




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (7): 113-121.DOI: 10.13304/j.nykjdb.2022.0105
• ANIMAL AND PLANT HEALTH • Previous Articles
Shuai LUO1( ), Yaqin WEN1, Hua ZHU2, Rong ZHANG2, Xiaowen WANG2, Lili LIU2, Jianya ZHU2(
), Yaqin WEN1, Hua ZHU2, Rong ZHANG2, Xiaowen WANG2, Lili LIU2, Jianya ZHU2( )
)
Received:2022-02-15
Accepted:2022-06-21
Online:2023-07-15
Published:2023-08-25
Contact:
Jianya ZHU
罗帅1( ), 问亚琴1, 朱华2, 张蓉2, 王晓雯2, 刘丽丽2, 朱建亚2(
), 问亚琴1, 朱华2, 张蓉2, 王晓雯2, 刘丽丽2, 朱建亚2( )
)
通讯作者:
朱建亚
作者简介:罗帅 E-mail: bj18010005109@163.com;
基金资助:CLC Number:
Shuai LUO, Yaqin WEN, Hua ZHU, Rong ZHANG, Xiaowen WANG, Lili LIU, Jianya ZHU. Effect of ECF-Sig16 on Avermectin Production in Streptomyces avermitilis[J]. Journal of Agricultural Science and Technology, 2023, 25(7): 113-121.
罗帅, 问亚琴, 朱华, 张蓉, 王晓雯, 刘丽丽, 朱建亚. 阿维链霉菌中ECF-Sig16对阿维菌素合成的影响[J]. 中国农业科技导报, 2023, 25(7): 113-121.
| 类型Type | 名称Name | 特征Character | 来源Source or reference |
|---|---|---|---|
| 质粒Plasmid | pKC1139 | 大肠杆菌-链霉菌穿梭质粒,多拷贝,温敏型E.coli-Streptomyces shuttle vector, multi-copy and temperature-sensitive | [ |
| pSET152 | 大肠杆菌-链霉菌穿梭质粒,整合型E.coli-Streptomyces shuttle vector, integrative | [ | |
| pET-28a(+) | 蛋白大量表达载体 Vector for heterologous protein overexpression | Novegen | |
| pJL117 | 携带红霉素抗性基因启动子的pIJ2925衍生质粒 pIJ2925 derivative carrying promoter with erythromycin resistance | [ | |
| 菌株Strain | 阿维链霉菌 S. avermitilis | 野生型阿维链霉菌Wild-type (WT) S. avermitilis | 实验室保存 Laboratary stock |
| 大肠杆菌E. coli | JM109 | 基因克隆宿主菌 General cloning host | Novegen |
| ET12567 | 甲基缺陷型菌株 MetHylation-defective strain | [ | |
| BL21(DE3) | 蛋白异源表达宿主菌 Host for protein heterologous overexpression | Novegen |
Table 1 Plasmids and strains used in the study
| 类型Type | 名称Name | 特征Character | 来源Source or reference |
|---|---|---|---|
| 质粒Plasmid | pKC1139 | 大肠杆菌-链霉菌穿梭质粒,多拷贝,温敏型E.coli-Streptomyces shuttle vector, multi-copy and temperature-sensitive | [ |
| pSET152 | 大肠杆菌-链霉菌穿梭质粒,整合型E.coli-Streptomyces shuttle vector, integrative | [ | |
| pET-28a(+) | 蛋白大量表达载体 Vector for heterologous protein overexpression | Novegen | |
| pJL117 | 携带红霉素抗性基因启动子的pIJ2925衍生质粒 pIJ2925 derivative carrying promoter with erythromycin resistance | [ | |
| 菌株Strain | 阿维链霉菌 S. avermitilis | 野生型阿维链霉菌Wild-type (WT) S. avermitilis | 实验室保存 Laboratary stock |
| 大肠杆菌E. coli | JM109 | 基因克隆宿主菌 General cloning host | Novegen |
| ET12567 | 甲基缺陷型菌株 MetHylation-defective strain | [ | |
| BL21(DE3) | 蛋白异源表达宿主菌 Host for protein heterologous overexpression | Novegen |
引物 Primer | 引物序列 Primer sequence (5’-3’) | 用途 Usage |
|---|---|---|
| LS63 | CCC | 缺失sig16基因 Deletion of sig16 |
| LS64 | ATGGCCGTCGGTGTCGAGAGTGAACCGCTACACGCTCA | |
| LS65 | TGAGCGTGTAGCGGTTCACTCTCGACACCGACGGCCAT | |
| LS66 | G | |
| LS67 | CCCAAGGTGCAGGAGGTGTA | 检测sig16基因是否缺失成功 Confirmation of sig16 deletion |
| LS68 | GCATCACGTCGTTGAGCCC | |
| LS69 | TCCAGGACGCCTTCACC | |
| LS70 | GAAGATGAGGTAGACGACGG | |
| LS81 | CCC | 扩增sig16基因开放阅读框用于构建回补和过表达菌株 Amplification of sig16 open reading frame |
| LS82 | GA | |
| LS89 | GGAATTC | 扩增sig16基因开放阅读框用于大量表达His6-Sig16蛋白 Amplification of sig16 open reading frame for overexpression of His6-Sig16 |
| LS90 | CG | |
| GJ105 | GGTATTCCATTCGGTGTTGC | 探针aveRp |
| GJ106 | TGTTATGAATTTGCCCTGGTG | Probe aveRp |
| LS52 | ATGGTCGGGAACCTCCGCAA | 探针aveA1p |
| LS53 | CTGTGTCCTCACCGCTAGGC | Probe aveA1p |
| GJ91 | CCAAGGGCTACAAGTTCTCC | 探针hrdB |
| GJ92 | TTGATGACCTCGACCATGTG | Probe hrdB |
| LS207 | CGCCTCGATACTGCTCGC | 探针 SAV1196p |
| LS208 | TGTGTCCTCCTCGGTGATCT | Probe SAV1196p |
| LS209 | ACCGTGAACCAGTCGGGC | 探针 SAV1188p |
| LS210 | GGCATCCTGCTCCTCATCC | Probe SAV1188p |
| LS211 | TCCACAGTTCCCGCCCTT | 探针 SAV1190p |
| LS212 | CGCTGGGACCGCTGGACA | Probe SAV1190p |
Table 2 Primer sequences in this study
引物 Primer | 引物序列 Primer sequence (5’-3’) | 用途 Usage |
|---|---|---|
| LS63 | CCC | 缺失sig16基因 Deletion of sig16 |
| LS64 | ATGGCCGTCGGTGTCGAGAGTGAACCGCTACACGCTCA | |
| LS65 | TGAGCGTGTAGCGGTTCACTCTCGACACCGACGGCCAT | |
| LS66 | G | |
| LS67 | CCCAAGGTGCAGGAGGTGTA | 检测sig16基因是否缺失成功 Confirmation of sig16 deletion |
| LS68 | GCATCACGTCGTTGAGCCC | |
| LS69 | TCCAGGACGCCTTCACC | |
| LS70 | GAAGATGAGGTAGACGACGG | |
| LS81 | CCC | 扩增sig16基因开放阅读框用于构建回补和过表达菌株 Amplification of sig16 open reading frame |
| LS82 | GA | |
| LS89 | GGAATTC | 扩增sig16基因开放阅读框用于大量表达His6-Sig16蛋白 Amplification of sig16 open reading frame for overexpression of His6-Sig16 |
| LS90 | CG | |
| GJ105 | GGTATTCCATTCGGTGTTGC | 探针aveRp |
| GJ106 | TGTTATGAATTTGCCCTGGTG | Probe aveRp |
| LS52 | ATGGTCGGGAACCTCCGCAA | 探针aveA1p |
| LS53 | CTGTGTCCTCACCGCTAGGC | Probe aveA1p |
| GJ91 | CCAAGGGCTACAAGTTCTCC | 探针hrdB |
| GJ92 | TTGATGACCTCGACCATGTG | Probe hrdB |
| LS207 | CGCCTCGATACTGCTCGC | 探针 SAV1196p |
| LS208 | TGTGTCCTCCTCGGTGATCT | Probe SAV1196p |
| LS209 | ACCGTGAACCAGTCGGGC | 探针 SAV1188p |
| LS210 | GGCATCCTGCTCCTCATCC | Probe SAV1188p |
| LS211 | TCCACAGTTCCCGCCCTT | 探针 SAV1190p |
| LS212 | CGCTGGGACCGCTGGACA | Probe SAV1190p |
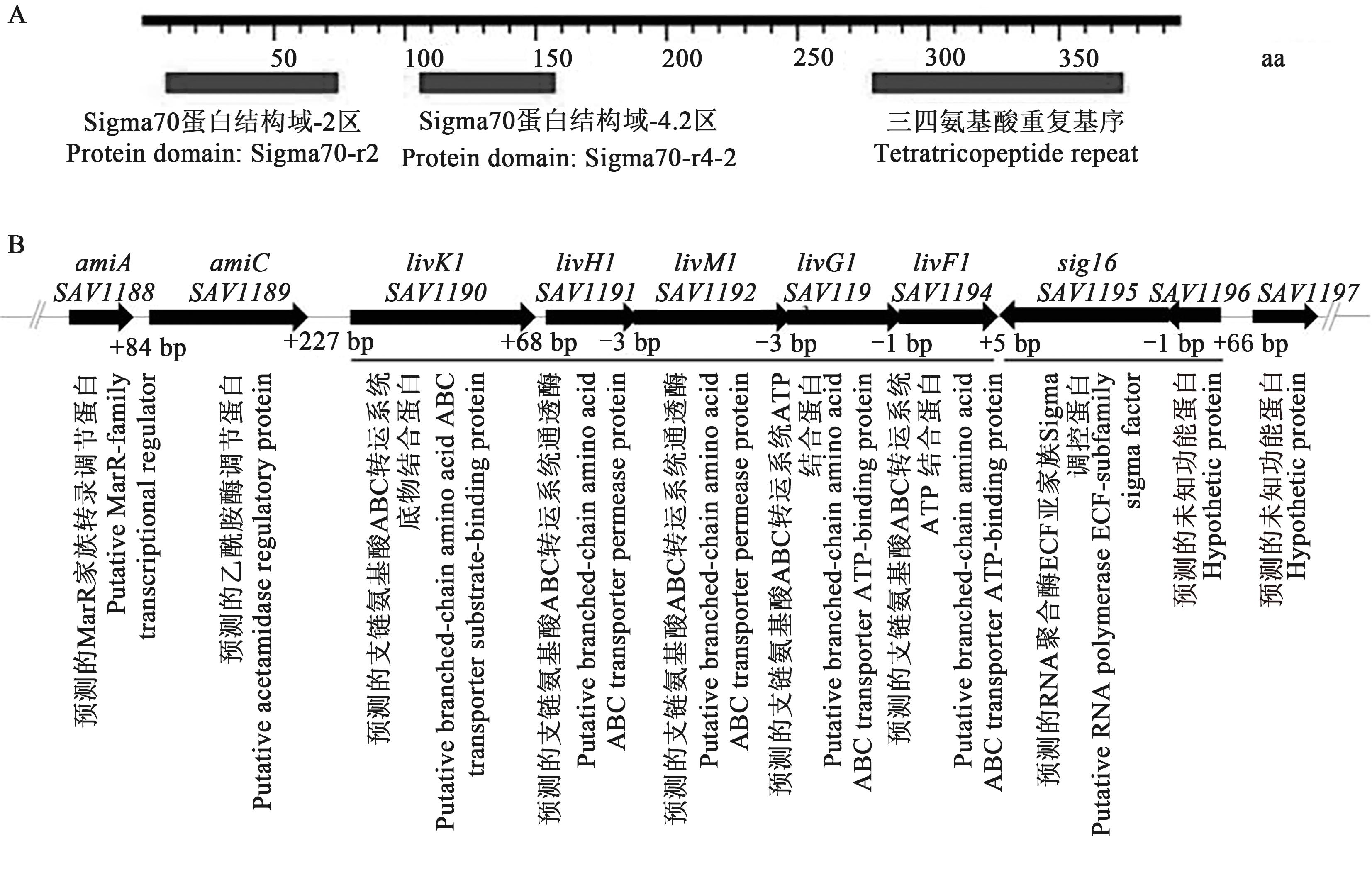
Fig. 1 Conservative domain of Sig16 and organization of sig16 and its adjacent genesA: Conservative domain of Sig16; B: Organization of sig16 and its adjacent genes; Black bars at bottom is transcriptional units
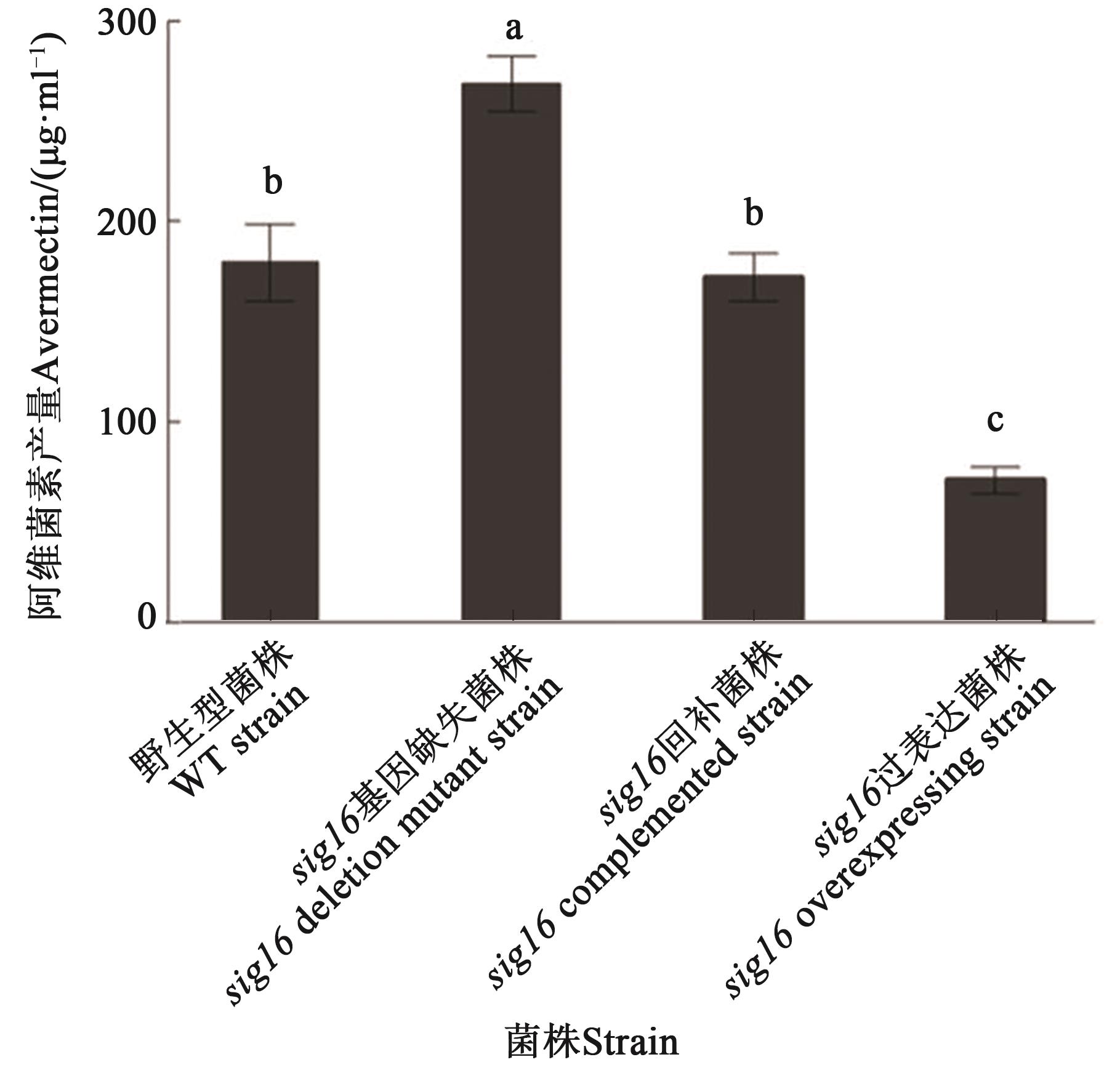
Fig. 2 Avermectin production in WT and sig16 mutant strainsNote: Different lowercase letters indicate significant differences between different strains at P<0.05 level.
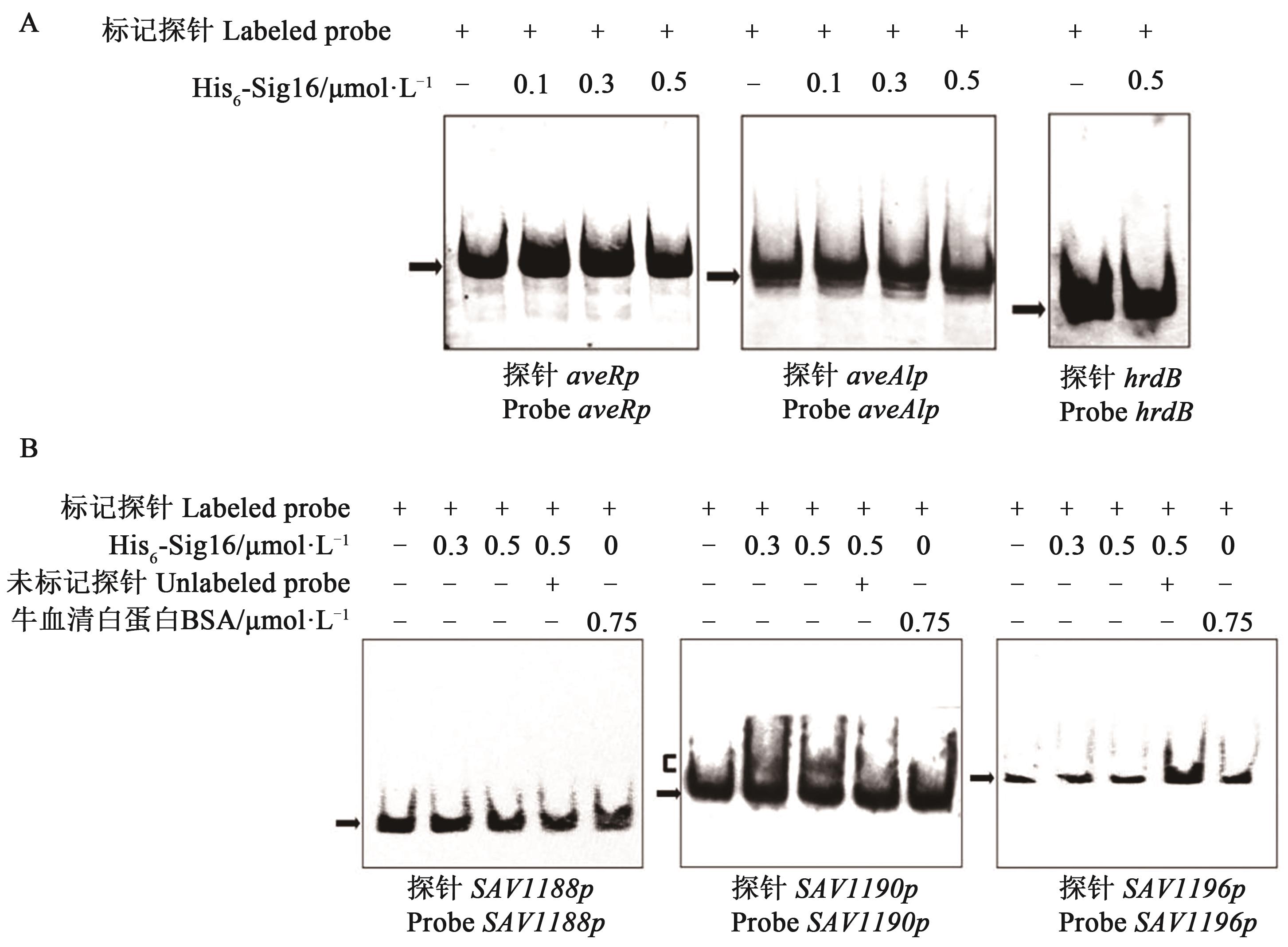
Fig. 4 EMSA assays of His6-Sig16 with probes aveRp, aveA1p, SAV1188p, SAV1190p and SAV1196pA: EMSA assays of the interaction of Probe aveRp, Probe aveA1p with His6-Sig16; B: EMSA assays of the interaction of Probe SAV1188p, Probe SAV1190p and Probe SAV1196p with His6-Sig16; Each reaction contained 0.15 nmol·L-1 DIG-labeled probe, ~100 fold excess of unlabeled specific probe was used for competition assays. Labeled probe hrdB or BSA protein was used as a negative control respectively for probe or His6-Sig16. Concentration of His6-Sig16 for probe hrdB was 0.5 μmol·L-1; arrowhead shows free probes; bracket shows Sig16-DNA complex
| 1 | FENICAL W, JENSEN P R. Developing a new resource for drug discovery: marine actinomycete bacteria [J] Nat. Chem. Biol., 2006, 2(12):666-673. |
| 2 | YANG S, SUN W, TANG C, et al.. Phylogenetic diversity of actinobacteria associated with soft coral Alcyonium gracllimum and stony coral Tubastraea coccinea in the East China Sea [J]. Microb. Ecol., 2013, 66(1):189-199. |
| 3 | FOULSTON L, BIBB M. Feed-forward regulation of microbisporicin biosynthesis in Microbispora coralline [J]. J. Bacteriol., 2011, 193(12):3064-3071. |
| 4 | BUSCHE T, SILAR R, PICMANOVA M, et al.. Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum [J/OL]. BMC Genomics, 2012, 13: 445 [2022-01-20]. . |
| 5 | WANG T T, GAO F, KANG Y W, et al.. Mycothiol peroxidase MPx protects Corynebacterium glutamicum against acid stress by scavenging ROS [J]. Biotechnol. Lett., 2016, 38(7):1221-1228. |
| 6 | WHITE M J, HE H J, PENOSKE R M, et al.. PepD participates in the mycobacterial stress response mediated through MprAB and SigE [J]. J. Bacteriol., 2010, 192(6):1498-1510. |
| 7 | HELMANN J D. The extracytoplasmic function (ECF) sigma factors [J]. Adv. Microb. Physiol., 2002, 46:47-110. |
| 8 | KALLIFIDAS D, THOMAS D, DOUGHTY P, et al.. The SigR regulon of Streptomyces coelicolor A3(2) reveals a key role in protein quality control during disulphide stress [J]. Microbiology, 2010, 156(Pt6): 1661-1672. |
| 9 | YOO J S, OH G S, RYOO S W, et al.. Induction of a stable sigma factor SigR by translation-inhibiting antibiotics confers resistance to antibiotics [J/OL]. Sci. Rep., 2016, 6:28628 [2022-01-20]. . |
| 10 | MAO X M, ZHOU Z, CHENG L Y, et al.. Involvement of SigT and RstA in the differentiation of Streptomyces coelicolor [J]. FEBS Lett., 2009, 583(19):3145-3150. |
| 11 | FENG W H, MAO X M, LIU Z H, et al.. The ECF sigma factor SigTregulates actinorhodin production in response to nitrogen stress in Streptomyces coelicolor [J]. Appl. Microbiol. Biotechnol., 2011, 92(5):1009-1021. |
| 12 | TRAN N T, HUANG X L, HONG H J, et al.. Defining the regulon of genes controlled by SigE, a key regulator of the cell envelope stress response in Streptomyces coelicolor [J]. Mol. Microbiol., 2019, 112(2):461-481. |
| 13 | LOPEZ-GARCIA M T, YAGUE P, GONZALEZ-QUINONEZ N, et al.. The SCO4117 ECF sigma factor pleiotropically controls secondary metabolism and morphogenesis in Streptomyces coelicolor [J/OL]. Front. Microbiol. , 2018, 9:312 [2022-01-20]. . |
| 14 | SEIPKE R F, PATRICK E, HUTCHINGS M I. Regulation of antimycin biosynthesis by the orphan ECF RNA polymerase sigma factor SigAntA [J/OL]. Peer J., 2014, 2:e253 [2022-01-20]. . |
| 15 | ZHANG K P, MOHSIN A, DAI Y C, et al.. Role of a two-component signal transduction system RspA1/A2 in regulating the biosynthesis of salinomycin in Streptomyces albus [J]. Appl. Biochem. Biotech., 2021, 193(5):1296-1310. |
| 16 | OLIVEIRA R, BUSH M J, PIRES S, et al.. The novel ECF56 SigG1-RsfG system modulates morphological differentiation and metal-ion homeostasis in Streptomyces tsukubaensis [J/OL]. Sci. Rep., 2020, 10:21728 [2022-01-20]. . |
| 17 | LEE S K, YANG S H, KANG C M, et al.. Overexpression of the putative extracytoplasmic function sigma (sigma) factor FujE enhances FK506 production in Streptomyces sp strain KCCM 11116P [J]. Can. J. Microbiol., 2014, 60(6):363-369. |
| 18 | DEVELOUX M. Ivermectin [J]. Ann. Dermatol. Venereal., 2004, 131(6-7 Pt1):561-570. |
| 19 | KAUR H, SHEKHAR N, SHARMA S, et al.. Ivermectin as a potential drug for treatment of COVID-19, an in-sync review with clinical and computational attributes [J]. Pharmacol. Rep., 2021, 73(3):736-749. |
| 20 | GUO J, ZHAO J L, LI L L, et al.. The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively regulates oligomycin biosynthesis [J]. Mol. Genet. Genomics, 2010, 283(2):123-133. |
| 21 | HE F, LIU W S, SUN D, et al.. Engineering of the TetR family transcriptional regulator SAV151 and its target genes increases avermectin production in Streptomyces avermilitis [J]. Appl. Microbiol. Biotechnol., 2014, 98(1):399-409. |
| 22 | GUO J, ZHANG X, CHEN Z, et al.. Two adjacent and similar TetR family transcriptional regulator genes, SAV 577 and SAV576, co-regulate avermectin production in Streptomyces avermitilis [J/OL]. PLoS One, 2014, 9(6):e99224 [2022-01-20]. . |
| 23 | ZHU J Y, SUN D, LIU W S, et al.. AvaR2, a pseudo γ-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth [J]. Mol. Microbiol., 2016, 102(4):562-578. |
| 24 | SUN D, ZHU J Y, CHEN Z, et al.. SAV 742, a novel AraC-family regulator from Streptomyces avermitilis, controls avermectin biosynthesis, cell growth and development [J/OL]. Sci. Rep., 2016, 6:36915 [2-22-01-20]. . |
| 25 | YAN H, LU X R, SUN D, et al.. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species [J]. Mol. Microbiol., 2020, 113(1):123-142. |
| 26 | LU X R, LIU X C, CHEN Z, et al.. The ROK-family regulator Rok7B7 directly controls carbon catabolite repression, antibiotic biosynthesis, and morphological development in Streptomyces avermitilis [J]. Environ. Microbiol., 2020, 22(12):5090-5108. |
| 27 | ZHUO Y, ZHANG W Q, CHEN D F, et al.. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis [J]. Proc. Natl. Acad. Sci. USA, 2010, 107(5):11250-11254. |
| 28 | JIANG L B, LIU Y P, WANG P, et al.. Inactivation of extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis [J]. Biotechnol. Lett., 2011, 33(10):1955-1961. |
| 29 | LUO S, SUN D, ZHU J Y, et al.. An extracytoplasmic function sigma factor, Sig25, differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis [J]. Appl. Microbiol. Biotechnol., 2014, 98(16):7097-7112. |
| 30 | 罗帅,孙地,陈芝,等.ECF-Sig5因子参与阿维链霉菌中阿维菌素合成和环境胁迫的研究[J].微生物学报,2016,56(3):471-484. |
| LUO S, SUN D, CHEN Z, et al.. ECF-Sig5 in Sreptomyces avermitilis is involved in regulation of avermectin biosynthesis and stress response [J]. Acta Microbiol. Sin., 2016, 56(3):471-484. | |
| 31 | WU G, CULLEY D E, ZHANG W W. Predicted highly expressed genes in the genomes of Streptomyces coelicolor and Streptomyces avermitilis and the implications for their metabolism [J]. Microbiology (Reading), 2005, 151(Pt7):2175-2187. |
| 32 | BIERMAN M, LOGAN R, O’BRIEN K, et al.. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. [J]. Gene, 1992, 116(1):43-49. |
| 33 | LI L L, GUO J, WEN Y, et al.. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains [J]. J. Ind. Microbiol. Biotechnol., 2010, 37(7):673-679. |
| 34 | MACNEI L, KLAPKO L M. Transformation of Streptomyces avermitilis by plasmid DNA [J]. J. Ind. Microbiol., 1987, 2(4):209-218. |
| 35 | HOPWOOD D A. Genetic manipulation of Streptomyces [J] Lab. Manua, 1986, 56(3):383-399. |
| 36 | IKEDA H, KOTAKI H, TANAKA H, et al.. Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis [J]. Antimicrob. Agents Chemother., 1988, 32(2):282-284. |
| 37 | CHEN Z, WEN J, SONG Y, et al.. Enhancement and selective production of avermectin B by recombinants of Streptomyces avermitilis via intraspecific protoplast fusion [J]. Chin. Sci. Bull., 2007, 52(5):616-622. |
| 38 | MASCHER T. Signaling diversity and evolution of extracytoplasmic function (ECF) sigma factors [J]. Curr. Opin. Microbiol., 2013, 16(2):148-155. |
| 39 | LIU J, CHEN Y F, WANG W W, et al.. Engineering of an Lrp family regulator SACE_Lrp improves erythromycin production in Saccharopolyspora erythraea [J]. Metab. Eng., 2017, 39:29-37. |
| 40 | IKEDA H, OMURA S. Avermectin biosynthesis [J]. Chem. Rev., 1997, 97(7):2591-2610. |
| [1] | LI Meng1, ZHANG Xuan2, WEN Ying2*, SONG Yuan2. Effects of Maltose Transport Related Gene Expression on Avermectins Production in Streptomyces avermitilis [J]. , 2014, 16(1): 71-75. |
| [2] |
GU Xiao-jun, TIAN Su-fen, LIU Wen-jing.
Indoor Toxicity Assessment of Avermectin and Fipronil on 3rd Instar Larvae of Diamondback Moth by Means of Leaf Protection Rates and Corrected Mortality Rate [J]. , 2009, 11(2): 98-105. |
| [3] |
ZHAO Zi-hua1,2, WANG Fang2, HE Da-han1, ZHANG Rong2.
The Dynamics of Effective Component Betaine in Lycium barbarum L. after Applying |Avermectins and Pyidaben [J]. , 2008, 10(3): 111-115. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号