripheral circadian clocks in mammals [J]. Annu. Rev. Neurosci., 2012, 35: 445- 462. |
| 60 | LEE Y, RIO D C. Mechanisms and regulation of alternative pre-mRNA splicing [J]. Annu. Rev. Biochem., 2015, 84: 291- 323. |
| 61 | GENOV N, BASTI A, ABREU M, et al.. A bioinformatic analysis identifies circadian expression of splicing factors and time-dependent alternative splicing events in the HD-MY-Z cell line [J/OL]. Sci. Rep., 2019, 9( 1): 11062 [ 2023-01-20]. . |
| 62 | SHAKHMANTSIR I, NAYAK S, GRANT G R, et al.. Spliceosome factors target timeless (tim) mRNA to control clock protein accumulation and circadian behavior in Drosophila [J/OL]. Elife, 2018, 7: 39821 [ 2023-01-20]. . |
| 63 | SANCHEZ S E, PETRILLO E, BECKWITH E J, et al.. A methyl transferase links the circadian clock to the regulation of alternative splicing [J]. Nature, 2010, 468( 7320): 112- 116. |
| 64 | PETRILLO E, SANCHEZ S E, KORNBLIHTT A R, et al. Alternative splicing adds a new loop to the circadian clock [J]. Comm. Integ. Biol., 2011, 4( 3): 284- 286. |
| 65 | MCGLINCY N J, VALOMON A, CHESHAM J E, et al.. Regulation of alternative splicing by the circadian clock and food related cues [J/OL]. Genome Biol., 2012, 13( 6):R 54 [ 2023-01-20]. . |
| 66 | HUGHES M E, GRANT G R, PAQUIN C, et al.. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain [J]. Genome Res., 2012, 22( 7): 1266- 1281. |
| 67 | PREUßNER M, WILHELMI I, SCHULTZ A S, et al.. Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice [J]. Mol. Cell, 2014, 54( 4): 651- 662. |
| [1] | 许鑫, 孟昆, 蔡红英, 杨培龙, 蒋显仁. 乳酸菌在猪生产中的应用研究进展[J]. 中国农业科技导报, 2023, 25(2): 19-26. |
| [2] | 翟利敏, 李文通, 冯政, 李华, 裴杨莉. 基因编辑猪的研究现状[J]. 中国农业科技导报, 2022, 24(8): 25-34. |
| [3] | 曾俊棋,岳万福*. 454焦磷酸测序技术对不同猪种肠道菌群差异的分析(英文)[J]. 中国农业科技导报, 2015, 17(6): 44-49. |
| [4] | 刘小红1,王健2,刘长春3,赵云翔1,杨军香3,梁菲菲1,黄萌萌3, 何耀棋1,胡斌1,闻丽君1,姚钟骁1,陈瑶生1*. 我国生猪标准化养殖模式和技术水平分析[J]. , 2013, 15(6): 72-77. |
| [5] | 李文刚1,申超1,隋超1,刘晓青2,吴志娟1,焦福林1,闫益波1. 现代化养猪新特点及发展趋势[J]. , 2012, 14(6): 133-138. |
| [6] | 张小波1,2*,吴潇2,3*,何慧1,朱连龙2,3,唐雪明2,3. 基于SNPs标记的猪肉DNA溯源技术的研究[J]. , 2011, 13(3): 85-91. |
| [7] | 张军民,李秋菊. 我国生猪适宜养殖模式的探讨[J]. , 2008, 10(6): 23-28. |
| [8] | 任登魁. 现代农资物流与供应链管理[J]. , 2005, 7(1): 72-75. |
| [9] | 荆继忠. 21世纪种猪经济及科技创新特点和可持续发展策略[J]. , 2002, 4(1): 70-75. |
| [10] | 王九峰 杨立彬. 不同类型的日粮纤维对怀孕母猪胃内pH值、微生物状态以及胃排空速度的影响研[J]. , 2001, 3(1): 28-33. |
| [11] | 顾宪红. 断奶仔猪日粮蛋白质需要量及低蛋白日粮对仔猪的影响[J]. , 2001, 3(1): 34-37. |
| [12] | 郑春田 杨立彬. 低蛋白质日粮补充异亮氨酸对仔猪全身蛋白质周转代谢的影响[J]. , 2001, 3(1): 38-42. |
| [13] | 张丽英 谯仁彦 等. 大豆寡糖对断奶仔猪生产性能、腹泻及相关生化指标影响的研究[J]. , 2001, 3(1): 43-48. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号