




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (4): 75-84.DOI: 10.13304/j.nykjdb.2021.0768
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Xiang HUANG1,2( ), Guangming CHU1, Xinkai ZHENG1, Jintao CHENG1, Jianhao CHEN1, Yingchun XU3, Qijiang JIN3, Meihua YANG1,2(
), Guangming CHU1, Xinkai ZHENG1, Jintao CHENG1, Jianhao CHEN1, Yingchun XU3, Qijiang JIN3, Meihua YANG1,2( )
)
Received:2021-09-01
Accepted:2021-10-20
Online:2022-04-15
Published:2022-04-19
Contact:
Meihua YANG
黄祥1,2( ), 楚光明1, 郑新开1, 程锦涛1, 陈健豪1, 徐迎春3, 金奇江3, 杨梅花1,2(
), 楚光明1, 郑新开1, 程锦涛1, 陈健豪1, 徐迎春3, 金奇江3, 杨梅花1,2( )
)
通讯作者:
杨梅花
作者简介:黄祥 E-mail:shzu_hx@163.com;
基金资助:CLC Number:
Xiang HUANG, Guangming CHU, Xinkai ZHENG, Jintao CHENG, Jianhao CHEN, Yingchun XU, Qijiang JIN, Meihua YANG. Analysis of Codon Usage Bias and Phylogenetic of Chloroplast Genome in Nymphaea[J]. Journal of Agricultural Science and Technology, 2022, 24(4): 75-84.
黄祥, 楚光明, 郑新开, 程锦涛, 陈健豪, 徐迎春, 金奇江, 杨梅花. 睡莲属叶绿体基因组密码子偏好性及系统发育分析[J]. 中国农业科技导报, 2022, 24(4): 75-84.
编号 Number | 物种 Species | 登录号 Genbank ID | 密码子适应指数 CAI | 有效密码子数 ENC | GC1/% | GC2/% | GC3/% | GCall/% |
|---|---|---|---|---|---|---|---|---|
| 1 | 澳洲巨花睡莲 N. gigantea | MT107637.1 | 0.170 | 51.468 | 46.11 | 38.66 | 32.61 | 39.13 |
| 2 | 白睡莲 N. alba | NC_006050.1 | 0.169 | 51.483 | 46.23 | 38.69 | 32.54 | 39.15 |
| 3 | 齿叶睡莲 N. lotus | NC_041238.1 | 0.168 | 51.365 | 46.34 | 38.66 | 32.4 | 39.13 |
| 4 | 厄德洛睡莲 N. heudelotii | MW802266.1 | 0.170 | 51.401 | 46.22 | 38.61 | 32.57 | 39.13 |
| 5 | 非洲睡莲 N. capensis | NC_040167.1 | 0.169 | 51.446 | 46.25 | 38.75 | 32.6 | 39.20 |
| 6 | 黄睡莲 N. mexicana | NC_024542.1 | 0.169 | 51.403 | 46.19 | 38.69 | 32.43 | 39.10 |
Table 1 Codon usage bias analysis of total 22 Nymphaea
编号 Number | 物种 Species | 登录号 Genbank ID | 密码子适应指数 CAI | 有效密码子数 ENC | GC1/% | GC2/% | GC3/% | GCall/% |
|---|---|---|---|---|---|---|---|---|
| 1 | 澳洲巨花睡莲 N. gigantea | MT107637.1 | 0.170 | 51.468 | 46.11 | 38.66 | 32.61 | 39.13 |
| 2 | 白睡莲 N. alba | NC_006050.1 | 0.169 | 51.483 | 46.23 | 38.69 | 32.54 | 39.15 |
| 3 | 齿叶睡莲 N. lotus | NC_041238.1 | 0.168 | 51.365 | 46.34 | 38.66 | 32.4 | 39.13 |
| 4 | 厄德洛睡莲 N. heudelotii | MW802266.1 | 0.170 | 51.401 | 46.22 | 38.61 | 32.57 | 39.13 |
| 5 | 非洲睡莲 N. capensis | NC_040167.1 | 0.169 | 51.446 | 46.25 | 38.75 | 32.6 | 39.20 |
| 6 | 黄睡莲 N. mexicana | NC_024542.1 | 0.169 | 51.403 | 46.19 | 38.69 | 32.43 | 39.10 |
编号 Number | 物种 Species | 登录号 Genbank ID | 密码子适应指数 CAI | 有效密码子数 ENC | GC1/% | GC2/% | GC3/% | GCall/% |
|---|---|---|---|---|---|---|---|---|
| 均值Mean | 0.169 | 51.401 | 46.26 | 38.67 | 32.54 | 35.16 | ||
| 8 | 蓝星睡莲 N. colorata | MT107631.1 | 0.170 | 51.466 | 46.13 | 38.61 | 32.65 | 39.13 |
| 9 | 鲁吉娜睡莲 N. rudgeana | MW802268.1 | 0.169 | 51.320 | 46.25 | 38.53 | 32.41 | 39.06 |
| 10 | 睡莲(子午莲) N. tetragona | MT107634.1 | 0.169 | 51.577 | 46.82 | 39.24 | 32.88 | 39.65 |
| 11 | 特尼日洼睡莲 N. tenerinervia | MW802269.1 | 0.170 | 51.317 | 46.21 | 38.54 | 32.39 | 39.05 |
| 12 | 细瓣睡莲 N. gracilis | MW802265.1 | 0.170 | 51.391 | 46.21 | 38.57 | 32.53 | 39.10 |
| 13 | 香睡莲 N. oborata | MT107636.1 | 0.170 | 51.500 | 47.03 | 39.33 | 33.00 | 39.79 |
| 14 | 小花睡莲 N. micrantha | MT107635.1 | 0.170 | 51.466 | 46.11 | 38.65 | 32.61 | 39.12 |
| 15 | 小腺睡莲 N. glandulifera | MW802264.1 | 0.170 | 51.303 | 46.19 | 38.52 | 32.37 | 39.03 |
| 16 | 亚马孙睡莲 N. amazonum | MW802262.1 | 0.170 | 51.317 | 46.21 | 38.53 | 32.41 | 39.05 |
| 17 | 延药睡莲 N. stellata | MT991061.1 | 0.170 | 51.410 | 46.19 | 38.60 | 32.57 | 39.12 |
| 18 | 夜雪睡莲 N. potamophila | MT107633.1 | 0.170 | 51.365 | 46.07 | 38.60 | 32.46 | 39.04 |
| 19 | 印度红睡莲 N. rubra | MT107632.1 | 0.169 | 51.399 | 46.17 | 38.51 | 32.44 | 39.04 |
| 20 | 永恒睡莲 N. immutabilis | MW802267.1 | 0.170 | 51.349 | 46.2 | 38.59 | 32.52 | 39.10 |
| 21 | 詹姆森睡莲 N. jamesoniana | NC_031826.2 | 0.169 | 51.362 | 46.25 | 38.67 | 32.44 | 39.12 |
| 22 | 侏儒卢旺达睡莲 N. thermarum | MW143076.1 | 0.170 | 51.419 | 46.19 | 38.61 | 32.60 | 39.13 |
Table 1 Codon usage bias analysis of total 22 Nymphaea
编号 Number | 物种 Species | 登录号 Genbank ID | 密码子适应指数 CAI | 有效密码子数 ENC | GC1/% | GC2/% | GC3/% | GCall/% |
|---|---|---|---|---|---|---|---|---|
| 均值Mean | 0.169 | 51.401 | 46.26 | 38.67 | 32.54 | 35.16 | ||
| 8 | 蓝星睡莲 N. colorata | MT107631.1 | 0.170 | 51.466 | 46.13 | 38.61 | 32.65 | 39.13 |
| 9 | 鲁吉娜睡莲 N. rudgeana | MW802268.1 | 0.169 | 51.320 | 46.25 | 38.53 | 32.41 | 39.06 |
| 10 | 睡莲(子午莲) N. tetragona | MT107634.1 | 0.169 | 51.577 | 46.82 | 39.24 | 32.88 | 39.65 |
| 11 | 特尼日洼睡莲 N. tenerinervia | MW802269.1 | 0.170 | 51.317 | 46.21 | 38.54 | 32.39 | 39.05 |
| 12 | 细瓣睡莲 N. gracilis | MW802265.1 | 0.170 | 51.391 | 46.21 | 38.57 | 32.53 | 39.10 |
| 13 | 香睡莲 N. oborata | MT107636.1 | 0.170 | 51.500 | 47.03 | 39.33 | 33.00 | 39.79 |
| 14 | 小花睡莲 N. micrantha | MT107635.1 | 0.170 | 51.466 | 46.11 | 38.65 | 32.61 | 39.12 |
| 15 | 小腺睡莲 N. glandulifera | MW802264.1 | 0.170 | 51.303 | 46.19 | 38.52 | 32.37 | 39.03 |
| 16 | 亚马孙睡莲 N. amazonum | MW802262.1 | 0.170 | 51.317 | 46.21 | 38.53 | 32.41 | 39.05 |
| 17 | 延药睡莲 N. stellata | MT991061.1 | 0.170 | 51.410 | 46.19 | 38.60 | 32.57 | 39.12 |
| 18 | 夜雪睡莲 N. potamophila | MT107633.1 | 0.170 | 51.365 | 46.07 | 38.60 | 32.46 | 39.04 |
| 19 | 印度红睡莲 N. rubra | MT107632.1 | 0.169 | 51.399 | 46.17 | 38.51 | 32.44 | 39.04 |
| 20 | 永恒睡莲 N. immutabilis | MW802267.1 | 0.170 | 51.349 | 46.2 | 38.59 | 32.52 | 39.10 |
| 21 | 詹姆森睡莲 N. jamesoniana | NC_031826.2 | 0.169 | 51.362 | 46.25 | 38.67 | 32.44 | 39.12 |
| 22 | 侏儒卢旺达睡莲 N. thermarum | MW143076.1 | 0.170 | 51.419 | 46.19 | 38.61 | 32.60 | 39.13 |
| 参数Parameter | ENC | GC1 | GC2 | GC12 | GC3 | GC3s |
|---|---|---|---|---|---|---|
| GC1 | 0.04 | |||||
| GC2 | 0.77** | 0.38 | ||||
| GC12 | 0.52 | 0.74** | 0.82** | |||
| GC3 | 0.90** | -0.01 | 0.64* | 0.42 | ||
| GC3s | 0.89** | -0.04 | 0.63* | 0.40 | 0.97** | |
| GCall | 0.83** | 0.46 | 0.86** | 0.82** | 0.78** | 0.74** |
Table 2 Correlation analysis of GC contents and related parameters in codons of Nymphaea
| 参数Parameter | ENC | GC1 | GC2 | GC12 | GC3 | GC3s |
|---|---|---|---|---|---|---|
| GC1 | 0.04 | |||||
| GC2 | 0.77** | 0.38 | ||||
| GC12 | 0.52 | 0.74** | 0.82** | |||
| GC3 | 0.90** | -0.01 | 0.64* | 0.42 | ||
| GC3s | 0.89** | -0.04 | 0.63* | 0.40 | 0.97** | |
| GCall | 0.83** | 0.46 | 0.86** | 0.82** | 0.78** | 0.74** |
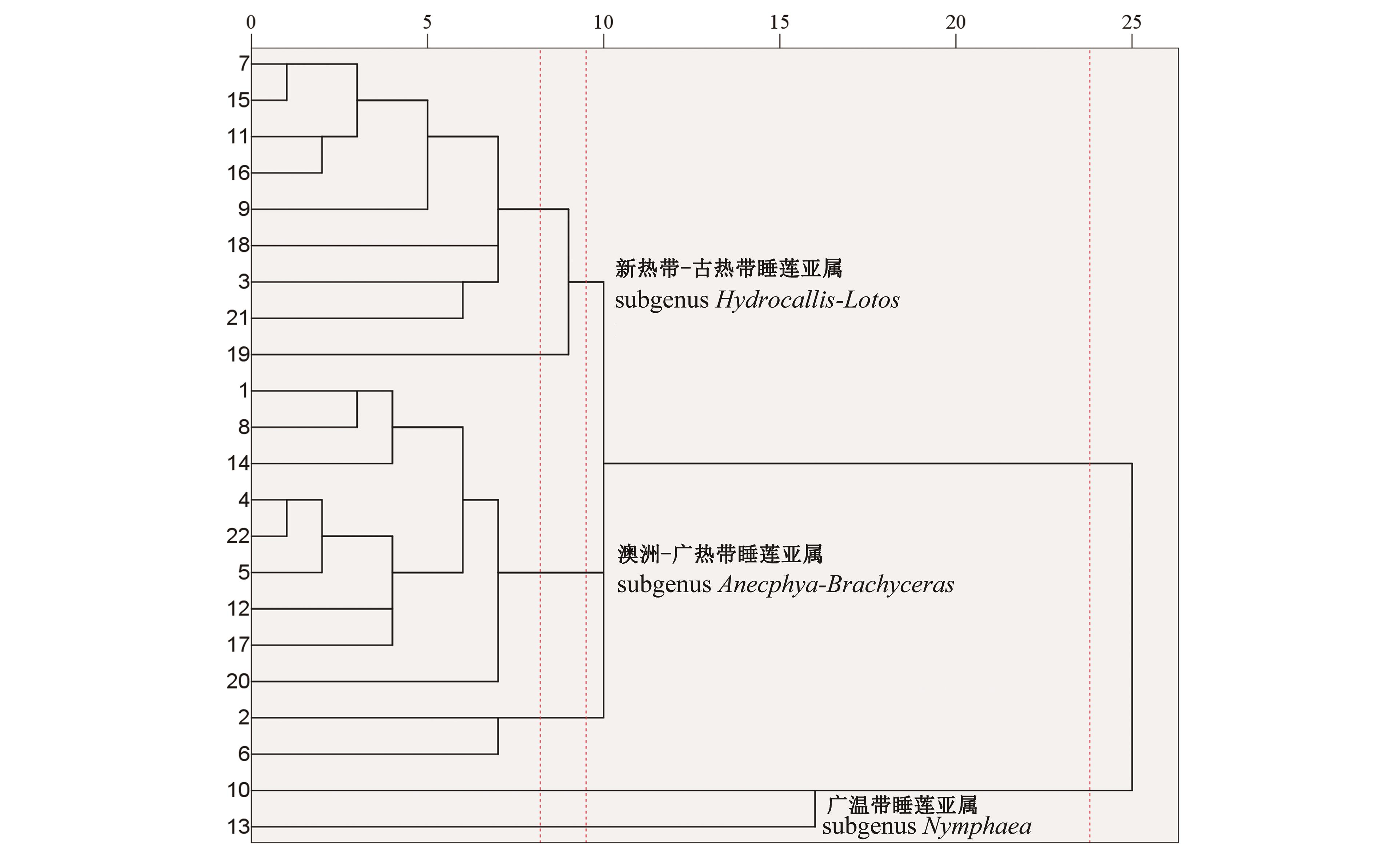
Fig.5 Cluster of total 22 Nymphaea species based on RSCU value of chloroplast codonsNote: 1~22 correspond to the number of total 22 Nymphaea species in Table 1.
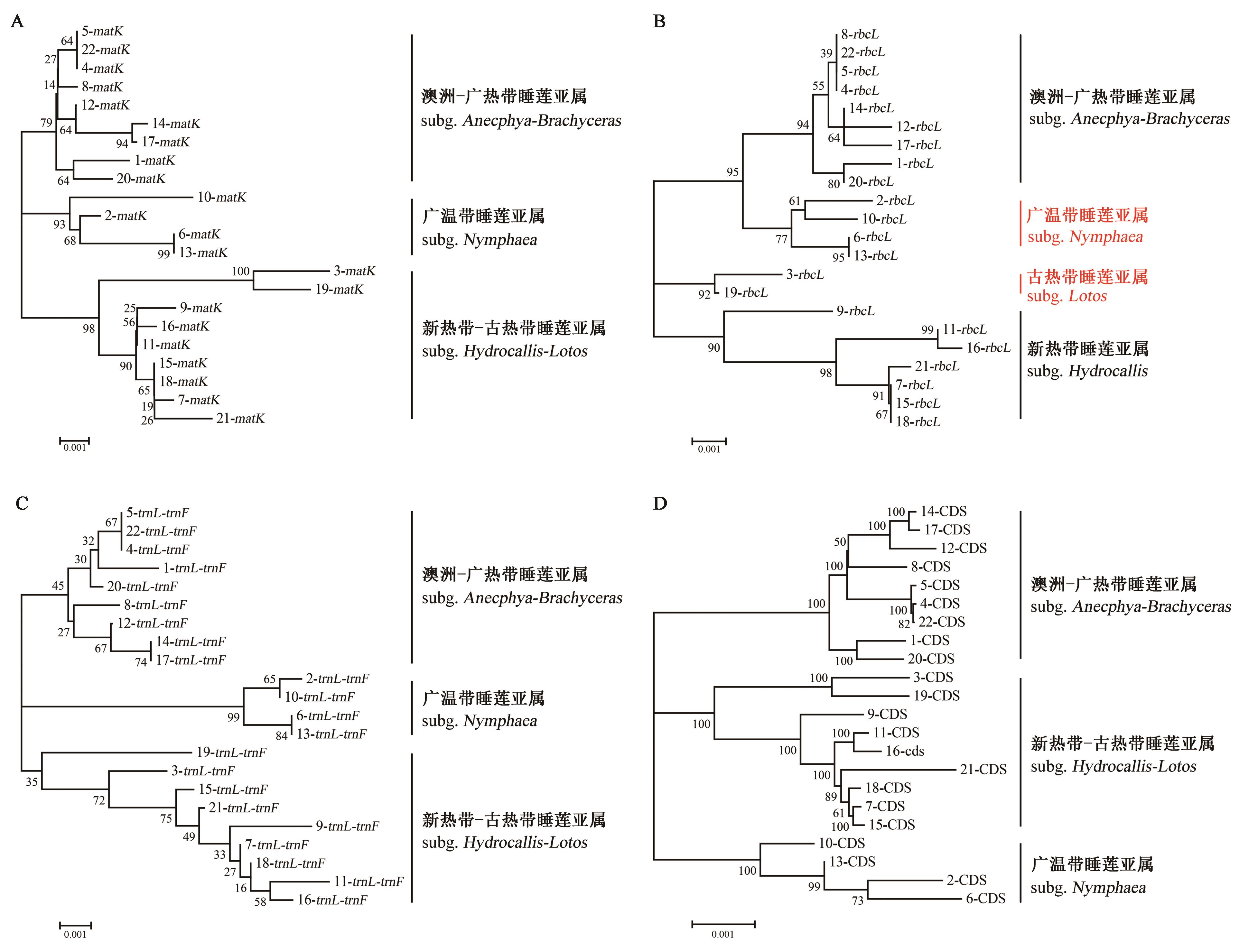
Fig. 6 Phylogenetic tree of total 22 chloroplasts from NymphaeaA: Based on chloroplast matK gene sequences; B: Based on chloroplast rbcL gene sequences; C: Based on chloroplast trnL?trnF gene sequences; D: Based on chloroplast CDS sequences
| 1 | GUAN D L, MA L B, MUHAMMAD S K, et al.. Analysis of codon usage patterns in Hirudinaria manillensis reveals a preference for GC-ending codons caused by dominant selection constraints [J/OL]. BMC Genomics, 2018, 19(1):542 [2021-08-19]. . |
| 2 | LIU H, HUANG Y, DU X, et al.. Patterns of synonymous codon usage bias in the model grass Brachypodium distachyon [J/OL]. Genet. Mol. Res.: GMR, 2012, 11(AOP):4695 [2021-08-19]. . |
| 3 | 刘慧,王梦醒,岳文杰,等.糜子叶绿体基因组密码子使用偏性的分析[J].植物科学学报,2017,35(3):362-371. |
| LIU H, WANG M X, YUE W J, et al.. Analysis of codon usage in the chloroplast genome of Broomcorn millet (Panicum miliaceum) [J]. Plant Sci. J., 2017, 35(3):362-371. | |
| 4 | PEDEN J F. Analysis of codon usage [D]. Nottingham: University of Nottingham, 1999. |
| 5 | ROMERO H, ZAVALA A, MUSTO H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces [J]. Nucleic Acids Res., 2000, 10:2084-2090. |
| 6 | XU C, CAI X, CHEN Q, et al.. Factors affecting synonymous codon usage bias in chloroplast genome of Oncidium gower ramsey [J]. Evol. Bioinform., 2011, 7:271-278. |
| 7 | KARLIN S, MRAZEK J. What drives codon choices in human genes? [J]. J. Mol. Biol., 1996, 262(4):459-472. |
| 8 | NIE X J, DENG P C, FENG K W, et al.. Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family [J]. Plant Mol. Biol. Rep., 2013, 32(4):828-840. |
| 9 | KAWABE A, MIYASHITA N T. Patterns of codon usage bias in three dicot and four monocot plant species [J]. Genes Genet. Syst., 2003, 78(5):343-352. |
| 10 | WANG L, ROOSSINCK M J. Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants [J]. Plant Mol. Biol., 2006, 61(4-5):699-710. |
| 11 | 季凯凯,宋希强,陈春国,等.木兰科叶绿体基因组的密码子使用特征分析[J].中国农业科技导报,2020,22(11):52-62. |
| JI K K, SONG X Q, CHEN C G, et al.. Codon usage profiling of chloroplast genome in Magnoliaceae [J]. J. Agric. Sci. Technol., 2020, 22(11):52-62. | |
| 12 | 陈哲,胡福初,王祥和,等.菠萝密码子使用偏好性分析[J].果树学报,2017(8):24-33. |
| CHEN Z, HU F C, WANG X H, et al.. Analysis of codon usage bias of Ananas comosus with genome sequencing data [J]. J. Fruit Sci., 2017(8):24-33. | |
| 13 | 黄国振.睡莲[M].北京:中国林业出版社,2008:1-15. |
| HUANG G Z. The Waterlilies [M]. Beijing: China Forestry Press, 2008:1-15. | |
| 14 | SUN C, CHEN F, TENG N, et al.. Comparative analysis of the complete chloroplast genome of seven Nymphaea species [J]. Aquat. Bot., 2021, 170(1):1-7. |
| 15 | ZHANG L, CHEN F, ZHANG X, et al.. The water lily genome and the early evolution of flowering plants [J]. Nature, 2020, 577(7788):1-6. |
| 16 | BORSCH T, HILU K W, WIERSEMA J H, et al.. Phylogeny of Nymphaea (Nymphaeaceae): evidence from substitutions and microstructural changes in the chloroplast trnT⁃trnF region [J]. Int. J. Plant Sci., 2007, 168(5):639-671. |
| 17 | LOHNE C, BORSCH T, WIERSEMA J H. Phylogenetic analysis of Nymphaeales using fast-evolving and noncoding chloroplast markers [J]. Bot. J. Linn. Soc., 2010, 154(2):141-163. |
| 18 | TANG D, FAN W, KASHIF M H, et al.. Analysis of chloroplast differences in leaves of rice isonuclear alloplasmic lines [J]. Protoplasma, 2018, 255(1):1-9. |
| 19 | 王艇,苏应娟,朱建明.叶绿体rbcL基因序列在植物系统学研究中的应用[J].植物科学学报,1999,17(S1):8-14. |
| WANG C, SU Y J, ZHU J M. Application of chloroplast rbcL gene sequence in plant systematics [J]. J. Plant Sci., 1999,17 (S1):8-14. | |
| 20 | AOKI S, ITO M. Molecular phylogeny of Nicotiana (Solanaceae) based on the nucleotide sequence of the matK Gene [J]. Plant Biol., 2010, 2(3):316-324. |
| 21 | JENA S N, KUMAR S, NAIR N K. Molecular phylogeny in Indian Citrus L. (Rutaceae) inferred through PCR-RFLP and trnL-trnF sequence data of chloroplast DNA [J]. Sci. Hortic., 2009, 119(4):403-416. |
| 22 | GOREMYKIN V V, HIRSCH K I, STEFAN W, et al.. The chloroplast genome of nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm [J/OL]. Mol. Biol. Evol., 2004(7):7 [2021-08-19]. . |
| 23 | KIM Y, MIN J, KWON W, et al.. The complete chloroplast genome sequence of the Nymphaea capensis Thumb. (Nymphaeaceae) [J]. Mitochondrial DNA B, 2019, 4(1):401-402. |
| 24 | 尚明照,刘方,华金平,等.陆地棉叶绿体基因组密码子使用偏性的分析[J].中国农业科学,2011,44(2):245-253. |
| SHANG M Z, LIU F, HUA J P, et al.. Analysis on codon usage of chloroplast genome of Gossypium hirsutum [J]. Agric. Sci. China, 2011, 44 (2):245-253. | |
| 25 | 叶友菊,倪州献,白天道,等.马尾松叶绿体基因组密码子偏好性分析[J].基因组学与应用生物学,2018,37(10):4464-4471. |
| YE Y J, NI Z X, BAI T D, et al.. Analysis of codon preference in the chloroplast genome of Masson Pine [J]. Genom. Appl. Bio., 2018, 37(10):4464-4471. | |
| 26 | 杨国锋,苏昆龙,赵恰然,等.蒺蔡首蓿叶绿体密码子偏好性分析[J].草业学报,2015,24(12):171-179. |
| YANG G F, SY K L, ZHAO Y R, et al.. Analysis of codon usage in the chloroplast genome of Medicago truncatula [J]. Acta Pratac. Sin., 2015, 24(12): 171-179. | |
| 27 | 吴完明,吴松锋,任大明,等.密码子偏性的分析方法及相关研究进展[J].遗传,2007,29(4):420-426. |
| WU W M, WU S F, REN D M, et al.. Analysis methods and related research progress of codon bias [J]. Hereditas, 2007, 29(4):420-426. | |
| 28 | KOICHIRO T, JOEL D, MASATOSHI N, et al.. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 [J/OL]. Mol. Biol. Evol., 2007, (8): 1596 [2021-08-19]. . |
| 29 | 王晓双,杨芳,罗茜,等.石斛属叶绿体基因组密码子使用偏性及系统发育分析[J/OL].分子植物育种,2021:1-22 [2021-08-19]. . |
| WANG X A, YANG F, LUO X, et al.. Codon usage bias and phylogenetic analysis of chloroplast genome of Dendrobium [J/OL]. Mol. Plant Breed., 2021:1-22 [2021-08-19]. . | |
| 30 | 赵森,邓力华,陈芬.秋茄叶绿体基因组密码子使用偏好性分析[J].森林与环境学报,2020,40(5):534-541. |
| ZHAO S, DENG L H, CHEN F. Analysis of codon use preference in the chloroplast genome of Kandelia [J]. J. Forest Environ., 2020, 40(5):534-541. | |
| 31 | MARAIS G, MOUCHIROU D, DURET L. Neutral effect of recombination on base composition in Drosophila [J]. Genet. Res., 2003, 81(2):79-87. |
| 32 | SHARP P M, LI W H. An evolutionary perspective on synonymous codon usage in unicellular organisms [J]. J. Mol. Evol., 1986, 24(1):28-38. |
| 33 | 尹为治,方正,黄良鸿,等.海南2种龙脑香科植物叶绿体基因组密码子偏好性分析[J].林业调查规划,2020,45(6):25-32. |
| YIN W Z, FANG Z, HUANG L H, et al.. Codon preference analysis of chloroplast genome of two species of Dipterocarpaceae in Hainan [J]. Forestry Surv. Plan., 2020, 45(6):25-32. | |
| 34 | ZHOU H, WANG H, HUANG L F, et al.. Heterogeneity in codon usages of sobemovirus genes [J]. Arch. Virol., 2005, 150(8):1591-1605. |
| 35 | 李蓉,谢析颖,王雪晶,等.兰科植物FNR基因的密码子偏好性分析[J].热带作物学报,2018,39(6):1137-1145. |
| LI R, XIE X Y, WANG X J, et al.. Codon preference analysis of FNR gene in orchids [J]. J. Trop. Crops, 2018, 39 (6):1137-1145. | |
| 36 | 晁岳恩,常阳,王美芳,等.7种作物叶绿体基因的密码子偏好性及聚类分析[J].华北农学报,2012(4):60-64. |
| CHAO Y E, CHANG Y, WANG M F, et al.. Codon preference and cluster analysis of chloroplast genes in 7 crops [J]. Acta Agric. Boreali-Sin., 2012 (4):60-64. |
| [1] | Lihong HAN, Xinnan JIA, Renhuan CHEN, Chao LIU. Analysis of Codon Usage Bias of the Genome of Medicinal Fungus Wolfiporia cocos [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 83-90. |
| [2] | SU Yue§, LIU Juanjuan§, WAN Bin, ZHANG Pengju, CHEN Zhenggen, SU Junji, WANG Caixiang. Chloroplast Genome Structure Characteristic and Phylogenetic Analysis of Mulgedium tataricum [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 33-42. |
| [3] | JI Kaikai1, SONG Xiqiang1, CHEN Chunguo2, LI Ge2, XIE Shangqian1*. Codon Usage Profiling of Chloroplast Genome in Magnoliaceae [J]. Journal of Agricultural Science and Technology, 2020, 22(11): 52-62. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号