




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (10): 133-142.DOI: 10.13304/j.nykjdb.2021.0891
• ANIMAL AND PLANT HEALTH • Previous Articles
Zhijian LIN1( ), Changjiang CHEN2, Ting ZHOU3, Gang GU3(
), Changjiang CHEN2, Ting ZHOU3, Gang GU3( ), Fangping HU1, Chunying LI3, Xueqing CAI1(
), Fangping HU1, Chunying LI3, Xueqing CAI1( )
)
Received:2021-10-20
Accepted:2021-12-09
Online:2022-10-15
Published:2022-10-25
Contact:
Gang GU,Xueqing CAI
林志坚1( ), 陈长江2, 周挺3, 顾钢3(
), 陈长江2, 周挺3, 顾钢3( ), 胡方平1, 李春英3, 蔡学清1(
), 胡方平1, 李春英3, 蔡学清1( )
)
通讯作者:
顾钢,蔡学清
作者简介:林志坚 E-mail:lzj06050@163.com
基金资助:CLC Number:
Zhijian LIN, Changjiang CHEN, Ting ZHOU, Gang GU, Fangping HU, Chunying LI, Xueqing CAI. Control Effect of Ralstonia Phage RPZH6 Strain on Tobacco Bacterial Wilt and Its Complete Genome Analysis[J]. Journal of Agricultural Science and Technology, 2022, 24(10): 133-142.
林志坚, 陈长江, 周挺, 顾钢, 胡方平, 李春英, 蔡学清. 青枯菌噬菌体RPZH6株系对烟草青枯病的生防效果及全基因组测序分析[J]. 中国农业科技导报, 2022, 24(10): 133-142.
| 处理 Treatment | 7 d | 14 d | 21 d | 28 d | 35 d | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | |
| RPZH6+TBRS12 | 4.17 b | 57.10 a | 19.44 c | 58.83 a | 22.22 b | 60.98 a | 30.56 b | 55.10 a | 33.33 c | 53.85 a |
| Cu(OH)2+TBRS12 | 5.55 b | 42.90 a | 36.11 b | 23.53 b | 54.17 a | 4.86 b | 62.50 a | 8.17 b | 65.28 b | 9.61 b |
| TBRS12 | 9.72 a | — | 47.22 a | — | 56.94 a | — | 68.06 a | — | 72.22 a | — |
| CK | 0.00 | — | 0.00 | — | 0.00 | — | 0.00 | — | 0.00 | — |
Table 1 Control effect of phage RPZH6 on tobacco bacterial wilt
| 处理 Treatment | 7 d | 14 d | 21 d | 28 d | 35 d | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | 病情指数Disease index | 防治效果Control effect/% | |
| RPZH6+TBRS12 | 4.17 b | 57.10 a | 19.44 c | 58.83 a | 22.22 b | 60.98 a | 30.56 b | 55.10 a | 33.33 c | 53.85 a |
| Cu(OH)2+TBRS12 | 5.55 b | 42.90 a | 36.11 b | 23.53 b | 54.17 a | 4.86 b | 62.50 a | 8.17 b | 65.28 b | 9.61 b |
| TBRS12 | 9.72 a | — | 47.22 a | — | 56.94 a | — | 68.06 a | — | 72.22 a | — |
| CK | 0.00 | — | 0.00 | — | 0.00 | — | 0.00 | — | 0.00 | — |
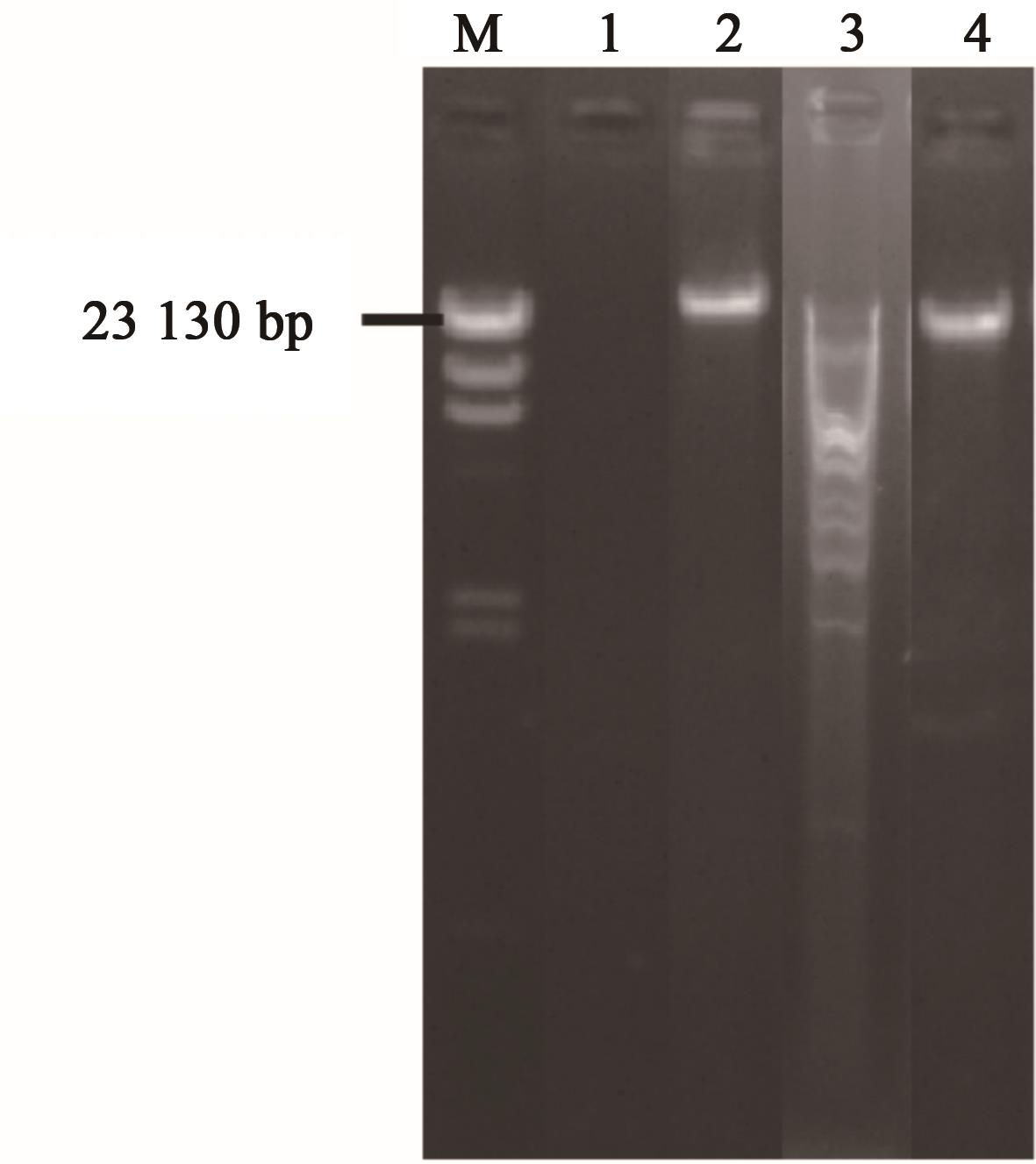
Fig. 1 Enzyme digestion of nuclear acid of phage RPZH6Note:M—λ HindⅢ DNA marker; 1—DNase Ⅰ treatment; 2—RNase A treatment; 3—EcoRⅠ treatment; 4—RPZH6 nucleic acid.
| 开放阅读框ORF | 开放阅读框位置 Position of ORF/bp | 蛋白大小 Protein size/aa | 链 Strand | 起始密码子 Start codon | 功能预测 Possible function |
|---|---|---|---|---|---|
| 1 | 1~262 | 86 | + | GTG | 转录调节蛋白 Putative transcription regulator protein |
| 2 | 296~478 | 60 | - | ATG | 含甘氨酸拉链2TM结构域的蛋白质 Glycine zipper 2TM domain-containing protein |
| 4 | 1 069~1 542 | 157 | - | ATG | 病毒粒子相关蛋白 Virion associated protein |
| 5 | 1 620~2 729 | 369 | - | ATG | 主要衣壳蛋白 Major capsid protein |
| 6 | 2 831~3 283 | 150 | - | ATG | 含AP2结构域蛋白AP2 domain-containing protein |
| 7 | 3 328~3 525 | 65 | - | ATG | 碳储存调节器 Carbon storage regulator |
| 11 | 4 844~7 189 | 781 | - | GTG | 门脉蛋白 Portal protein |
| 14 | 8 170~9 915 | 581 | - | ATG | 末端酶大亚基蛋白 Terminase large subunit protein |
| 16 | 10 578~10 769 | 63 | - | ATG | 辅酶A酯裂解酶 CoA ester lyase |
| 17 | 10 769~10 867 | 32 | - | ATG | 类连接蛋白 Connectin-like protein |
| 24 | 13 090~13 326 | 78 | - | ATG | 转录调节因子 Transcriptional regulator |
| 32 | 15 480~15 725 | 81 | - | GTG | 末端酶小亚基蛋白 Terminase small subunit protein |
| 34 | 16 836~17 324 | 162 | - | ATG | HNH归巢核酸内切酶Ⅱ HNH homing endonuclease Ⅱ |
| 41 | 19 861~20 667 | 268 | - | ATG | DnaC类蛋白 DnaC-like protein |
| 42 | 20 667~21 437 | 102 | - | TTG | 复制蛋白 Replication protein |
| 46 | 24 667~25 026 | 119 | - | ATG | DUF1364家族蛋白 DUF1364 family protein |
| 50 | 26 150~26 731 | 193 | - | ATG | 单链DNA结合蛋白 Single-stranded DNA-binding protein |
| 52 | 27 410~27 778 | 122 | - | ATG | 转录调节因子 Transcriptional regulator |
| 53 | 27 805~28 497 | 230 | + | ATG | 转录调节因子 Transcriptional regulator |
| 54 | 28 570~29 016 | 148 | + | TTG | 转录调节因子 Transcriptional regulator |
| 55 | 29 183~30 211 | 342 | + | ATG | RecB类核酸内切酶 RecB-like endonuclease |
| 56 | 30 250~31 203 | 317 | + | ATG | RecT类蛋白酶 RecT-like protein |
| 57 | 31 209~31 436 | 75 | + | ATG | DNA结合蛋白 DNA-binding protein |
| 58 | 31 461~31 907 | 148 | + | ATG | DNA结合蛋白 DNA-binding protein |
| 59 | 31 932~32 996 | 354 | + | ATG | DNA聚合酶Ⅲ β亚基 DNA polymerase Ⅲ subunit beta |
| 61 | 34 378~34 650 | 90 | + | ATG | Pyocin激活蛋白 PrtN Pyocin activator protein PrtN |
| 63 | 35 571~36 764 | 397 | + | ATG | 酪氨酸重组酶 Tyrosine recombinase |
| 64 | 36 838~37 005 | 55 | + | TTG | 酪氨酸重组酶 Tyrosine recombinase |
| 65 | 37 017~37 553 | 178 | - | ATG | Rz类裂解蛋白 Rz-like lysis protein |
| 67 | 37 929~38 309 | 126 | - | ATG | 趋化蛋白 Chemotaxis protein |
| 69 | 38 626~39 291 | 221 | - | TTG | 含DUF3380结构域蛋白 DUF3380 domain-containing protein |
| 72 | 40 455~46 457 | 2 000 | - | ATG | DarB类抗抑制蛋白 DarB-like antirestriction protein |
| 73 | 46 436~50 134 | 1 232 | - | GTG | DarB类抗抑制蛋白 DarB-like antirestriction protein |
| 74 | 50 192~51 736 | 514 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 75 | 51 733~52 458 | 241 | - | ATG | PAPS还原酶类蛋白 PAPS reductase-like protein |
| 76 | 52 470~53 303 | 277 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 77 | 53 293~53 850 | 185 | - | ATG | 酰基辅酶A N-酰基转移酶 Acyl-CoA N-acyltransferase |
| 79 | 54 246~55 979 | 577 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 80 | 55 991~56 977 | 328 | - | ATG | 尾纤蛋白 Tail fiber protein |
| 81 | 57 063~58 382 | 439 | - | ATG | 推定尾纤蛋白 Putative tail fiber protein |
| 82 | 58 382~58 663 | 93 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 84 | 60 366~61 343 | 325 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 85 | 61 350~61 772 | 140 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 88 | 62 505~63 167 | 220 | - | ATG | 病毒粒子相关蛋白 Virion associated protein |
| 90 | 63 713~64 150 | 145 | - | GTG | DarB类抗抑制蛋白 DarB-like antirestriction protein |
| 91 | 64 147~64 512 | 121 | - | GTG | 钼蝶呤结合蛋白 Molybdopterin-binding protein |
| 92 | 64 534~64 657 | 40 | - | TAT | 细胞色素B Cytochrome B |
Table 2 ORF function prediction of the phage RPZH6 genome
| 开放阅读框ORF | 开放阅读框位置 Position of ORF/bp | 蛋白大小 Protein size/aa | 链 Strand | 起始密码子 Start codon | 功能预测 Possible function |
|---|---|---|---|---|---|
| 1 | 1~262 | 86 | + | GTG | 转录调节蛋白 Putative transcription regulator protein |
| 2 | 296~478 | 60 | - | ATG | 含甘氨酸拉链2TM结构域的蛋白质 Glycine zipper 2TM domain-containing protein |
| 4 | 1 069~1 542 | 157 | - | ATG | 病毒粒子相关蛋白 Virion associated protein |
| 5 | 1 620~2 729 | 369 | - | ATG | 主要衣壳蛋白 Major capsid protein |
| 6 | 2 831~3 283 | 150 | - | ATG | 含AP2结构域蛋白AP2 domain-containing protein |
| 7 | 3 328~3 525 | 65 | - | ATG | 碳储存调节器 Carbon storage regulator |
| 11 | 4 844~7 189 | 781 | - | GTG | 门脉蛋白 Portal protein |
| 14 | 8 170~9 915 | 581 | - | ATG | 末端酶大亚基蛋白 Terminase large subunit protein |
| 16 | 10 578~10 769 | 63 | - | ATG | 辅酶A酯裂解酶 CoA ester lyase |
| 17 | 10 769~10 867 | 32 | - | ATG | 类连接蛋白 Connectin-like protein |
| 24 | 13 090~13 326 | 78 | - | ATG | 转录调节因子 Transcriptional regulator |
| 32 | 15 480~15 725 | 81 | - | GTG | 末端酶小亚基蛋白 Terminase small subunit protein |
| 34 | 16 836~17 324 | 162 | - | ATG | HNH归巢核酸内切酶Ⅱ HNH homing endonuclease Ⅱ |
| 41 | 19 861~20 667 | 268 | - | ATG | DnaC类蛋白 DnaC-like protein |
| 42 | 20 667~21 437 | 102 | - | TTG | 复制蛋白 Replication protein |
| 46 | 24 667~25 026 | 119 | - | ATG | DUF1364家族蛋白 DUF1364 family protein |
| 50 | 26 150~26 731 | 193 | - | ATG | 单链DNA结合蛋白 Single-stranded DNA-binding protein |
| 52 | 27 410~27 778 | 122 | - | ATG | 转录调节因子 Transcriptional regulator |
| 53 | 27 805~28 497 | 230 | + | ATG | 转录调节因子 Transcriptional regulator |
| 54 | 28 570~29 016 | 148 | + | TTG | 转录调节因子 Transcriptional regulator |
| 55 | 29 183~30 211 | 342 | + | ATG | RecB类核酸内切酶 RecB-like endonuclease |
| 56 | 30 250~31 203 | 317 | + | ATG | RecT类蛋白酶 RecT-like protein |
| 57 | 31 209~31 436 | 75 | + | ATG | DNA结合蛋白 DNA-binding protein |
| 58 | 31 461~31 907 | 148 | + | ATG | DNA结合蛋白 DNA-binding protein |
| 59 | 31 932~32 996 | 354 | + | ATG | DNA聚合酶Ⅲ β亚基 DNA polymerase Ⅲ subunit beta |
| 61 | 34 378~34 650 | 90 | + | ATG | Pyocin激活蛋白 PrtN Pyocin activator protein PrtN |
| 63 | 35 571~36 764 | 397 | + | ATG | 酪氨酸重组酶 Tyrosine recombinase |
| 64 | 36 838~37 005 | 55 | + | TTG | 酪氨酸重组酶 Tyrosine recombinase |
| 65 | 37 017~37 553 | 178 | - | ATG | Rz类裂解蛋白 Rz-like lysis protein |
| 67 | 37 929~38 309 | 126 | - | ATG | 趋化蛋白 Chemotaxis protein |
| 69 | 38 626~39 291 | 221 | - | TTG | 含DUF3380结构域蛋白 DUF3380 domain-containing protein |
| 72 | 40 455~46 457 | 2 000 | - | ATG | DarB类抗抑制蛋白 DarB-like antirestriction protein |
| 73 | 46 436~50 134 | 1 232 | - | GTG | DarB类抗抑制蛋白 DarB-like antirestriction protein |
| 74 | 50 192~51 736 | 514 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 75 | 51 733~52 458 | 241 | - | ATG | PAPS还原酶类蛋白 PAPS reductase-like protein |
| 76 | 52 470~53 303 | 277 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 77 | 53 293~53 850 | 185 | - | ATG | 酰基辅酶A N-酰基转移酶 Acyl-CoA N-acyltransferase |
| 79 | 54 246~55 979 | 577 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 80 | 55 991~56 977 | 328 | - | ATG | 尾纤蛋白 Tail fiber protein |
| 81 | 57 063~58 382 | 439 | - | ATG | 推定尾纤蛋白 Putative tail fiber protein |
| 82 | 58 382~58 663 | 93 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 84 | 60 366~61 343 | 325 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 85 | 61 350~61 772 | 140 | - | ATG | 病毒粒子相关噬菌体蛋白 Virion-associated phage protein |
| 88 | 62 505~63 167 | 220 | - | ATG | 病毒粒子相关蛋白 Virion associated protein |
| 90 | 63 713~64 150 | 145 | - | GTG | DarB类抗抑制蛋白 DarB-like antirestriction protein |
| 91 | 64 147~64 512 | 121 | - | GTG | 钼蝶呤结合蛋白 Molybdopterin-binding protein |
| 92 | 64 534~64 657 | 40 | - | TAT | 细胞色素B Cytochrome B |
| 1 | 乔俊卿,陈志谊,刘邮洲,等.茄科作物青枯病研究进展[J].植物病理学报, 2013, 43(1): 1-10. |
| QIAO J Q, CHEN Z Y, LIU Y Z, et al.. Research progress on bacterial wilt of nightshade family [J]. Acta Phytopathol. Sin., 2013, 43(1): 1-10. | |
| 2 | 黎妍妍,刘海龙,郑露,等.我国植物青枯菌遗传多样性研究进展[J].安徽农业科学, 2015, 43(14): 107-110, 112. |
| LI Y Y, LIU H L, ZHENG L, et al.. Research progress on genetic diversity of Ralstonia Solanacearum in China [J]. J. Anhui Agric. Sci., 2015, 43(14): 107-110, 112. | |
| 3 | 徐进,冯洁.植物青枯菌遗传多样性及致病基因组学研究进展[J].中国农业科学, 2013, 46(14): 2902-2909. |
| XU J, FENG J. Advances in research of genetic diversity and pathogenome of Ralstonia solanacearum species complex [J]. Sci. Agric. Sin., 2013, 46(14): 2902-2909. | |
| 4 | VOS M, BIRKETT P J, BIRCH E, et al.. Local adaptation of bacteriophages to their bacterial hosts in soil [J]. Science, 2009, 325(14): 833. |
| 5 | 苏靖芳,于浩,刘俊杰,等.青枯雷尔氏菌噬菌体研究进展[J].土壤与作物, 2017, 6(1): 61-66. |
| SU J F, YU H, LIU J J, et al.. Research progress of bacteriophages infecting Ralstonia solanacearum [J]. Soil Crop, 2017, 6(1): 61-66. | |
| 6 | 胡重怡,蔡刘体.噬菌体治疗作物细菌性病害的研究进展[J].贵州农业科学, 2011, 39(3): 101-103. |
| HU C Y, CAI L T. Research progress on phage therapy of bacterial plant disease [J]. Guizhou Agric. Sci., 2011, 39(3): 101-103. | |
| 7 | ADDY H S, AHMAD A A, QI H. Molecular and biological characterization of ralstonia phage RsoM1USA, a new species of P2virus, isolated in the United States [J/OL]. Front. Microbiol., 2019, 10:267 [2021-11-22]. . |
| 8 | WANG R, CONG Y, MI Z, et al.. Characterization and complete genome sequence analysis of phage GP4, a novel lytic Bcep22-like podovirus [J]. Archives Virol., 2019, 164(9): 2339-2343. |
| 9 | VAN TRUONG T B, KHANH N H P, NAMIKAWA R, et al.. Genomic characterization of Ralstonia solanacearum phage varphiRS138 of the family Siphoviridae [J]. Archives Virol., 2016, 161(2): 483-486. |
| 10 | AHMAD A A, STULBERG M J, MERSHON J P, et al.. Molecular and biological characterization of varphiRs 551, a filamentous bacteriophage isolated from a race 3 biovar 2 strain of Ralstonia solanacearum [J/OL]. PLoS One, 2017, 12(9): e0185034 [2021-11-22]. . |
| 11 | 林志坚,夏志辉,顾钢,等.繁殖青枯菌噬菌体无毒菌株的筛选及应用[J].中国生物防治学报, 2018, 34(6): 906-913. |
| LIN Z J, XIA Z H, GU G, et al.. Screening avirulent Ralstonia solanacearum strain to culture bacteriophage and its application [J]. Chin. J. Biol. Control, 2018, 34(6): 906-913. | |
| 12 | 林志坚, 吴秀琴, 梁颁捷,等.青枯病菌噬菌体P3株系的生物学特性及其应用研究[J]. 中国生物防治学报, 2020, 36(4): 611-618. |
| LIN Z J, WU X Q, LIANG B J, et al.. The biological characteristics of ralstonia phage P3 strain and its application [J]. Chin. J. Biol. Control, 2020, 36(4): 611-618. | |
| 13 | 方中达.植病研究方法[M]. 第3版.北京:中国农业出版社, 1998: 182-183, 224. |
| 14 | 高苗,杨金广,刘旭,等.一株裂解性青枯雷尔氏菌噬菌体的分离及生物学特性分析[J].中国农业科学, 2015, 48(7): 1330-1338. |
| GAO M, YANG J G, LIU X, et al.. Isolation and biological properties of a lytic phage infecting Ralstonia solanacearum [J]. Sci. Agric. Sin., 2015, 48(7): 1330-1338. | |
| 15 | 王新,吴新儒,王卫锋,等.烟草抗青枯病突变体的室内接种鉴定[J].分子植物育种,2018, 16(19): 6468-6475. |
| WANG X, WU X R, WANG W F, et al.. Indoor inoculation identification of tobacco mutants resistant to bacterial wilt [J]. Mol. Plant Breeding, 2018, 16(19): 6468-6475. | |
| 16 | SAMBROOK J, RUSSELL D W. Molecular Cloning a Laboratory Manual [M]. 3rd Ed n. Beijing: Science Press. 2002. |
| 17 | 苏靖芳,刘俊杰,于浩,等.一株烟草青枯雷尔氏菌烈性噬菌体RS-PII-1的分离及全基因组分析[J]. 病毒学报, 2017, 33(3): 441-449. |
| SU J F, YU H, LIU J J, et al.. Research progress of bacteriophages infecting Ralstonia solanacearum [J]. Chin. J. Virol., 2017, 33(3): 441-449. | |
| 18 | JIN J, LI Z J, WANG S W, et al.. Isolation and characterization of ZZ 1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates [J/OL]. BMC Microbiol., 2012, 12(1): 156 [2021-11-22]. . |
| 19 | BOLGER A M, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for Illumina sequence data [J]. Bioinformatics, 2014, 30(15): 2114-2120. |
| 20 | HAHN C, BACHMANN L, CHEVREUX B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach [J/OL]. Nucleic Acids Res., 2013, 41(13): e129 [2021-11-22]. . |
| 21 | HALL T A. BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT [J]. Nucleic Acids Symposium Series, 1999, 41(41): 95-98. |
| 22 | BESEMER J, LOMSADZE A, BORODOVSKY M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. implications for finding sequence motifs in regulatory regions [J]. Nucleic Acids Res., 2001, 29(12): 2607-2618. |
| 23 | STOTHARD P, WISHART D S. Circular genome visualization and exploration using CGView [J]. Bioinformatics, 2005, 21(4): 537-539. |
| 24 | LOWE T M, CHAN P P. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes [J]. Nucleic Acids Res., 2016, 44(W1): W54-W57. |
| 25 | REESE M G. Computational prediction of gene structure and regulation in the genome of Drosophila melanogaster [D]. Baden, Germany: University of Hohenheim, 2000. |
| 26 | HOFACKER I L, FONTANA W, STADLER P F, et al.. Fast folding and comparison of RNA secondary structures [J]. Monatshefte Für Chemie 1994, 125(2): 167-188. |
| 27 | SULLIVAN M J, PETTY N K, BEATSON S A. Easyfig: a genome comparison visualizer [J]. Bioinformatics, 2011, 27(7): 1009-1010. |
| 28 | ZHANG D, GAO F, JAKOVLIĆ I, et al.. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies [J]. Mol. Ecol. Resour., 2020, 20(1): 348-355. |
| 29 | NGUYEN L T, SCHMIDT H A, VON HAESELER A, et al.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies [J]. Mol. Biol. Evol., 2015, 32(1): 268-274. |
| 30 | MISAWA K, KATOH K, KUMA K I, et al.. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform [J]. Nucleic Acids Res., 2002, 30(14): 3059-3066. |
| 31 | BREITBART M, ROHWER F. Here a virus, there a virus, everywhere the same virus? [J]. Trends Microbiol., 2005, 13(6): 278-284. |
| 32 | TANAKA H, NEGISHI H, MAEDA H. Control of tobacco bacterial wilt by an avirulent strain of Pseudomonas solanacearum M4S and its bacteriophage [J]. Jap. J. Phytopathol. 1990, 56(2): 243-246. |
| 33 | WEI C, LIU J, MAINA A N, et al.. Developing a bacteriophage cocktail for biocontrol of potato bacterial wilt [J]. Virologica Sin., 2017, 32(6): 476-484. |
| 34 | 冯烨,刘军,孙洋,等.噬菌体最新分类与命名[J].中国兽医学报, 2013, 33(12): 1954-1958. |
| FENG Y, LIU J, SUN Y, et al.. An Introduction to current classification and nomenclature of bacterial viruses [J]. Chin. J. Veterin. Sci., 2013, 33(12): 1954-1958. | |
| 35 | SILVA XAVIER A DA, SILVA F P DA, VIDIGAL P M P, et al.. Genomic and biological characterization of a new member of the genus Phikmvvirus infecting phytopathogenic Ralstonia bacteria [J]. Archs Virol., 2018, 163(12): 3275-3290. |
| 36 | BHUNCHOTH A, BLANC-MATHIEU R, MIHARA T, et al.. Two asian jumbo phages, ϕRSL2 and ϕRSF1, infect Ralstonia solanacearum and show common features of ϕKZ-related phages [J]. Virology, 2016, 494: 56-66. |
| [1] | Hui JIN, Wei WANG, Chendong YAN, Wei WANG, Xiying LI. Isolation, Identification and Adaptability of Trichoderma spp. for Biocontrol of Rice Sheath Blight [J]. Journal of Agricultural Science and Technology, 2022, 24(9): 139-148. |
| [2] |
ZHAO Xingli1, TAO Gang2,3*, LOU Xuan4, GU Jingang5*.
Colonization Dynamics of Trichoderma hamatum in Pepper Rhizosphere and Its Biological Control Against Pepper Phytophthora Blight
[J]. Journal of Agricultural Science and Technology, 2020, 22(5): 106-114.
|
| [3] | ZHOU Hongzi1, ZHOU Fangyuan1, ZHAO Xiaoyan1, WU Cuixia2, ZHANG Guangzhi1, YUAN Weiwei3, WU Xiaoqing1, XIE Xueying1, FAN Susu1, ZHANG Xinjian1*. Screening of Biocontrol Agents Against Wheat Fusarium Head Blight and Its Field Control Experiment [J]. Journal of Agricultural Science and Technology, 2020, 22(1): 67-77. |
| [4] | LU Lu1,2, ZHANG Mengli2, DI Yilin2, ZHU Kai1*, SHI Baojun2*. Insecticidal Effects of Thymol Against Caenorhabditis elegans at Different Stages [J]. Journal of Agricultural Science and Technology, 2019, 21(9): 97-103. |
| [5] | WU Xiaoqing1, ZHAO Xiaoyan1, XU Yuanzhang2, WANG Jianing1, ZHOU Fangyuan1, ZHOU Hongzi1, ZHANG Guangzhi1, XIE Xueying1, YAN Kun3, ZHANG Xinjian1*. Research Progress on Precision Application Technology of Biological Control [J]. Journal of Agricultural Science and Technology, 2019, 21(3): 13-21. |
| [6] | CHENG Liang1,2, GUO Qing\|yun1,2*. Potential Research of Fusarium avenaceum Isolate GD\|2 as a Bioherbicide Agent for Wild Oats(Avena fatua L.) [J]. , 2014, 16(3): 70-80. |
| [7] | QIU De-wen. Development Strategy for Bio-pesticide and Biological Control [J]. , 2011, 13(5): 88-92. |
| [8] | YAN Pei-Sheng, CAO Li-Xin, WANG Kai, WANG Zhuo. Research Progress on Biological Control of Mycotoxin Contamination [J]. , 2008, 10(6): 89-94. |
| [9] | SONG Xiao-yan, SUN Cai-yun, CHEN Xiu-lan, ZHANG Yu-zhong. Research Advances on Mechanism of Trichoderma in Biological Control [J]. , 2006, 8(6): 20-25. |
| [10] | JIANG Pei-zeng, LI Hong-yuan, CHEN Tie-bao . Advance in Paecilmyces Lilacinus Research for |Plant Parasitic Nematodes Control [J]. , 2006, 8(6): 38-41. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号