




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (8): 73-79.DOI: 10.13304/j.nykjdb.2024.1030
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Yiyang ZHANG1,2( ), Tuoyu LIU2, Yuan WANG3, Jian TIAN3, Feifei GUAN2(
), Tuoyu LIU2, Yuan WANG3, Jian TIAN3, Feifei GUAN2( )
)
Received:2024-12-12
Accepted:2025-02-26
Online:2025-08-15
Published:2025-08-26
Contact:
Feifei GUAN
张铱洋1,2( ), 刘拓宇2, 王苑3, 田健3, 关菲菲2(
), 刘拓宇2, 王苑3, 田健3, 关菲菲2( )
)
通讯作者:
关菲菲
作者简介:张铱洋 E-mail:13251619660@163.com;
基金资助:CLC Number:
Yiyang ZHANG, Tuoyu LIU, Yuan WANG, Jian TIAN, Feifei GUAN. Expression and Thermostability Modification of Alkaline Xylanase[J]. Journal of Agricultural Science and Technology, 2025, 27(8): 73-79.
张铱洋, 刘拓宇, 王苑, 田健, 关菲菲. 碱性木聚糖酶表达及耐热性改造[J]. 中国农业科技导报, 2025, 27(8): 73-79.
| 木聚糖酶Xylanase | 突变位点Mutation site | 模型评分Model scoring |
|---|---|---|
| Pm10868 | — | 0.26 |
| Pm10868-117 | T17V,T51E,V107Q,S124E,A300E,N437S,T493K | 0.49 |
| Pm10868-14 | G15F,A214E,N246H,T324D,S464D | 0.46 |
| Pm10868-153 | G24H,G41D,K43S,L45K,A218K,L419N,E445K | 0.45 |
Table 1 Screening criteria and mutation sites for wild-type and its mutant proteins
| 木聚糖酶Xylanase | 突变位点Mutation site | 模型评分Model scoring |
|---|---|---|
| Pm10868 | — | 0.26 |
| Pm10868-117 | T17V,T51E,V107Q,S124E,A300E,N437S,T493K | 0.49 |
| Pm10868-14 | G15F,A214E,N246H,T324D,S464D | 0.46 |
| Pm10868-153 | G24H,G41D,K43S,L45K,A218K,L419N,E445K | 0.45 |
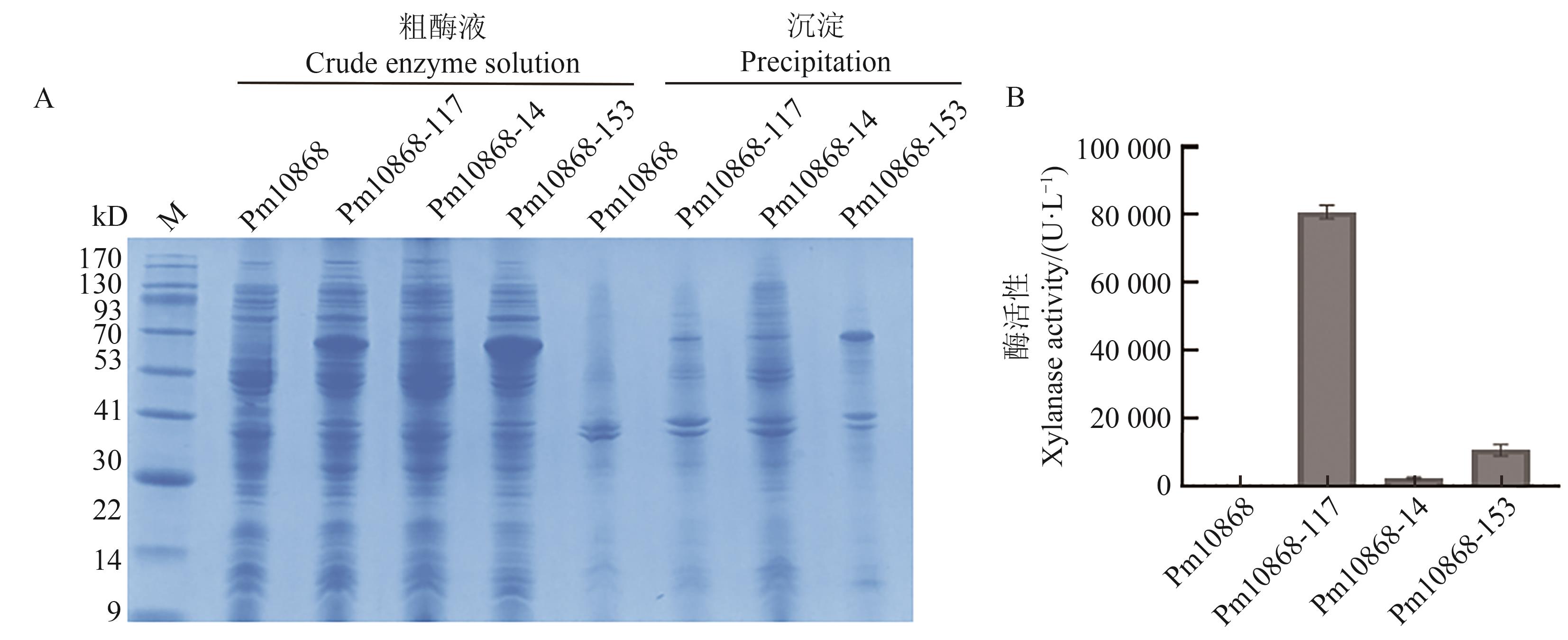
Fig. 1 Expression and crude enzyme activity of Pm10868 and its mutants in Escherichia coliA: SDS-PAGE analysis of soluble expression of Pm10868 and its mutants; B: Enzymatic activity of Pm10868 and its mutants

Fig. 2 Determination of expression of purification and relative activity of Pm10868-117 under different temperatures and pHA: Detection of Pm10868-117 enzyme solution; B: Relative activity of different temperatures; C: Relative activity of different pH

Fig. 3 Stability of Pm10868 and Pm10868-117 under different conditionsA: Temperature stability at 45 ℃; B: Temperature stability at 50 ℃; C: pH stability after incubation for 1 h

Fig. 4 Specific activity and kinetic parameter fitting curve of Pm10868 and Pm10868-117A: Enzyme specific activity under optimal condition; B: Kinetic parameter fitting curve of Pm10868; C: Kinetic parameter fitting curve of Pm10868-117
| 参数Parameter | Pm10868 | Pm10868-117 |
|---|---|---|
| Km/(mg·mL-1) | 3.48±0.50 | 3.63±0.18 |
| kcat/min-1 | 663.87±22.25 | 828.33±6.75 |
| kcat/Km/(mL·min-1·mg-1) | 190.67±19.89 | 228.09±11.63 |
Table 2 Kinetic parameters of Pm10868 and Pm10868-117
| 参数Parameter | Pm10868 | Pm10868-117 |
|---|---|---|
| Km/(mg·mL-1) | 3.48±0.50 | 3.63±0.18 |
| kcat/min-1 | 663.87±22.25 | 828.33±6.75 |
| kcat/Km/(mL·min-1·mg-1) | 190.67±19.89 | 228.09±11.63 |

Fig. 5 Structural information and dynamic simulation analysisA: 3D structure of Pm10868-17 protein, purple is CBM structure, blue is binding pocket, black labels is fluctuation sites; B: RMSD analysis of Pm10868 and Pm10868-117; C: RMSF analysis of Pm10868 and Pm10868-117
| [1] | MENDONÇA M, BARROCA M, COLLINS T. Endo-1, 4-β- xylanase-containing glycoside hydrolase families: characteristics, singularities and similarities [J/OL]. Biotechnol. Adv., 2023, 65: 108148 [2024-11-20]. . |
| [2] | ZARAFETA D, GALANOPOULOU A P, LENI M E, et al.. XynDZ5: a new thermostable GH10 xylanase [J/OL]. Front. Microbiol., 2020, 11: 545 [2024-11-20]. . |
| [3] | GUPTA G K, DIXIT M, KAPOOR R K, et al.. Xylanolytic enzymes in pulp and paper industry: new technologies and perspectives [J]. Mol. Biotechnol., 2022, 64(2): 130-143. |
| [4] | BASIT A, LIU J, RAHIM K, et al.. Thermophilic xylanases: from bench to bottle [J]. Crit. Rev. Biotechnol., 2018, 38(7): 989-1002. |
| [5] | MALHOTRA G, CHAPADGAONKAR S S. Thermo-alkali stable bacterial xylanase for deinking of copier paper [J/OL]. J. Genet. Eng. Biotechnol., 2023, 21(1): 107 [2024-11-20]. . |
| [6] | XING H, ZOU G, LIU C, et al.. Improving the thermostability of a GH11 xylanase by directed evolution and rational design guided by B-factor analysis [J/OL]. Enzyme Microb. Technol., 2021, 143: 109720 [2024-11-20]. . |
| [7] | LI G, ZHOU X, LI Z, et al.. Significantly improving the thermostability of a hyperthermophilic GH10 family xylanase XynAF1 by semi-rational design [J]. Appl. Microbiol. Biotechnol., 2021, 105(11): 4561-4576. |
| [8] | CHEN Q, XIAO Y, SHAKHNOVICH E I, et al.. Semi-rational design and molecular dynamics simulations study of the thermostability enhancement of cellobiose 2-epimerases [J]. Int. J. Biol. Macromol., 2020, 154: 1356-1365. |
| [9] | KHAKZAD H, IGASHOV I, SCHNEUING A, et al.. A new age in protein design empowered by deep learning [J]. Cell Syst., 2023, 14(11): 925-939. |
| [10] | QIU Y, WEI G W. Artificial intelligence-aided protein engineering: from topological data analysis to deep protein language models [J]. ArXiv, 2023, 23(7):14587-14596. |
| [11] | CHOWDHURY R, BOUATTA N, BISWAS S, et al.. Single-sequence protein structure prediction using a language model and deep learning [J]. Nat. Biotechnol., 2022, 40(11): 1617-1623. |
| [12] | JISNA V A, JAYARAJ P B. Protein structure prediction: conventional and deep learning perspectives [J]. Protein J., 2021, 40(4): 522-544. |
| [13] | LIU T, ZHANG Y, LI Y, et al.. Effective gene expression prediction and optimization from protein sequences [J/OL]. Adv. Sci. (Weinh), 2025, 12(8): e2407664 [2024-11-20]. . |
| [14] | 李恒, 唐双焱. 碳水化合物结合结构域研究进展[J].微生物学报, 2017, 57(8): 1160-1167. |
| LI H, TANG S Y. Carbohydrate-binding modules: assisted polysaccharide recognition [J]. Acta Microbiol. Sin., 2017, 57(8): 1160-1167. | |
| [15] | MEHMOOD A, NAWAB S, JIN Y, et al.. Mutational impacts on the N and C terminal domains of the MUC5B protein: a transcriptomics and structural biology study [J]. ACS Omega, 2023, 8(4): 3726-3735. |
| [16] | DEVILLARD E, BERA-MAILLET C, FLINT H J, et al.. Characterization of XYN10B, a modular xylanase from the ruminal protozoan Polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose [J]. Biochem. J., 2003, 373(Pt 2): 495-503. |
| [17] | 杜坤,甘一如,黄鹤.活性位点邻近的?-loop对胰蛋白酶热稳定性和活性的影响[J].高校化学工程学报,2017,31(3):657-662. |
| DU K, GAN Y R, HUANG H. Effects of?-loop near active sites on the stability and activity of trypsin [J]. J. Chem. Eng. Chin. Univ., 2017, 31(3): 657-662. | |
| [18] | FURUITA K, JEE J, FUKADA H, et al.. Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies [J]. J. Biol. Chem., 2010, 285(17): 12961-12970. |
| [19] | HETZ C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond [J]. Nat. Rev. Mol. Cell Biol., 2012, 13(2): 89-102. |
| [20] | HOU Q, BOURGEAS R, PUCCI F, et al.. Computational analysis of the amino acid interactions that promote or decrease protein solubility [J/OL]. Sci. Rep., 2018, 8(1): 14661 [2024-11-20]. . |
| [21] | TRETYACHENKO V, VYMĚTAL J, NEUWIRTHOVÁ T, et al.. Modern and prebiotic amino acids support distinct structural profiles in proteins [J/OL]. Open Biol., 2022, 12(6): 220040 [2024-11-20]. . |
| [22] | RAJ A, KUMAR S, SINGH S K. A highly thermostable xylanase from Stenotrophomonas maltophilia: purification and partial characterization [J/OL]. Enzyme Res., 2013, 2013: 429305 [2024-11-20]. . |
| [1] | Wei WANG, Linnan DU, Yuanyuan XU, Hongwei YU. Three-level Mid-infrared Spectroscopy Study of Tubers Starch Crystal/amorphous Structure and Thermostability [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 148-157. |
| [2] | Taotao MAO, Xiaoqiang ZHAO, Xiaodong BAI, Bin YU. Effect of Low Temperature Stress on Photosynthetic Performance, Antioxidant Enzyme System and Related Gene Expression in Maize Seedlings [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 49-60. |
| [3] | Nan WANG, Caifeng YANG, Huakang PENG, Wenfang GUO, Mengqi WANG, Gangqiang LI, Dehu LIU. N-glycosylation in the Propeptide Improved Thermostability of Camel Prochymosin and Enhanced Its Secretion in Pichia pastoris [J]. Journal of Agricultural Science and Technology, 2022, 24(10): 71-78. |
| [4] | ZHENG Yongxing, LI Gezi*, KANG Guozhang*. Physiological Response and Expression of Genes Encoding Ascorbate-glutathione Synthesis Enzymes to Cu 2+ Stress in Wheat [J]. Journal of Agricultural Science and Technology, 2021, 23(1): 21-29. |
| [5] | DUAN Yihong, YAN Yuanyuan, CHEN Liting, LI Qing, ZHANG Dongmei, SUN Zhengwen, ZHANG Yan, MA Zhiying, WANG Xingfen*. Cloning and Functional Analysis of GhMYB44 Related to Flowering Time in Gossypium hirsutum#br# [J]. Journal of Agricultural Science and Technology, 2020, 22(12): 29-38. |
| [6] | WEI Qi-chao1,2, ZHANG Rui1, MENG Zhi-gang1, SUN Guo-qing1, ZHOU Tao1, ZHAO Lei-l. Construction of Plant Expression Vector of shTRAIL and its Transient Expression [J]. Journal of Agricultural Science and Technology, 2016, 18(1): 40-45. |
| [7] | LUO Xiao-liang1§, GU Xing-lu2§, TIAN Jian1, CHU Xiao-yu1*, WU Ning-feng1. Studies on Enhancing Thermostability of Methyl Parathion Hydrolase by Combinatorial Mutagenesis [J]. , 2015, 17(1): 167-172. |
| [8] | YANG Wenhan, GUO Xiaojing, CHEN Yiqun, LV Junnan, XIE Fei, CAO Yunhe*. Studies on Secreted Expression of an Acidic Xylanase in Pichia pastoris and Evaluation of its in vitro Activity [J]. , 2013, 15(5): 59-66. |
| [9] | LI Yanan1, YU Lihong1, LI Yunkai1, MA Wenkang, ZHANG Wei2*. Gene Cloning and Analysis of Enzymology Property of an Thermophilic Xylanase from Humicola insolens Y1 [J]. , 2013, 15(4): 121-128. |
| [10] | CHENG Fei-fei1,2, ZHAO Jun-qi2, SHI Peng-jun2, LI Jiang1, YAO Bin2. Cloning, Enzymology Characteristics Analysis of a Xylanase Gene from Phialophora sp. P13 [J]. , 2012, 14(1): 85-90. |
| [11] | ZHANG Fan1,2, SHI Peng-jun2, MIAO Li-hong1, YAO Bin2. Studies on Cloning of Acid Xylanase from Phialophora sp. G5 and its Characteristics [J]. , 2011, 13(3): 60-66. |
| [12] | WANG Jian-she1,2, BAI Ying-guo2, YIN Jun1, YAO Bin2. Cloning, Expression and Characterization Studies on Xylanase Gene, xynA15, from Alicyclobacillus sp.A15 [J]. , 2010, 12(6): 114-119. |
| [13] | LUO Jian-jie1, WANG Ya-ru1, YUAN Tie-zheng1, BAI Ying-guo1, HUANG Huo-qing1, LUO. High-efficiency Expression of Xylanase XYN-W with High Specific Activity in Pichia pastoris [J]. , 2010, 12(5): 80-85. |
| [14] | KUI Hong1, LUO Hui-ying2, DONG Shou-liang1, YAO Bin2. Gene Cloning, Expression and Characterization of |an Acidic Xylanase from Bispora betulina [J]. , 2010, 12(5): 109-115. |
| [15] | LIU Xin, SHI Peng-jun, YANG Pei-long, YAO Bin. Research Progress on Bifunctional Xylanases [J]. , 2010, 12(2): 50-56. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号