




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (4): 63-74.DOI: 10.13304/j.nykjdb.2021.0926
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Meili LI1( ), Junji SU1, Yonglin YANG2, Jianghong QIN2, Xianxian LI1, Delong YANG1(
), Junji SU1, Yonglin YANG2, Jianghong QIN2, Xianxian LI1, Delong YANG1( ), Qi MA3(
), Qi MA3( ), Caixiang WANG1(
), Caixiang WANG1( )
)
Received:2021-11-02
Accepted:2021-12-09
Online:2022-04-15
Published:2022-04-19
Contact:
Delong YANG,Qi MA,Caixiang WANG
李美丽1( ), 宿俊吉1, 杨永林2, 秦江鸿2, 李鲜鲜1, 杨德龙1(
), 宿俊吉1, 杨永林2, 秦江鸿2, 李鲜鲜1, 杨德龙1( ), 马麒3(
), 马麒3( ), 王彩香1(
), 王彩香1( )
)
通讯作者:
杨德龙,马麒,王彩香
作者简介:李美丽 E-mail: 2538268277@qq.com
基金资助:CLC Number:
Meili LI, Junji SU, Yonglin YANG, Jianghong QIN, Xianxian LI, Delong YANG, Qi MA, Caixiang WANG. Identification of COI Family Genes and Their Expression in Gossypium hirsutum L. Under Drought and Salt Stress[J]. Journal of Agricultural Science and Technology, 2022, 24(4): 63-74.
李美丽, 宿俊吉, 杨永林, 秦江鸿, 李鲜鲜, 杨德龙, 马麒, 王彩香. 陆地棉COI家族基因鉴定及在干旱和盐胁迫下的表达分析[J]. 中国农业科技导报, 2022, 24(4): 63-74.
引物名称 Primer name | 正向引物 Forward primer(5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| GhCOI1⁃D01 | GTGACTTGACCGATGCAGGA | GAAAGCGCAAGAACCACAGG |
| GhCOI1⁃D06 | CTCGTTTCGATGCTGTCACG | CGAGTCAAATTCCGGCAACG |
| GhCOI3⁃D04 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI3⁃A05 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI5⁃A05 | CAGCTCTGCGTTCTGGTTTG | AGCAATCTCCTGGCAACCAT |
| GhCOI5⁃A06 | TGGCGGAGCAAATGTCTGAT | ACTCGGAATGACCCGAAACC |
| GhCOI7⁃A07 | CCGGCGACAGTAACAAAGGA | GGACATCCAAAGGGATCGCA |
| GhCOI9⁃D08 | AGCTCGTAAGCCAGTTTCCC | CCTCAACGGCCAACAATTCG |
| GhCOI10⁃A09 | TCAAGTTCCTCGCTCAACGG | TCATCTCTGCGACGCCTTTT |
| GhCOI10⁃D09 | TTTGGCAACTGAGTGTGGGA | GCCTCCAGTGATTTGTCGGT |
| GhActin | ATCCTCCGTCTTGACCTTG | TGTCCGTCAGGCAACTCAT |
Table 1 Primer sequence for qRT?PCR
引物名称 Primer name | 正向引物 Forward primer(5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| GhCOI1⁃D01 | GTGACTTGACCGATGCAGGA | GAAAGCGCAAGAACCACAGG |
| GhCOI1⁃D06 | CTCGTTTCGATGCTGTCACG | CGAGTCAAATTCCGGCAACG |
| GhCOI3⁃D04 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI3⁃A05 | CAACCCGGTATGTCCGATGT | CACCGCCTGCAAACTAGAGA |
| GhCOI5⁃A05 | CAGCTCTGCGTTCTGGTTTG | AGCAATCTCCTGGCAACCAT |
| GhCOI5⁃A06 | TGGCGGAGCAAATGTCTGAT | ACTCGGAATGACCCGAAACC |
| GhCOI7⁃A07 | CCGGCGACAGTAACAAAGGA | GGACATCCAAAGGGATCGCA |
| GhCOI9⁃D08 | AGCTCGTAAGCCAGTTTCCC | CCTCAACGGCCAACAATTCG |
| GhCOI10⁃A09 | TCAAGTTCCTCGCTCAACGG | TCATCTCTGCGACGCCTTTT |
| GhCOI10⁃D09 | TTTGGCAACTGAGTGTGGGA | GCCTCCAGTGATTTGTCGGT |
| GhActin | ATCCTCCGTCTTGACCTTG | TGTCCGTCAGGCAACTCAT |
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI1⁃A01 | GH_A01G1190 | 534 | 57.36 | 8.7 | 叶绿体、细胞核Chloroplast, nucleus |
| GhCOI1⁃D01 | GH_D01G1258 | 534 | 57.54 | 8.82 | 细胞核Nucleus |
| GhCOI1⁃A05 | GH_A05G2065 | 534 | 57.38 | 8.86 | 细胞核Nucleus |
| GhCOI1⁃D05 | GH_D05G2095 | 535 | 57.44 | 8.83 | 细胞核Nucleus |
| GhCOI1⁃A06 | GH_A06G0210 | 528 | 56.82 | 8.74 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI1⁃D06 | GH_D06G0195 | 528 | 56.7 | 8.86 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI2⁃A02 | GH_A02G1803 | 544 | 58.97 | 8.49 | 细胞核Nucleus |
| GhCOI2⁃D03 | GH_D03G0256 | 514 | 55.87 | 7.87 | 细胞质Cytoplasm |
| GhCOI2⁃A10 | GH_A10G0913 | 545 | 59.31 | 8.6 | 细胞质Cytoplasm |
| GhCOI2⁃D10 | GH_D10G1048 | 545 | 59.21 | 8.59 | 细胞质Cytoplasm |
| GhCOI3⁃D04 | GH_D04G0696 | 591 | 66.99 | 5.98 | 细胞质Cytoplasm |
| GhCOI3⁃A05 | GH_A05G3439 | 589 | 66.74 | 5.81 | 细胞质Cytoplasm |
| GhCOI4⁃A05 | GH_A05G0481 | 362 | 39.99 | 6.32 | 细胞质Cytoplasm |
| GhCOI4⁃D05 | GH_D05G0480 | 362 | 40.08 | 6.96 | 细胞质Cytoplasm |
| GhCOI5⁃A05 | GH_A05G2217 | 611 | 68.38 | 5.97 | 细胞核Nucleus |
| GhCOI5⁃D05 | GH_D05G2242 | 611 | 68.32 | 5.88 | 叶绿体Chloroplast |
| GhCOI5⁃A06 | GH_A06G0343 | 615 | 69.03 | 5.67 | 细胞核Nucleus |
| GhCOI5⁃D06 | GH_D06G0328 | 615 | 69.11 | 5.54 | 细胞核Nucleus |
| GhCOI6⁃A06 | GH_A06G1509 | 701 | 78.94 | 6.57 | 叶绿体 Chloroplast |
| GhCOI6⁃D06 | GH_D06G1549 | 428 | 47.46 | 8.17 | 细胞质Cytoplasm |
| GhCOI6⁃A10 | GH_A10G0382 | 650 | 72.69 | 6.75 | 叶绿体Chloroplast |
| GhCOI6⁃D10 | GH_D10G0398 | 650 | 72.59 | 6.75 | 细胞核Nucleus |
| GhCOI6⁃A12 | GH_A12G1052 | 702 | 79.13 | 5.83 | 细胞核Nucleus |
| GhCOI6⁃D12 | GH_D12G1027 | 702 | 79.06 | 5.95 | 细胞核Nucleus |
| GhCOI7⁃A07 | GH_A07G2611 | 579 | 65.23 | 7.93 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI7⁃D07 | GH_D07G2552 | 572 | 64.35 | 8.23 | 细胞核Nucleus |
Table 2 Characteristics and subcellular localization prediction of GhCOI genes
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI1⁃A01 | GH_A01G1190 | 534 | 57.36 | 8.7 | 叶绿体、细胞核Chloroplast, nucleus |
| GhCOI1⁃D01 | GH_D01G1258 | 534 | 57.54 | 8.82 | 细胞核Nucleus |
| GhCOI1⁃A05 | GH_A05G2065 | 534 | 57.38 | 8.86 | 细胞核Nucleus |
| GhCOI1⁃D05 | GH_D05G2095 | 535 | 57.44 | 8.83 | 细胞核Nucleus |
| GhCOI1⁃A06 | GH_A06G0210 | 528 | 56.82 | 8.74 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI1⁃D06 | GH_D06G0195 | 528 | 56.7 | 8.86 | 叶绿体、线粒体Chloroplast, mitochondria |
| GhCOI2⁃A02 | GH_A02G1803 | 544 | 58.97 | 8.49 | 细胞核Nucleus |
| GhCOI2⁃D03 | GH_D03G0256 | 514 | 55.87 | 7.87 | 细胞质Cytoplasm |
| GhCOI2⁃A10 | GH_A10G0913 | 545 | 59.31 | 8.6 | 细胞质Cytoplasm |
| GhCOI2⁃D10 | GH_D10G1048 | 545 | 59.21 | 8.59 | 细胞质Cytoplasm |
| GhCOI3⁃D04 | GH_D04G0696 | 591 | 66.99 | 5.98 | 细胞质Cytoplasm |
| GhCOI3⁃A05 | GH_A05G3439 | 589 | 66.74 | 5.81 | 细胞质Cytoplasm |
| GhCOI4⁃A05 | GH_A05G0481 | 362 | 39.99 | 6.32 | 细胞质Cytoplasm |
| GhCOI4⁃D05 | GH_D05G0480 | 362 | 40.08 | 6.96 | 细胞质Cytoplasm |
| GhCOI5⁃A05 | GH_A05G2217 | 611 | 68.38 | 5.97 | 细胞核Nucleus |
| GhCOI5⁃D05 | GH_D05G2242 | 611 | 68.32 | 5.88 | 叶绿体Chloroplast |
| GhCOI5⁃A06 | GH_A06G0343 | 615 | 69.03 | 5.67 | 细胞核Nucleus |
| GhCOI5⁃D06 | GH_D06G0328 | 615 | 69.11 | 5.54 | 细胞核Nucleus |
| GhCOI6⁃A06 | GH_A06G1509 | 701 | 78.94 | 6.57 | 叶绿体 Chloroplast |
| GhCOI6⁃D06 | GH_D06G1549 | 428 | 47.46 | 8.17 | 细胞质Cytoplasm |
| GhCOI6⁃A10 | GH_A10G0382 | 650 | 72.69 | 6.75 | 叶绿体Chloroplast |
| GhCOI6⁃D10 | GH_D10G0398 | 650 | 72.59 | 6.75 | 细胞核Nucleus |
| GhCOI6⁃A12 | GH_A12G1052 | 702 | 79.13 | 5.83 | 细胞核Nucleus |
| GhCOI6⁃D12 | GH_D12G1027 | 702 | 79.06 | 5.95 | 细胞核Nucleus |
| GhCOI7⁃A07 | GH_A07G2611 | 579 | 65.23 | 7.93 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI7⁃D07 | GH_D07G2552 | 572 | 64.35 | 8.23 | 细胞核Nucleus |
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI7⁃A11 | GH_A11G0655 | 572 | 64.46 | 7.42 | 细胞核Nucleus |
| GhCOI7⁃D11 | GH_D11G0684 | 572 | 64.28 | 8.2 | 细胞核Nucleus |
| GhCOI8A⁃A08 | GH_A08G0483 | 587 | 65.9 | 5.8 | 细胞核Nucleus |
| GhCOI8B⁃A08 | GH_A08G0792 | 586 | 65.92 | 6.13 | 细胞核Nucleus |
| GhCOI8C⁃A08 | GH_A08G1389 | 586 | 65.62 | 5.5 | 叶绿体、细胞质Chloroplast, cytoplasm |
| GhCOI8A⁃D08 | GH_D08G0497 | 587 | 65.67 | 5.61 | 细胞核Nucleus |
| GhCOI8B⁃D08 | GH_D08G0784 | 586 | 65.91 | 5.97 | 细胞质Cytoplasm |
| GhCOI8C⁃D08 | GH_D08G1411 | 586 | 65.57 | 5.51 | 叶绿体Chloroplast |
| GhCOI8⁃A11 | GH_A11G1227 | 585 | 65.64 | 6.54 | 细胞质Cytoplasm |
| GhCOI8⁃D11 | GH_D11G1257 | 585 | 65.49 | 5.76 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI9⁃A08 | GH_A08G2517 | 467 | 52.62 | 7.74 | 细胞核Nucleus |
| GhCOI9⁃D08 | GH_D08G2524 | 467 | 52.6 | 6.91 | 细胞质Cytoplasm |
| GhCOI9A⁃A11 | GH_A11G1519 | 395 | 43.46 | 6.63 | 细胞质Cytoplasm |
| GhCOI9B⁃A11 | GH_A11G3019 | 654 | 72.04 | 8.25 | 叶绿体、细胞核、细胞质Chloroplast, nucleus, cytoplasm |
| GhCOI9A⁃D11 | GH_D11G1550 | 395 | 43.43 | 6.98 | 细胞质Cytoplasm |
| GhCOI9B⁃D11 | GH_D11G3049 | 654 | 72.27 | 8.17 | 细胞核Nucleus |
| GhCOI10⁃A09 | GH_A09G2525 | 592 | 64.35 | 7.91 | 细胞质Cytoplasm |
| GhCOI10⁃D09 | GH_D09G2461 | 607 | 65.93 | 8.11 | 细胞质Cytoplasm |
| GhCOI10⁃A12 | GH_A12G1492 | 411 | 44.68 | 5.8 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI10⁃D12 | GH_D12G1505 | 411 | 44.61 | 5.95 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI11⁃A10 | GH_A10G0227 | 586 | 65.71 | 5.97 | 细胞核Nucleus |
| GhCOI11⁃D10 | GH_D10G0239 | 586 | 65.87 | 5.92 | 细胞核Nucleus |
Table 2 Characteristics and subcellular localization prediction of GhCOI genes
基因名称 Gene name | 基因编号 Gene ID | 氨基酸数目 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点pI | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| GhCOI7⁃A11 | GH_A11G0655 | 572 | 64.46 | 7.42 | 细胞核Nucleus |
| GhCOI7⁃D11 | GH_D11G0684 | 572 | 64.28 | 8.2 | 细胞核Nucleus |
| GhCOI8A⁃A08 | GH_A08G0483 | 587 | 65.9 | 5.8 | 细胞核Nucleus |
| GhCOI8B⁃A08 | GH_A08G0792 | 586 | 65.92 | 6.13 | 细胞核Nucleus |
| GhCOI8C⁃A08 | GH_A08G1389 | 586 | 65.62 | 5.5 | 叶绿体、细胞质Chloroplast, cytoplasm |
| GhCOI8A⁃D08 | GH_D08G0497 | 587 | 65.67 | 5.61 | 细胞核Nucleus |
| GhCOI8B⁃D08 | GH_D08G0784 | 586 | 65.91 | 5.97 | 细胞质Cytoplasm |
| GhCOI8C⁃D08 | GH_D08G1411 | 586 | 65.57 | 5.51 | 叶绿体Chloroplast |
| GhCOI8⁃A11 | GH_A11G1227 | 585 | 65.64 | 6.54 | 细胞质Cytoplasm |
| GhCOI8⁃D11 | GH_D11G1257 | 585 | 65.49 | 5.76 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI9⁃A08 | GH_A08G2517 | 467 | 52.62 | 7.74 | 细胞核Nucleus |
| GhCOI9⁃D08 | GH_D08G2524 | 467 | 52.6 | 6.91 | 细胞质Cytoplasm |
| GhCOI9A⁃A11 | GH_A11G1519 | 395 | 43.46 | 6.63 | 细胞质Cytoplasm |
| GhCOI9B⁃A11 | GH_A11G3019 | 654 | 72.04 | 8.25 | 叶绿体、细胞核、细胞质Chloroplast, nucleus, cytoplasm |
| GhCOI9A⁃D11 | GH_D11G1550 | 395 | 43.43 | 6.98 | 细胞质Cytoplasm |
| GhCOI9B⁃D11 | GH_D11G3049 | 654 | 72.27 | 8.17 | 细胞核Nucleus |
| GhCOI10⁃A09 | GH_A09G2525 | 592 | 64.35 | 7.91 | 细胞质Cytoplasm |
| GhCOI10⁃D09 | GH_D09G2461 | 607 | 65.93 | 8.11 | 细胞质Cytoplasm |
| GhCOI10⁃A12 | GH_A12G1492 | 411 | 44.68 | 5.8 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI10⁃D12 | GH_D12G1505 | 411 | 44.61 | 5.95 | 细胞质、细胞核Cytoplasm, nucleus |
| GhCOI11⁃A10 | GH_A10G0227 | 586 | 65.71 | 5.97 | 细胞核Nucleus |
| GhCOI11⁃D10 | GH_D10G0239 | 586 | 65.87 | 5.92 | 细胞核Nucleus |
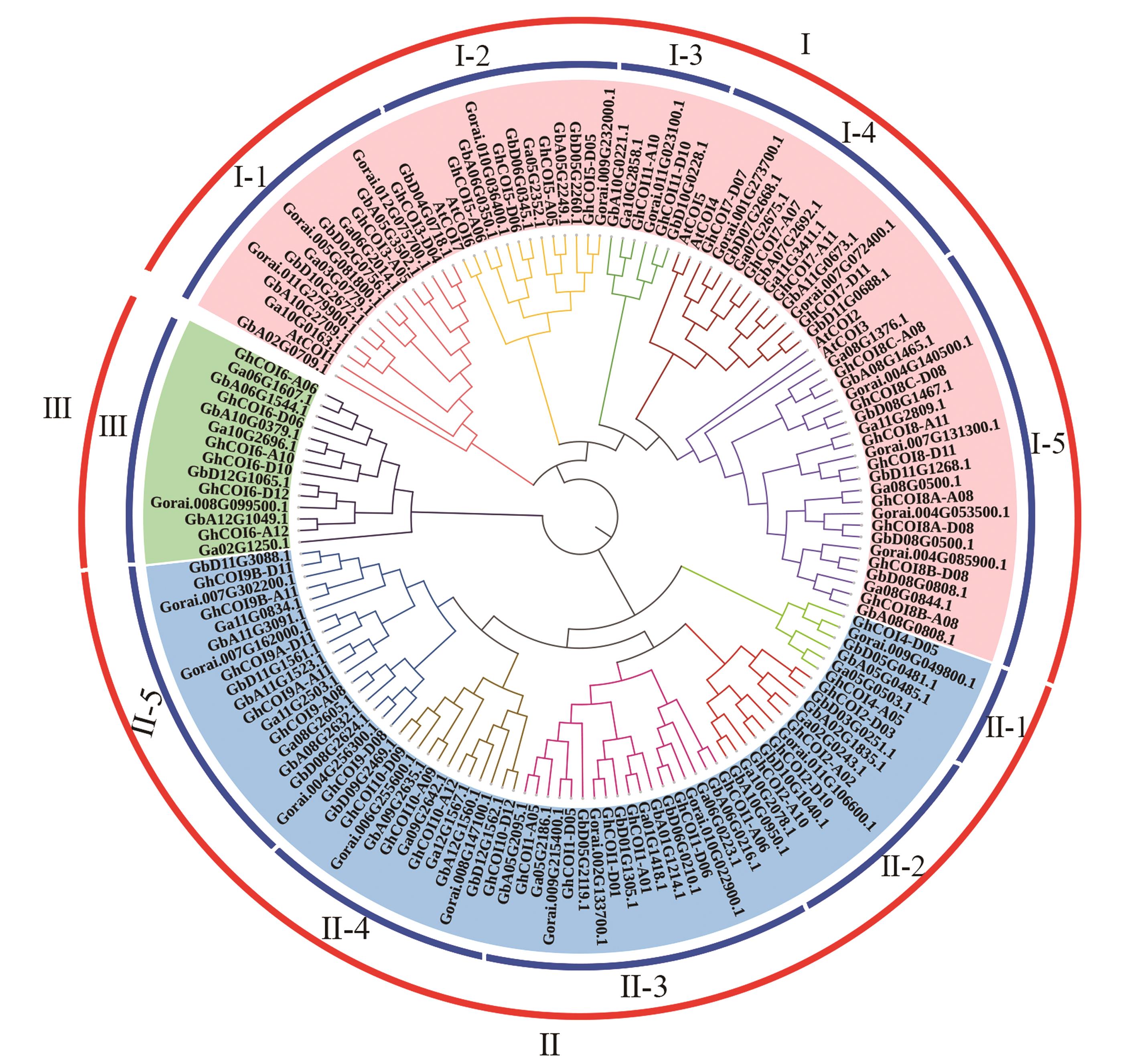
Fig.2 Phylogeny analysis of COI genesNote: Gb represents Gossypium barbadense, Gh represents Gossypium hirsutum, Ga represents Gossypium arboreum, Gorai represents Gossypium raimondii, and At represents Arabidopsis thaliana.

Fig.5 cis?acting elements and expression analysis of GhCOI genesA: ,cis-acting elements of GhCOIs promoter; B: Expression analysis of GhCOIs under NaCl and PEG stress; C: Expression analysis of GhCOIs under cold and high-temperature stress

Fig. 6 Expression analysis of 9 GhCOIs under NaCl and PEG stressA: Expression analysis of 9 GhCOIs under NaCl stress; B: Expression analysis of 9 GhCOIs under PEG stress
| 1 | CREELMAN R A, MULLET J E. Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress [J]. Proc. Natl. Acad. Sci. USA, 1995, 92(10): 4114-4119. |
| 2 | YAN J, ZHANG C, GU M, et al.. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor [J]. Plant Cell, 2009, 21(8): 2220-2236. |
| 3 | XIE D X, FEYS B F, JAMES S, et al.. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility [J]. Science, 1998, 280(5366): 1091-1094. |
| 4 | SKOWYRA D, CRAIG K L, TYERS M, et al.. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex [J]. Cell, 1997, 91(2): 209-219. |
| 5 | CASTILLO M C, LEÓN J. Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis [J]. J. Exp. Bot., 2008, 59(8): 2171-2179. |
| 6 | SHAN X, WANG J, CHUA L, et al.. The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence [J]. Plant Physiol., 2011, 155(2): 751-764. |
| 7 | REINBOTHE C, SPRINGER A, SAMOL I, et al.. Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence [J]. FEBS J., 2009, 276(17): 4666-4681. |
| 8 | UEDA J, KATO J. Isolation and identification of a senescence-promoting substance from Wormwood (Artemisia absinthium L.) [J]. Plant Physiol., 1980, 66(2): 246-249. |
| 9 | KIM J, DOTSON B, REY C, et al.. New clothes for the jasmonic acid receptor COI1: delayed abscission, meristem arrest and apical dominance [J/OL]. PLoS One, 2013, 8(4): e60505 [ 2021-11-17 ]. . |
| 10 | 李娟,阳文龙,刘冬成,等.小麦K-型细胞质雄性不育系COI1基因的克隆与表达分析[J].分子植物育种,2015,13(3):518-530. |
| LI J, YANG W L, LIU D C, et al.. Cloning and expression analysis of COI1 gene from a wheat K-type cytoplasmic male sterility line [J]. Mol. Plant Breed., 2015, 13(3): 518-530. | |
| 11 | YE M, LUO S M, XIE J F, et al.. Silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense [J/OL]. PLoS One, 2012, 7(4): e36214 [2021-11-17 ]. . |
| 12 | 王文静,杨小川,丁永强,等.甘蓝型油菜COI1的调控功能分析[J].中国农业科学,2015,48(10):1882-1891. |
| WANG W J, YANG X C, DING Y Q, et al.. Functional analysis of COI1 genes in Oilseed Rape(Brassica napus L.)[J]. Sci. Agric. Sin., 2015, 48(10): 1882-1891. | |
| 13 | 段龙飞.茉莉酸信号途径上关键基因家族COI/JAZ/MYC分子进化分析[D].陕西杨凌:西北农林科技大学,2013. |
| DUAN L F. Molecular evolutionary analysis of the key gene families COI/JAZ/MYC in jasmonic acid signaling pathway [D]. Shaanxi Yangling: Northwest A&F University, 2013. | |
| 14 | HU Y, CHEN J, FANG L, et al.. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton [J]. Nat. Genet., 2019, 51(4): 739-748. |
| 15 | ZHU T, LIANG C, MENG Z, et al.. CottonFGD: an integrated functional genomics database for cotton [J]. BMC Plant Biol., 2017, 17(1): 101-109. |
| 16 | BERARDINI T Z, REISER L, LI D, et al.. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome [J]. Genesis, 2015, 53(8): 474-485. |
| 17 | EL-GEBALI S, MISTRY J, BATEMAN A, et al.. The Pfam protein families database in 2019 [J]. Nucleic Acids Res., 2019, 47(D1): D427-D432. |
| 18 | LETUNIC I, BORK P. 20 years of the SMART protein domain annotation resource [J]. Nucleic Acids Res., 2018, 46(D1): D493-D496. |
| 19 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002, 30(1): 325-327. |
| 20 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2-ΔΔ CT method [J]. Methods, 2001, 25(4): 402-408. |
| 21 | 宋云,李林宣,卓凤萍,等.茉莉酸信号传导在植物抗逆性方面研究进展[J].中国农业科技导报, 2015,17(2): 17-24. |
| SONG Y, LI L X, ZHUO F P, et al.. Progress on jasmonic acid signaling in plant sress resistant [J]. J. Agric. Sci. Technol., 2015, 17(2): 17-24. | |
| 22 | MOORE R C, PURUGGANAN M D. The early stages of duplicate gene evolution [J]. Proc. Natl. Acad. Sci. USA, 2003, 100(26): 15682-15687. |
| 23 | CANNON S B, MITRA A, BAUMGARTEN A, et al.. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J]. BMC Plant Biol., 2004, 4(1): 10-30. |
| 24 | BAI J F, WANG Y K, WANG P, et al.. Genome-wide identification and analysis of the COI gene family in wheat (Triticum aestivum L.) [J]. BMC Genomics, 2018, 19(1): 754-770. |
| 25 | 杨秀,许艳超,杨芳芳,等.棉花CML基因家族成员鉴定与功能分析[J].棉花学报,2019, 31(4):307-318. |
| YANG X, XU Y C, YANG F F, et al.. Identification and functional analysis of CML gene family in cotton [J]. Cotton Sci., 2019, 31(4): 307-318. | |
| 26 | ZHU Y, WU N, SONG W, et al.. Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies [J]. BMC Plant Biol., 2014, 14(1): 93-111. |
| 27 | 段龙飞,李文燕,慕小倩,等.茉莉酸信号受体COI蛋白家族的分子进化与基因表达分析[J].生物信息学,2016,14(3):146-155. |
| DUAN L F, LI W Y, MU X Y, et al.. Molecular evolution and gene expression of jasmonic acid receptor COI protein family [J]. Chin. J. Bioinform., 2016, 14(3): 146-155. |
| [1] | CUI Jianghui§, YANG Puyuan§, CHANG Jinhua*. Identification and Expression Analysis Under Abiotic Stress of GRF Gene Family in Sorghum [J]. Journal of Agricultural Science and Technology, 2021, 23(4): 37-46. |
| [2] | LUO Ping, PANG Bo, CUI Jinxin, YU Shuang, WANG Xiaonan, CHENG Ming, CHEN Yong, GAO Wenwei, HAO Zhuangfang. Gene Structural Characteristics and Regulation Prediction of SNAC Transcription Factors Against Stresses in Maize [J]. Journal of Agricultural Science and Technology, 2021, 23(10): 35-44. |
| [3] | LI Ziwei1, CHEN Simeng1, WANG Fazhan1, ZHANG Haoyang1, ZHANG Li2*, XU Zicheng1*. Mechanism Research Advances in Plant Abiotic Stress Resistance Regulated by Hydrogen Sulfide [J]. Journal of Agricultural Science and Technology, 2020, 22(4): 24-32. |
| [4] | CHEN Simeng, LI Ziwei, ZHANG Luxiang, WEI Yanqiu, ZHU Zhiwei, ZHANG Huanwei, XU Zicheng, HUANG Wuxing, SHAO Huifang*. Research Advances on the Role of Selenium in Plants Resistance to Stress [J]. Journal of Agricultural Science and Technology, 2020, 22(3): 6-13. |
| [5] | ZHOU Miaoyi, REN Wen, ZHAO Bingbing, LI Hanshuai, LIU Ya*. Advances on the MAPK Cascade Pathway in Response to Abiotic Stress in Plant [J]. Journal of Agricultural Science and Technology, 2020, 22(2): 22-29. |
| [6] | LIANG Guihong, HUA Yingpeng, ZHOU Ting, SONG Haixing, ZHANG Zhenhua*. Research Progress on Vacuolar H+-ATPase and H+-PPase in Plant [J]. Journal of Agricultural Science and Technology, 2020, 22(1): 19-27. |
| [7] | HAN Jiahui1, LIU Yingying1, JIANG Shijie2, CHEN Yun1, GENG Xiuxiu1,2, PING Shuzhen1, WANG Jin1,2*. Biological Identification of LEA3 Protein Dgl3 Under Abiotic Stress in Deinococcus gobiensis [J]. Journal of Agricultural Science and Technology, 2019, 21(9): 69-76. |
| [8] | XIE Zheng-wen1, WANG Lian-jun2, CHEN Jin-yang1, WANG Jiao1, SU Yi-jun1, . Studies on WRKY Transcription Factors and Their Biological Functions in Plants [J]. Journal of Agricultural Science and Technology, 2016, 18(3): 46-54. |
| [9] | LI Yong-liang1,2, DONG Xue-ni3, LEI Zhi2, YANG Pei-yan2, TAO Fei2, TANG Yi-xiong. Analysis of Stress Resistance Ability of Transgenic Cotton Expressing HhERF2 and PeDREB2a Genes [J]. , 2015, 17(3): 19-28. |
| [10] | ZHANG Shao-xuan1, LI Zhe2, LIU Xin1*, HUANG Rong-feng2. Expression Character of Rice SUPERNUMERARY BRACT (OsSNB) in Response to Abiotic Stress and Hormone [J]. , 2015, 17(1): 42-48. |
| [11] | LEI Zhi, ZHOU Mei\|liang, WU Yan\|min*. Advances of the Abiotic Stress\|related Genes in Cotton Stress Tolerance [J]. , 2014, 16(2): 35-43. |
| [12] | XIAN Xian\|yi1,2, JIANG Shi\|jie1,2, CUI Guang\|yan1,2, LIU Ying\|ying2, DAI Qi\. Functional Analysis of ktrA Gene Encoding Potassium Uptake Protein from Deinococcus radiodurans [J]. , 2014, 16(1): 76-81. |
| [13] | YANG De-xin, ZHOU Shi-rong, QUAN Rui-dang, HUANG Rong-feng*. Mechanism of Plant Response to Abiotic Stresses Regulated by AP2/ERF Proteins [J]. , 2012, 14(6): 23-29. |
| [14] | LIN Fa-zhuang1,2, ZHANG Rui1, SUN Guo-qing1, SUN Bao1, GUO San-dui1*. Molecular Cloning and its Functional Analysis of GhSRK2D Gene from Cotton [J]. , 2012, 14(6): 36-42. |
| [15] | ZHONG Gui-mai, WU Lin-tao, WANG Jian-mei, YANG Yi, LI Xu-feng*. Subcellular Localization and Expression Analysis of Transcription Factor AtWRKY28 under Biotic Stresses [J]. , 2012, 14(5): 57-63. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号