




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (6): 11-21.DOI: 10.13304/j.nykjdb.2021.0924
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Received:2021-11-01
Accepted:2022-01-18
Online:2023-06-01
Published:2023-07-28
Contact:
Yuanyuan YAN
通讯作者:
阎媛媛
作者简介:陈丽婷 E-mail:clt1158433820@163.com;
基金资助:CLC Number:
Liting CHEN, Yuanyuan YAN. Investigation of Regulatory Mechanism of Floral Integrators in Upland Cotton[J]. Journal of Agricultural Science and Technology, 2023, 25(6): 11-21.
陈丽婷, 阎媛媛. 陆地棉开花整合子调控机制解析[J]. 中国农业科技导报, 2023, 25(6): 11-21.
引物名称 Primer name | 引物序列 Primer sequence(5’-3’) | 用途 Purpose |
|---|---|---|
| GhLFY-2-F(PstI) | TT | 基因克隆 Gene cloning |
| GhLFY-2-R(SmaI) | TT | |
| GhSOC1-4-F(EcoRI) | CC | |
| GhSOC1.4-R(BamHI) | TT | |
| GhFT-2-F(EcoRI) | TT | |
| GhFT-2-R(XmaI) | TT | |
| GhLFY-2-RT-F | AATTCGAGGGTGGTGATGAT | qRT-PCR |
| GhLFY-2-RT-R | CTCCGTTACGATGAAAGGGT | |
| GhSOC1-5-RT-F | GGCGCATAGAGAACGATACA | |
| GhSOC1-5-RT-R | TTTGTATGCCGCCTATAACG | |
| GhFT-2-RT-F | GGGATGATCTGAGGACCTTCT | |
| GhFT-2-RT-R | CACCAGTTGTGGCTGGAATA | |
| AtTUB2-F | ATCCGTGAAGAGTACCCAGAT | |
| AtTUB2-R | AAGAACCATGCACTCATCAGC |
Table 1 Primer sequences in the study
引物名称 Primer name | 引物序列 Primer sequence(5’-3’) | 用途 Purpose |
|---|---|---|
| GhLFY-2-F(PstI) | TT | 基因克隆 Gene cloning |
| GhLFY-2-R(SmaI) | TT | |
| GhSOC1-4-F(EcoRI) | CC | |
| GhSOC1.4-R(BamHI) | TT | |
| GhFT-2-F(EcoRI) | TT | |
| GhFT-2-R(XmaI) | TT | |
| GhLFY-2-RT-F | AATTCGAGGGTGGTGATGAT | qRT-PCR |
| GhLFY-2-RT-R | CTCCGTTACGATGAAAGGGT | |
| GhSOC1-5-RT-F | GGCGCATAGAGAACGATACA | |
| GhSOC1-5-RT-R | TTTGTATGCCGCCTATAACG | |
| GhFT-2-RT-F | GGGATGATCTGAGGACCTTCT | |
| GhFT-2-RT-R | CACCAGTTGTGGCTGGAATA | |
| AtTUB2-F | ATCCGTGAAGAGTACCCAGAT | |
| AtTUB2-R | AAGAACCATGCACTCATCAGC |

Fig. 2 Phylogenetic analysis of plant floral integrator genesNote:Gh—Gossypium hirsutum; Gb—Gossypium barbadense; Ga—Gossypium arboreum; Gr—Gossypium raimondii; At—Arabidopsis thaliana; Pt—Populus tomentosa; Nt—Nicotiana tabacum; Vv—Vitis vinifera.
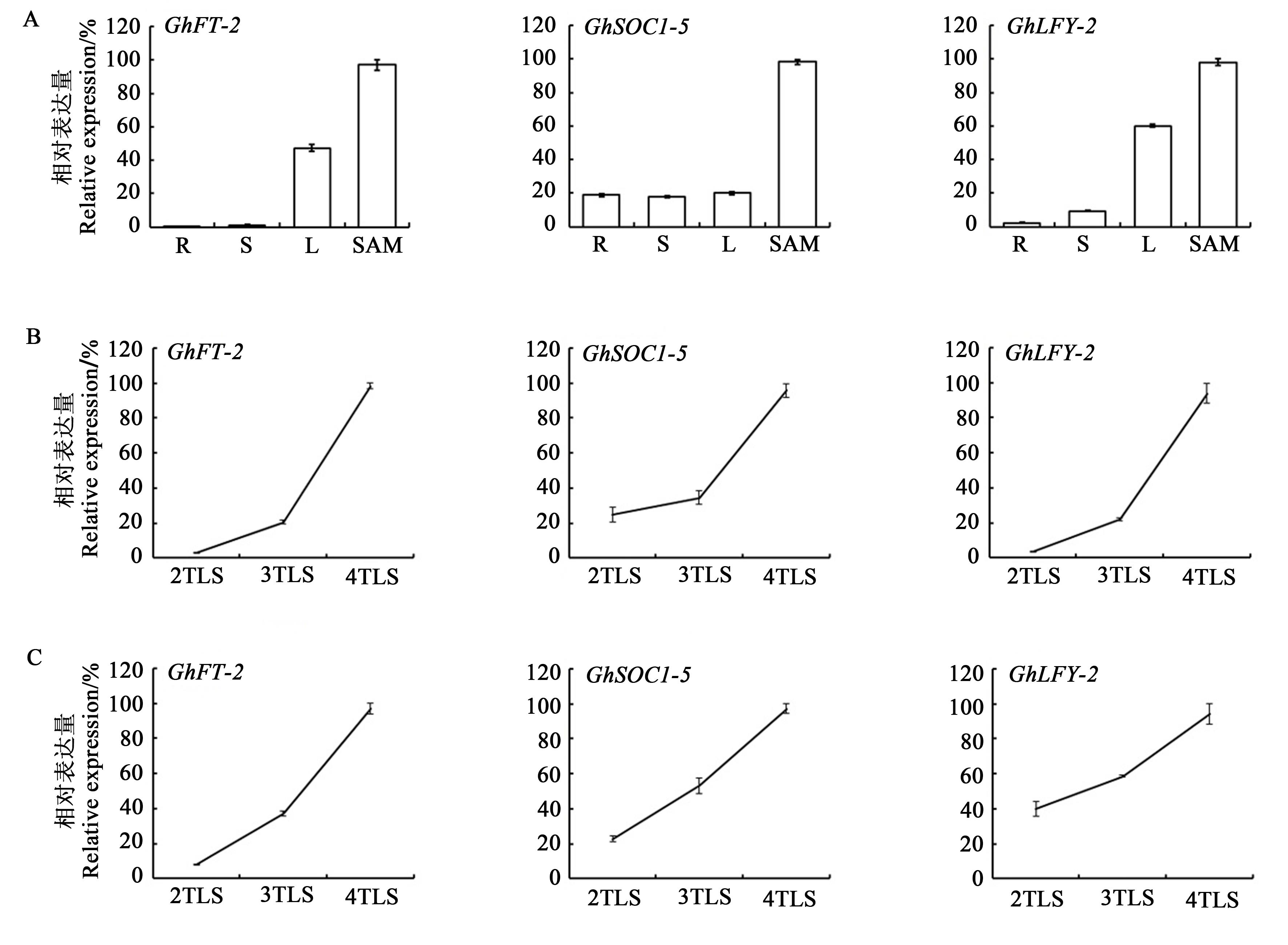
Fig. 3 Expression of floral integrators genes in Gossypium hirsutumA: Tissue-specific expression of flowering integrator of Gossypium hirsutum; B: Expression trends of flowering integrators in cotton leaves during seedling development; C: Expression trends of flowering integrators in cotton SAM during seedling development. R—Root; S—Stem; L—Leaf; TLS—True leaf stage

Fig. 4 Cotton flowering integrators promotes floweringA: Early flowering phenotype of transgenic plants; B: Comparison of gene expression and flowering time between WT and transgenic plants; C: Flowering time of WT and transgenic lines, ** indicates significant differences (P<0.01); D: Solitary flowers produced in GhLFY-2 transgenic plants

Fig. 5 Expression of floral integrator genes in transgenic Arabidopsis plantsNote:A, B and C shows relative expression of flowering integrators in GhFT-2, GhSOC1-5 and GhLFY-2 transgenic plants, respectively.
| 1 | 喻树迅,王寒涛,魏恒玲,等.棉花早熟性研究进展及其应用[J].棉花学报, 2017, 29(): 1-10. |
| YU S X, WANG H T, WEI H L, et al.. Research progress and application of early maturity in upland cotton [J]. Cotton Sci., 2017, 29(S1): 1-10. | |
| 2 | FORNARA F, MONTAIGU A D, COUPLAND G. SnapShot: control of flowering in Arabidopsis [J]. Cell, 2010, 141(3): 550-550. |
| 3 | WIGGE P. FT, a mobile developmental signal in plants [J]. Curr. Biol., 2011, 21(9): R374-R378. |
| 4 | KOBAYASHI Y, KAYA H, GOTO K, et al.. A pair of related genes with antagonistic roles in mediating flowering signals [J]. Science, 2000, 286(5446): 1960-1962. |
| 5 | ABE M, KOBAYASHI Y, YAMAMOTO S, et al.. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex [J]. Science, 2005, 309(5737): 1052-1056. |
| 6 | WIGGE P A, MIN C K, JAEGER K E, et al.. Integration of spatial and temporal information during floral induction in Arabidopsis [J]. Science, 2005, 309(5737): 1056-1059. |
| 7 | TAMAKI S, MATSUO S, WONG H L, et al.. Hd3a protein is a mobile flowering signal in rice [J]. Science, 2007, 316(5827): 1033-1036. |
| 8 | MCGARRY R C, PREWITT S, AYRE B G. Overexpression of FT in cotton affects architecture but not floral organogenesis [J/OL]. Plant Signal Behav., 2013, 8(4):e23602 [2021-12-03]. . |
| 9 | GUO D, LI C, DONG R, et al.. Molecular cloning and functional analysis of the FLOWERING LOCUS T(FT) homolog GhFT1 from Gossypium hirsutum [J]. J. Integr. Plant Biol., 2015, 57(6): 522-533. |
| 10 | LI C, ZHANG Y, ZHANG K, et al.. Promoting flowering, lateral shoot outgrowth, leaf development, and flower abscission in tobacco plants overexpressing cotton FLOWERING LOCUS T (FT)-like gene GhFT1 [J/OL]. Front. Plant Sci., 2015, 6: 454 [2021-12-03]. . |
| 11 | BLÁZQUEZ M A, WEIGEL D. Integration of floral inductive signals in Arabidopsis [J]. Nature, 2000, 404(6780): 889-892. |
| 12 | YOO S K, CHUNG K S, KIM J, et al.. Constant activates suppressor of overexpression of constant 1 through flowering locus T to promote flowering in Arabidopsis [J]. Plant Physiol., 2005, 139(2): 770-778. |
| 13 | ZHANG X, WEI J, FAN S, et al.. Functional characterization of GhSOC1 and GhMADS42 homologs from upland cotton (Gossypium hirsutum L.) [J]. Plant Sci., 2016, 242: 178-186. |
| 14 | CERDÁN P D, CHORY J. Regulation of flowering time by light quality [J]. Nature, 2003, 423(6942):881-885. |
| 15 | SVEN E, HENRIK B, THOMAS M, et al.. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation [J]. Plant Cell, 2006, 18(9): 2172-2181. |
| 16 | LILJEGREN S J, GUSTAFSON-BROWN C, PINYOPICH A, et al.. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate [J]. Plant Cell, 1999, 11(6): 1007-1018. |
| 17 | WAGNER D, SABLOWSKI R, MEYEROWITZT E M. Transcriptional activation of APETALA1 by LEAFY [J]. Science, 1999, 285(5427): 582-584. |
| 18 | HEMPEL F D, WEIGEL D, MANDEL M A, et al.. Floral determination and expression of floral regulatory genes in Arabidopsis [J]. Development, 1997, 124(19): 3845-3853. |
| 19 | BLAZQUEZ M A, SOOWAL L N, LEE I, et al.. LEAFY expression and flower initiation in Arabidopsis [J]. Development, 1997, 124(19): 3835-3844. |
| 20 | WEIGEL D, NILSSON O. A developmental switch sufficient for flower initiation in diverse plants [J]. Nature, 1995, 377(6549): 495-500. |
| 21 | AHEARN K P, JOHNSON H A, DETLEF W, et al.. NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristem initiation and floral structure [J]. Plant Cell Physiol., 2001, 10: 1130-1139. |
| 22 | KYOZUKA J, KONISHI S, NEMOTO K, et al.. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation [J]. Proc. Natl. Acad. Sci. USA, 1998, 95(5): 1979-1982. |
| 23 | LI J, FAN S L, SONG M Z, et al.. Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L [J]. Plant Cell Rep., 2013, 32(11): 1675-1686. |
| 24 | LEE J, OH M, PARK H, et al.. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY [J]. Plant J., 2008, 55(5): 832-843. |
| 25 | YAN Y, SHEN L, CHEN Y, et al.. A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis [J]. Dev. Cell, 2014, 30(4): 437-448. |
| 26 | HIGUCHI Y, NARUMI T, ODA A, et al.. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums [J]. Proc. Natl. Acad. Sci. USA, 2013, 110(42): 17137-17142. |
| 27 | SONG Q, ZHANG T, STELLY D M, et al.. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons [J/OL]. Genome Biol., 2017, 18(1): 99 [2021-12-03]. . |
| 28 | MA Z, ZHANG Y, WU L . et al ..High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement [J]. Nat. Genet., 2021, 53: 1385-1391. |
| 29 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method [J]. Methods, 2001, 25(4): 402-408. |
| 30 | SARAH M N, SINARA A, GUSTAVO M A, et al.. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii) [J]. Plant Physiol. Bioch., 2018, 127: 169-184. |
| 31 | REN Z Y, YU D Q, YANG Z E, et al.. Genome-wide identification of the MIKC-Type MADS-box gene family in Gossypium hirsutum L. unravels their roles in flowering [J/OL]. Front. Plant Sci., 2017, 8: 384 [2021-12-03]. . |
| 32 | PATIRANAGE D, ASARE E, MALDONADO-TAIPE N, et al.. Haplotype variations of major flowering time genes in quinoa unveil their role in the adaptation to different environmental conditions [J]. Plant Cell Environ., 2021, 44(8): 2565-2579. |
| 33 | 樊红娟,程雪敏,郑思,等.棉花GhFT基因的克隆及功能分析[J].分子植物育种, 2019, 18(17): 29-36. |
| FAN H J, CHENG X M, ZHENG S, et al.. The cloning and function analysis of GhFT gene in Gossypium hirsutum [J]. Mol. Plant Breed., 2020, 18(17): 29-36. | |
| 34 | JACK T. Molecular and genetic mechanisms of floral control [J]. Plant Cell, 2004, 16 (Supp. l): S1-S17. |
| 35 | MAIZEL A, BUSCH M A, TANAHASHI T, et al.. The floral regulator LEAFY evolves by substitutions in the DNA binding domain [J]. Science, 2005, 308(5719): 260-263. |
| 36 | KELLY A J, BONNLANDER M B, MEEKS-WAGNER D R. NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems [J]. Plant Cell, 1995, 7(2): 225-234. |
| 37 | HOFER J, TURNER L, HELLENS R, et al.. UNIFOLIATA regulates leaf and flower morphogenesis in pea [J]. Curr. Biol., 1997, 7(8): 581-587. |
| 38 | CARMONA M J, CUBAS P, MARTÍNEZ-ZAPATER J M. VFL, the grapevine FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate [J]. Plant Physiol., 2002, 130(1): 68-77. |
| [1] | ZHANG Quanyan, ZHANG Peigao, XU Chunxia, CONG Yining, LIU Li. Advances in Genetic Researches and Molecular Regulation Mechanism of Leaf Angle in Maize [J]. Journal of Agricultural Science and Technology, 2021, 23(10): 15-24. |
| [2] | WANG Heng, DING Jun. Mechanism of Polyunsaturated Fatty Acids Biosynthesis and Metabolism in Marine Invertebrates: A Review [J]. Journal of Agricultural Science and Technology, 2021, 23(10): 181-191. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号 