




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (12): 18-29.DOI: 10.13304/j.nykjdb.2024.0895
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Haixia LIU( ), Yinhui ZHANG, Lei ZHUANG, Mengjiao GUO, Li ZHAO, Meijuan WU, Jian HOU, Tian LI, Hongxia LIU, Xueyong ZHANG, Chenyang HAO(
), Yinhui ZHANG, Lei ZHUANG, Mengjiao GUO, Li ZHAO, Meijuan WU, Jian HOU, Tian LI, Hongxia LIU, Xueyong ZHANG, Chenyang HAO( )
)
Received:2024-10-31
Accepted:2024-11-15
Online:2024-12-15
Published:2024-12-17
Contact:
Chenyang HAO
刘海霞( ), 张寅辉, 庄蕾, 郭梦娇, 赵李, 吴美娟, 侯健, 李甜, 刘红霞, 张学勇, 郝晨阳(
), 张寅辉, 庄蕾, 郭梦娇, 赵李, 吴美娟, 侯健, 李甜, 刘红霞, 张学勇, 郝晨阳( )
)
通讯作者:
郝晨阳
作者简介:刘海霞E-mail: 82101222231@caas.cn;
基金资助:CLC Number:
Haixia LIU, Yinhui ZHANG, Lei ZHUANG, Mengjiao GUO, Li ZHAO, Meijuan WU, Jian HOU, Tian LI, Hongxia LIU, Xueyong ZHANG, Chenyang HAO. Discovering of Candidate Genes for Wheat SDS-Sedimentation Value Using Association Study and Development of KASP Marker[J]. Journal of Agricultural Science and Technology, 2024, 26(12): 18-29.
刘海霞, 张寅辉, 庄蕾, 郭梦娇, 赵李, 吴美娟, 侯健, 李甜, 刘红霞, 张学勇, 郝晨阳. 基于关联分析挖掘小麦SDS沉降值相关候选基因及KASP标记开发[J]. 中国农业科技导报, 2024, 26(12): 18-29.
| 标记 Marker | 引物 Primer | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| Sv-3D-KASP | F1-FAM (1): Allele-T | GAAGGTGACCAAGTTCATGCTCGGGTATCGATAGCAGAAGTCTTT |
| F2-HEX (2): Allele-C | GAAGGTCGGAGTCAACGGATTCGGGTATCGATAGCAGAAGTCTTC | |
| Common-primer-R (3) | TGACACTCCAGATCCCCTTTTT | |
| Sv-3D-dCAPS | dCAPS-F1 (1) | GATGGCACTGTCCCTTGG |
| dCAPS-R1 (2) | GGTAGAGTTTGTTGGATTGATT | |
| dCAPS-F2 (3) | ATCGGGTATCGATAGCAGAAGTCGA | |
| dCAPS-R2 (4) | GTCAAGCGACCCTTATGTTAT |
Table 1 Primer sequences of molecular markers developed based on significant associated loci of candidate gene
| 标记 Marker | 引物 Primer | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| Sv-3D-KASP | F1-FAM (1): Allele-T | GAAGGTGACCAAGTTCATGCTCGGGTATCGATAGCAGAAGTCTTT |
| F2-HEX (2): Allele-C | GAAGGTCGGAGTCAACGGATTCGGGTATCGATAGCAGAAGTCTTC | |
| Common-primer-R (3) | TGACACTCCAGATCCCCTTTTT | |
| Sv-3D-dCAPS | dCAPS-F1 (1) | GATGGCACTGTCCCTTGG |
| dCAPS-R1 (2) | GGTAGAGTTTGTTGGATTGATT | |
| dCAPS-F2 (3) | ATCGGGTATCGATAGCAGAAGTCGA | |
| dCAPS-R2 (4) | GTCAAGCGACCCTTATGTTAT |
| 环境Environment | 均值±标准差Mean±SD/mL | 最小值 Min/mL | 最大值 Max/mL | 变异系数 CV/% | 峰度 Kurtosis | 偏度 Skewness | 广义遗传力H2 /% |
|---|---|---|---|---|---|---|---|
| E1 | 18.44±2.98 | 11.30 | 25.40 | 16.16 | -0.08 | 0.10 | 94.10 |
| E2 | 20.81±3.62 | 12.10 | 29.00 | 17.38 | -0.30 | 0.18 | |
| E3 | 20.40±3.11 | 12.50 | 28.50 | 15.25 | -0.41 | -0.02 | |
| E4 | 22.74±4.39 | 13.00 | 34.10 | 19.30 | -0.25 | 0.29 | |
| E5 | 20.74±3.88 | 11.00 | 30.10 | 18.72 | -0.28 | 0.27 | |
| E6 | 19.80±3.21 | 13.00 | 27.10 | 16.19 | -0.42 | 0.04 | |
| E7 | 20.65±3.43 | 11.00 | 29.90 | 16.62 | -0.21 | 0.01 | |
| BLUP | 20.20±2.86 | 12.64 | 26.08 | 14.15 | -0.42 | -0.05 |
Table 2 Basic descriptive statistics of SDS-sedimentation value in multiple environments
| 环境Environment | 均值±标准差Mean±SD/mL | 最小值 Min/mL | 最大值 Max/mL | 变异系数 CV/% | 峰度 Kurtosis | 偏度 Skewness | 广义遗传力H2 /% |
|---|---|---|---|---|---|---|---|
| E1 | 18.44±2.98 | 11.30 | 25.40 | 16.16 | -0.08 | 0.10 | 94.10 |
| E2 | 20.81±3.62 | 12.10 | 29.00 | 17.38 | -0.30 | 0.18 | |
| E3 | 20.40±3.11 | 12.50 | 28.50 | 15.25 | -0.41 | -0.02 | |
| E4 | 22.74±4.39 | 13.00 | 34.10 | 19.30 | -0.25 | 0.29 | |
| E5 | 20.74±3.88 | 11.00 | 30.10 | 18.72 | -0.28 | 0.27 | |
| E6 | 19.80±3.21 | 13.00 | 27.10 | 16.19 | -0.42 | 0.04 | |
| E7 | 20.65±3.43 | 11.00 | 29.90 | 16.62 | -0.21 | 0.01 | |
| BLUP | 20.20±2.86 | 12.64 | 26.08 | 14.15 | -0.42 | -0.05 |

Fig. 1 Correlation analysis and distribution histogram of SDS-sedimentation value in multiple environmentsNote:*** indicates significant correlation at P<0.001 level.

Fig. 2 Candidate region on chromosome 3DL for SDS-sedimentation value identified by genome-wide association studyA: Manhattan plot of SDS-sedimentation value at the end of 3DL in multiple environments, and the dashed line indicates the threshold -log10(P)=6; B: Heat map of linkage disequilibrium in the interval from 587.523 to 589.514 Mb, and the black vertical lines represent the relative position of candidate genes; C: The expression pattern of the 28 candidate genes based on the expVIP database (https://www.wheat-expression.com/)
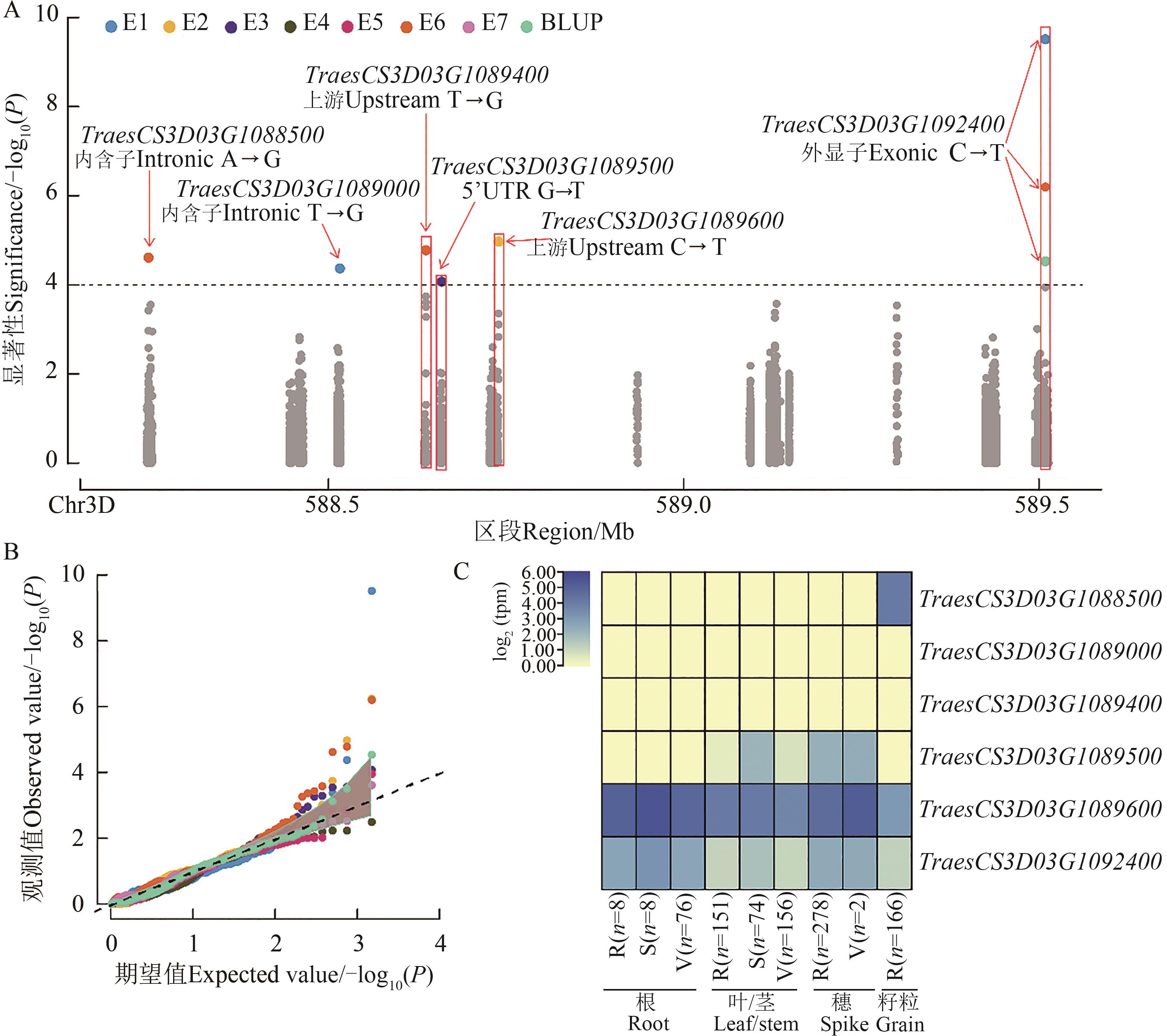
Fig. 3 Candidate gene association study on SDS-sedimentation value within the end of chromosome 3DL regionA and B are Manhattan plot and Q-Q plot of CGAS within a 3DL region in multiple environments, respectively; C is the expression patterns of 6 associated genes based on the expVIP database, R, S and V represent reproductive, seedling and vegetative, respectively
位置 Position/bp | P值 P value | 变异 类型 Variant type | 基因ID Gene ID | 参考碱基 REF SNP | 突变碱基 ALT SNP | 基因注释 Gene annotation |
|---|---|---|---|---|---|---|
| 589 508 971 | 9.51,6.23,6.19,4.52 | 外显子 Exonic | TraesCS3D03G1092400 | C | T | ARF-GTP 酶激活蛋白Arf GTPase activating protein |
| 588 739 600 | 4.96 | 上游 Upstream | TraesCS3D03G1089600 | C | T | 绒毛蛋白Villin |
| 588 637 344 | 4.78 | 上游 Upstream | TraesCS3D03G1089400 | T | G | 羟基肉桂酰-CoA莽草酸/奎宁酸羟基肉桂酰转移酶 Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase |
| 588 246 926 | 4.61 | 内含子 Intronic | TraesCS3D03G1088500 | A | G | 非特异性丝氨酸/苏氨酸蛋白激酶 Non-specific serine/threonine protein kinase |
| 588 516 258 | 4.37 | 内含子 Intronic | TraesCS3D03G1089000 | T | G | 羧基末端肽酶carboxyl-terminal peptidase |
| 588 659 614 | 4.07 | 5’-UTR | TraesCS3D03G1089500 | G | T | 葡萄糖醛酸氧酶4-O-甲基转移酶 Glucuronoxylan 4-O-methyltransferase |
Table 3 CGAS results of candidate genes for SDS-sedimentation value within the end of chromosome 3DL region
位置 Position/bp | P值 P value | 变异 类型 Variant type | 基因ID Gene ID | 参考碱基 REF SNP | 突变碱基 ALT SNP | 基因注释 Gene annotation |
|---|---|---|---|---|---|---|
| 589 508 971 | 9.51,6.23,6.19,4.52 | 外显子 Exonic | TraesCS3D03G1092400 | C | T | ARF-GTP 酶激活蛋白Arf GTPase activating protein |
| 588 739 600 | 4.96 | 上游 Upstream | TraesCS3D03G1089600 | C | T | 绒毛蛋白Villin |
| 588 637 344 | 4.78 | 上游 Upstream | TraesCS3D03G1089400 | T | G | 羟基肉桂酰-CoA莽草酸/奎宁酸羟基肉桂酰转移酶 Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase |
| 588 246 926 | 4.61 | 内含子 Intronic | TraesCS3D03G1088500 | A | G | 非特异性丝氨酸/苏氨酸蛋白激酶 Non-specific serine/threonine protein kinase |
| 588 516 258 | 4.37 | 内含子 Intronic | TraesCS3D03G1089000 | T | G | 羧基末端肽酶carboxyl-terminal peptidase |
| 588 659 614 | 4.07 | 5’-UTR | TraesCS3D03G1089500 | G | T | 葡萄糖醛酸氧酶4-O-甲基转移酶 Glucuronoxylan 4-O-methyltransferase |
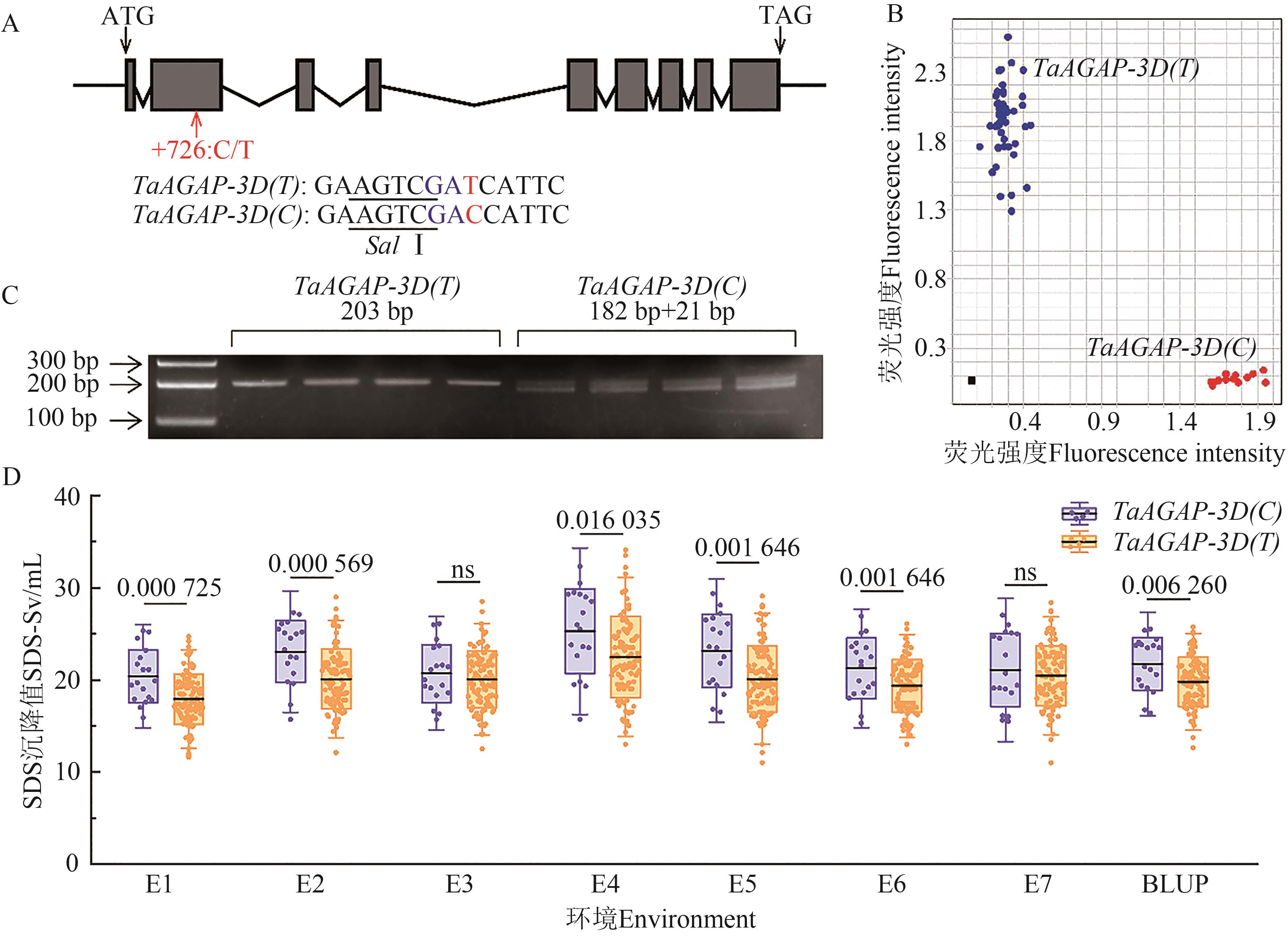
Fig. 4 Development and validation of KASP markers of TaAGAP-3D for SDS-sedimentation valueA: Gene structure and variations of TaAGAP-3D, the red letters and arrow indicate the significantly associated SNP site located at +726 bp, blue letters represent introduced variant loci, and the underline indicates the Sal Ⅰendonuclease site; B: Genotyping of KASP marker Sv-3D-KASP, blue dot represents the TaAGAP-3D(T) type material, red dot represents the TaAGAP-3D(C) type material, and black dots represent negative controls; C: Enzyme digestion results of dCAPS marker Sv-3D-dCAPS; D: Significance of difference analysis of 2 alleles of TaAGAP-3D with SDS-sedimentation value in multiple environments, ns indicates not significant
| 编号 Code | 材料 Material | KASP | dCAPS | 编号 Code | 材料 Material | KASP | dCAPS |
|---|---|---|---|---|---|---|---|
| 1 | 甘肃96 CI 12203 | TT | 203 bp | 27 | 豫麦2号 Yumai 2 | CC | 182 bp |
| 2 | 碧蚂1号 Bima 1 | TT | 203 bp | 28 | 扬麦5号 Yangmai 5 | TT | 203 bp |
| 3 | 石家庄54 Shijiazhuang 54 | TT | 203 bp | 29 | 济南13 Jinan 13 | TT | 203 bp |
| 4 | 济南2号 Jinan 2 | TT | 203 bp | 30 | 冀麦26 Jimai 26 | TT | 203 bp |
| 5 | 内乡5号 Neixiang 5 | TT | 203 bp | 31 | 鄂恩1号 Een 1 | TT | 203 bp |
| 6 | 博爱7023 Boai 7023 | TT | 203 bp | 32 | 鲁麦21 Lumai 21 | TT | 203 bp |
| 7 | 济南9号 Jinan 9 | TT | 203 bp | 33 | 烟农19 Yannong 19 | TT | 203 bp |
| 8 | 徐州14 Xuzhou 14 | TT | 203 bp | 34 | 济南16 Jinan 16 | TT | 203 bp |
| 9 | 郑引1号 St 1472/506 | CC | 182 bp | 35 | 济麦20 Jimai 20 | TT | 203 bp |
| 10 | 丰产3号 Fengchan 3 | TT | 203 bp | 36 | 扬麦158 Yangmai 158 | TT | 203 bp |
| 11 | 泰山1号 Taishan 1 | CC | 182 bp | 37 | 济南17 Jinan 17 | TT | 203 bp |
| 12 | 甘麦8号 Ganmai 8 | TT | 203 bp | 38 | 温麦6号 Wenmai 6 | TT | 203 bp |
| 13 | 鲁麦14 Lumai 14 | TT | 203 bp | 39 | 宁春4号 Ningchun 4 | TT | 203 bp |
| 14 | 冀麦30 Jimai 30 | TT | 203 bp | 40 | 周麦9号 Zhoumai 9 | TT | 203 bp |
| 15 | 豫麦13 Yumai 13 | TT | 203 bp | 41 | 豫麦18 Yumai 18 | TT | 203 bp |
| 16 | 豫麦7号 Yumai 7 | TT | 203 bp | 42 | 豫麦41 Yumai 41 | TT | 203 bp |
| 17 | 西安8号 Xi’an 8 | TT | 203 bp | 43 | 绵阳26号 Mianyang 26 | TT | 203 bp |
| 18 | 豫麦17 Yumai 17 | TT | 203 bp | 44 | 高优503 Gaoyou 503 | TT | 203 bp |
| 19 | 绵阳11 Mianyang 11 | TT | 203 bp | 45 | 冀麦38 Jimai 38 | TT | 203 bp |
| 20 | 绵阳15 Mianyang 15 | TT | 203 bp | 46 | 济麦19 Jimai 19 | TT | 203 bp |
| 21 | 绵阳20 Mianyang 20 | TT | 203 bp | 47 | 郑麦366 Zhengmai 366 | CC | 182 bp |
| 22 | 山农辐63 Shannongfu 63 | TT | 203 bp | 48 | 邯郸6172 Handan 6172 | TT | 203 bp |
| 23 | 川麦22 Chuanmai 22 | TT | 203 bp | 49 | 周麦18 Zhoumai 18 | TT | 203 bp |
| 24 | 新克旱9号 Xinkehan 9 | TT | 203 bp | 50 | 济麦22 Jimai 22 | TT | 203 bp |
| 25 | 百农3217 Bainong 3217 | TT | 203 bp | 51 | 石4185 Shi 4185 | TT | 203 bp |
| 26 | 陕农7859 Shaannong 7859 | TT | 203 bp | 52 | 郑麦9023 Zhengmai 9023 | CC | 182 bp |
Table 4 Genotyping data of KASP and dCAPS markers in 116 wheat materials used in this study
| 编号 Code | 材料 Material | KASP | dCAPS | 编号 Code | 材料 Material | KASP | dCAPS |
|---|---|---|---|---|---|---|---|
| 1 | 甘肃96 CI 12203 | TT | 203 bp | 27 | 豫麦2号 Yumai 2 | CC | 182 bp |
| 2 | 碧蚂1号 Bima 1 | TT | 203 bp | 28 | 扬麦5号 Yangmai 5 | TT | 203 bp |
| 3 | 石家庄54 Shijiazhuang 54 | TT | 203 bp | 29 | 济南13 Jinan 13 | TT | 203 bp |
| 4 | 济南2号 Jinan 2 | TT | 203 bp | 30 | 冀麦26 Jimai 26 | TT | 203 bp |
| 5 | 内乡5号 Neixiang 5 | TT | 203 bp | 31 | 鄂恩1号 Een 1 | TT | 203 bp |
| 6 | 博爱7023 Boai 7023 | TT | 203 bp | 32 | 鲁麦21 Lumai 21 | TT | 203 bp |
| 7 | 济南9号 Jinan 9 | TT | 203 bp | 33 | 烟农19 Yannong 19 | TT | 203 bp |
| 8 | 徐州14 Xuzhou 14 | TT | 203 bp | 34 | 济南16 Jinan 16 | TT | 203 bp |
| 9 | 郑引1号 St 1472/506 | CC | 182 bp | 35 | 济麦20 Jimai 20 | TT | 203 bp |
| 10 | 丰产3号 Fengchan 3 | TT | 203 bp | 36 | 扬麦158 Yangmai 158 | TT | 203 bp |
| 11 | 泰山1号 Taishan 1 | CC | 182 bp | 37 | 济南17 Jinan 17 | TT | 203 bp |
| 12 | 甘麦8号 Ganmai 8 | TT | 203 bp | 38 | 温麦6号 Wenmai 6 | TT | 203 bp |
| 13 | 鲁麦14 Lumai 14 | TT | 203 bp | 39 | 宁春4号 Ningchun 4 | TT | 203 bp |
| 14 | 冀麦30 Jimai 30 | TT | 203 bp | 40 | 周麦9号 Zhoumai 9 | TT | 203 bp |
| 15 | 豫麦13 Yumai 13 | TT | 203 bp | 41 | 豫麦18 Yumai 18 | TT | 203 bp |
| 16 | 豫麦7号 Yumai 7 | TT | 203 bp | 42 | 豫麦41 Yumai 41 | TT | 203 bp |
| 17 | 西安8号 Xi’an 8 | TT | 203 bp | 43 | 绵阳26号 Mianyang 26 | TT | 203 bp |
| 18 | 豫麦17 Yumai 17 | TT | 203 bp | 44 | 高优503 Gaoyou 503 | TT | 203 bp |
| 19 | 绵阳11 Mianyang 11 | TT | 203 bp | 45 | 冀麦38 Jimai 38 | TT | 203 bp |
| 20 | 绵阳15 Mianyang 15 | TT | 203 bp | 46 | 济麦19 Jimai 19 | TT | 203 bp |
| 21 | 绵阳20 Mianyang 20 | TT | 203 bp | 47 | 郑麦366 Zhengmai 366 | CC | 182 bp |
| 22 | 山农辐63 Shannongfu 63 | TT | 203 bp | 48 | 邯郸6172 Handan 6172 | TT | 203 bp |
| 23 | 川麦22 Chuanmai 22 | TT | 203 bp | 49 | 周麦18 Zhoumai 18 | TT | 203 bp |
| 24 | 新克旱9号 Xinkehan 9 | TT | 203 bp | 50 | 济麦22 Jimai 22 | TT | 203 bp |
| 25 | 百农3217 Bainong 3217 | TT | 203 bp | 51 | 石4185 Shi 4185 | TT | 203 bp |
| 26 | 陕农7859 Shaannong 7859 | TT | 203 bp | 52 | 郑麦9023 Zhengmai 9023 | CC | 182 bp |
| 编号 Code | 材料 Material | KASP | dCAPS | 编号 Code | 材料 Material | KASP | dCAPS |
|---|---|---|---|---|---|---|---|
| 53 | 淮麦20 Huaimai 20 | TT | 203 bp | 79 | 中国春 Chinese Spring | CC | 182 bp |
| 54 | 衡观35 Hengguan 35 | TT | 203 bp | 80 | 望水白 Wangshuibai | TT | 203 bp |
| 55 | 周麦22号 Zhoumai 22 | TT | 203 bp | 81 | 府麦 Fumai | TT | 182 bp |
| 56 | 百农AK58 Bainong AK 58 | TT | 203 bp | 82 | 丰德存麦5号 Fengdecunmai 5 | TT | 203 bp |
| 57 | 周麦16 Zhoumai 16 | TT | 203 bp | 83 | 川麦42 Chuanmai 42 | TT | 203 bp |
| 58 | 燕大1817 Yanda 1817 | TT | 203 bp | 84 | 师栾02-1 Shiluan 02-1 | TT | 182 bp |
| 59 | 五一麦 Wuyimai | TT | 203 bp | 85 | 良星66 Liangxing 66 | TT | 203 bp |
| 60 | 江东门 Jiangdongmen | CC | 182 bp | 86 | 良星99 Liangxing 99 | TT | 203 bp |
| 61 | 阿夫 Funo | TT | 203 bp | 87 | 烟农21号 Yannong 21 | TT | 203 bp |
| 62 | 南大2419 Mentana | TT | 203 bp | 88 | 新麦26 Xinmai 26 | TT | 203 bp |
| 63 | 早洋麦 Early Premium | TT | 203 bp | 89 | 百农4199 Bainong 4199 | TT | 203 bp |
| 64 | 碧蚂4号 Bima 4 | TT | 203 bp | 90 | 徐麦856 Xumai 856 | TT | 203 bp |
| 65 | 蚂蚱麦 Mazhamai | CC | 182 bp | 91 | 徐麦35 Xumai 35 | TT | 203 bp |
| 66 | 成都光头 Chengduguangtou | CC | 182 bp | 92 | 偃展4110 Yanzhan 4110 | TT | 203 bp |
| 67 | 北京8号 Beijing 8 | TT | 203 bp | 93 | 轮选987 Lunxuan 987 | TT | 203 bp |
| 68 | 欧柔 Orofen | CC | 182 bp | 94 | 小偃22 Xiaoyan 22 | TT | 203 bp |
| 69 | 阿勃 Abbondanza | TT | 203 bp | 95 | 石家庄8号 Shijiazhuang 8 | TT | 203 bp |
| 70 | 郑引4号 St 2422/464 | CC | 182 bp | 96 | 劳改大青芒 Laogaidaqingmang | TT | 203 bp |
| 71 | 洛夫林10号 Lovrin 10 | TT | 203 bp | 97 | 蔷薇麦 Qiangweimai | CC | 182 bp |
| 72 | 繁6 Fan 6 | TT | 203 bp | 98 | 呷托 Gatuo | TT | 203 bp |
| 73 | 墨巴66 Mexipak 66 | TT | 203 bp | 99 | 白山麦 Baishanmai | CC | 182 bp |
| 74 | 小偃6号 Xiaoyan 6 | CC | 182 bp | 100 | 蚰子头 Youzitou | CC | 182 bp |
| 75 | 周8425B Zhou 8425B | TT | 203 bp | 101 | 光头白糁麦 Guangtoubaishenmai | TT | 182 bp |
| 76 | 碧玉麦 Quality | TT | 203 bp | 102 | 蝉不吱 Chanbuzhi | CC | 182 bp |
| 77 | 威尔赫明那 Wilhelmina | TT | 203 bp | 103 | 紫秸红 Zijiehong | TT | 203 bp |
| 78 | 赤小麦 Akagomughi | TT | 203 bp | 104 | 尕老汉 Galaohan | TT | 203 bp |
Table 4 Genotyping data of KASP and dCAPS markers in 116 wheat materials used in this study
| 编号 Code | 材料 Material | KASP | dCAPS | 编号 Code | 材料 Material | KASP | dCAPS |
|---|---|---|---|---|---|---|---|
| 53 | 淮麦20 Huaimai 20 | TT | 203 bp | 79 | 中国春 Chinese Spring | CC | 182 bp |
| 54 | 衡观35 Hengguan 35 | TT | 203 bp | 80 | 望水白 Wangshuibai | TT | 203 bp |
| 55 | 周麦22号 Zhoumai 22 | TT | 203 bp | 81 | 府麦 Fumai | TT | 182 bp |
| 56 | 百农AK58 Bainong AK 58 | TT | 203 bp | 82 | 丰德存麦5号 Fengdecunmai 5 | TT | 203 bp |
| 57 | 周麦16 Zhoumai 16 | TT | 203 bp | 83 | 川麦42 Chuanmai 42 | TT | 203 bp |
| 58 | 燕大1817 Yanda 1817 | TT | 203 bp | 84 | 师栾02-1 Shiluan 02-1 | TT | 182 bp |
| 59 | 五一麦 Wuyimai | TT | 203 bp | 85 | 良星66 Liangxing 66 | TT | 203 bp |
| 60 | 江东门 Jiangdongmen | CC | 182 bp | 86 | 良星99 Liangxing 99 | TT | 203 bp |
| 61 | 阿夫 Funo | TT | 203 bp | 87 | 烟农21号 Yannong 21 | TT | 203 bp |
| 62 | 南大2419 Mentana | TT | 203 bp | 88 | 新麦26 Xinmai 26 | TT | 203 bp |
| 63 | 早洋麦 Early Premium | TT | 203 bp | 89 | 百农4199 Bainong 4199 | TT | 203 bp |
| 64 | 碧蚂4号 Bima 4 | TT | 203 bp | 90 | 徐麦856 Xumai 856 | TT | 203 bp |
| 65 | 蚂蚱麦 Mazhamai | CC | 182 bp | 91 | 徐麦35 Xumai 35 | TT | 203 bp |
| 66 | 成都光头 Chengduguangtou | CC | 182 bp | 92 | 偃展4110 Yanzhan 4110 | TT | 203 bp |
| 67 | 北京8号 Beijing 8 | TT | 203 bp | 93 | 轮选987 Lunxuan 987 | TT | 203 bp |
| 68 | 欧柔 Orofen | CC | 182 bp | 94 | 小偃22 Xiaoyan 22 | TT | 203 bp |
| 69 | 阿勃 Abbondanza | TT | 203 bp | 95 | 石家庄8号 Shijiazhuang 8 | TT | 203 bp |
| 70 | 郑引4号 St 2422/464 | CC | 182 bp | 96 | 劳改大青芒 Laogaidaqingmang | TT | 203 bp |
| 71 | 洛夫林10号 Lovrin 10 | TT | 203 bp | 97 | 蔷薇麦 Qiangweimai | CC | 182 bp |
| 72 | 繁6 Fan 6 | TT | 203 bp | 98 | 呷托 Gatuo | TT | 203 bp |
| 73 | 墨巴66 Mexipak 66 | TT | 203 bp | 99 | 白山麦 Baishanmai | CC | 182 bp |
| 74 | 小偃6号 Xiaoyan 6 | CC | 182 bp | 100 | 蚰子头 Youzitou | CC | 182 bp |
| 75 | 周8425B Zhou 8425B | TT | 203 bp | 101 | 光头白糁麦 Guangtoubaishenmai | TT | 182 bp |
| 76 | 碧玉麦 Quality | TT | 203 bp | 102 | 蝉不吱 Chanbuzhi | CC | 182 bp |
| 77 | 威尔赫明那 Wilhelmina | TT | 203 bp | 103 | 紫秸红 Zijiehong | TT | 203 bp |
| 78 | 赤小麦 Akagomughi | TT | 203 bp | 104 | 尕老汉 Galaohan | TT | 203 bp |
| 编号 Code | 材料 Material | KASP | dCAPS | 编号 Code | 材料 Material | KASP | dCAPS |
|---|---|---|---|---|---|---|---|
| 105 | 火里炎 Huoliyan | TT | 203 bp | 111 | 奥托诺米亚 Autonomia | TT | 203 bp |
| 106 | 白花麦 Baihuamai | TT | 203 bp | 112 | 科春14 Kechun 14 | TT | 203 bp |
| 107 | 跃进5号 Yuejin 5 | TT | 203 bp | 113 | 小偃96 Xiaoyan 96 | TT | 203 bp |
| 108 | 济宁3号 Jining 3 | TT | 203 bp | 114 | 百农791 Bainong 791 | TT | 203 bp |
| 109 | 偃师4号 Yanshi 4 | TT | 203 bp | 115 | 丰塔隆科 Fontarronco | TT | 203 bp |
| 110 | 豫麦47 Yumai 47 | CC | 182 bp | 116 | 孟县201 Mengxian 201 | CC | 182 bp |
Table 4 Genotyping data of KASP and dCAPS markers in 116 wheat materials used in this study
| 编号 Code | 材料 Material | KASP | dCAPS | 编号 Code | 材料 Material | KASP | dCAPS |
|---|---|---|---|---|---|---|---|
| 105 | 火里炎 Huoliyan | TT | 203 bp | 111 | 奥托诺米亚 Autonomia | TT | 203 bp |
| 106 | 白花麦 Baihuamai | TT | 203 bp | 112 | 科春14 Kechun 14 | TT | 203 bp |
| 107 | 跃进5号 Yuejin 5 | TT | 203 bp | 113 | 小偃96 Xiaoyan 96 | TT | 203 bp |
| 108 | 济宁3号 Jining 3 | TT | 203 bp | 114 | 百农791 Bainong 791 | TT | 203 bp |
| 109 | 偃师4号 Yanshi 4 | TT | 203 bp | 115 | 丰塔隆科 Fontarronco | TT | 203 bp |
| 110 | 豫麦47 Yumai 47 | CC | 182 bp | 116 | 孟县201 Mengxian 201 | CC | 182 bp |
| 1 | LIU Y X, LIN Y, GAO S, et al.. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces [J]. Plant J., 2017,91(5):861-873. |
| 2 | 范金平,吕国锋,张伯桥,等.SDS沉降值在小麦良种繁育中的应用[J].安徽农业科学, 2003(6):1043-1044. |
| FAN J P, LYU G F, ZHANG B Q, et al.. Application of SDS sedimentation value in wheat breeding [J]. J. Anhui Agric. Sci., 2003(6):1043-1044. | |
| 3 | 王峰.小麦品质性状的遗传变异和诱变研究[D].合肥:安徽农业大学,2013. |
| WANG F. Studies on genetic variation and mutation of quality traits in wheat [D]. Hefei: Anhui Agricultural University, 2013. | |
| 4 | 夏云祥,陈辉,司红起,等. 小麦微核心种质中育成品种和地方品种的SDS沉降值分析[J]. 粮食与饲料工业,2011(10):12-14. |
| XIA Y X, CHEN H, SI H Q,et al.. Analysis of SDS-sedimentation volume in bred and local varieties of Chinese wheat micro-core collections [J]. Cereal Feed. Ind., 2011(10):12-14. | |
| 5 | 马传喜,徐风,汪心乐.影响SDS沉降值的试验因素分析[J].安徽农业大学学报,1995, 22 (1):1-6. |
| MA C X, XU F. WANG X L. Analysis of experimental factors affecting SDS sedimentation value [J]. J. Anhui Agric. Univ., 1995,22(1) :1-6. | |
| 6 | FLINT-GARCIA S A, THORNSBERRY J M, BUCKLER E S. Structure of linkage disequilibrium in plants [J]. Annu. Rev. Plant Biol., 2003,54:357-374. |
| 7 | ZHU C S, GORE M, BUCKLER E S, et al.. Status and prospects of association mapping in plants [J]. Plant Genome, 2008, 1(1): 5-20. |
| 8 | 席甜甜,吴倩,杨建光,等. 337份小麦品种籽粒相关性状的全基因组关联分析[J]. 麦类作物学报,2024,44(5):547-558. |
| XI T T, WU Q, YANG J G, et al.. Genome-wide association studies of 337 wheat varieties for grain-related traits [J]. J. Triticeae Crops, 2024,44(5):547-558. | |
| 9 | 董一帆,任毅,程宇坤,等. 冬小麦籽粒主要品质性状的全基因组关联分析[J]. 中国农业科学,2023,56(11):2047-2063. |
| DONG Y F, REN Y, CHENG Y K, et al.. Genome-wide association study of grain main quality related traits in winter wheat [J]. Sci. Agric. Sin., 2023,56(11):2047-2063. | |
| 10 | 王荣焕,王天宇,黎裕.关联分析在作物种质资源分子评价中的应用[J].植物遗传资源学报,2007,8(3):366-372. |
| WANG R H, WANG T Y, LI Y. Application of association analysis in molecular evaluation of crop germplasm resources [J]. J. Plant Genet. Resour., 2007,8(3):366-372. | |
| 11 | EHRENREICH I M, HANZAWA Y, CHOU L, et al.. Candidate gene association mapping of Arabidopsis flowering time [J]. Genetics, 2009,183(1):325-335. |
| 12 | WILSON L M, WHITT S R, IBANEZ A M, et al. Dissection of maize kernel composition and starch production by candidate gene association [J]. Plant Cell, 2004, 16(10): 2719-2733. |
| 13 | HAO C Y, JIAO C Z, HOU J, et al.. Resequencing of 145 landmark cultivars reveals asymmetric sub-genome selection and strong founder genotype effects on wheat breeding in China [J]. Mol. Plant, 2020,13(12):1733-1751. |
| 14 | 高修吾,杨浩然,吴艳霞,等. 粮食、油料检验 水分测定法: [S].北京:中国标准出版社,1985. |
| 15 | BATES D, MACHLER M, BOLKER B, et al.. Fitting linear mixed-effects models using lme4 [J]. J. Stat. Software, 2015, 67(1):1-48. |
| 16 | ZHOU X, STEPHENS M. Genome-wide efficient mixed-model analysis for association studies [J]. Nat. Genet., 2012,44(7):821-824. |
| 17 | DONG S S, HE W M, JI J J, et al.. LDBlockShow:a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files [J/OL]. Brief. Bioinform., 2021,22(4):bbaa227 [2024-11-05]. . |
| 18 | WANG J B, ZHANG Z W. GAPIT version 3:boosting power and accuracy for genomic association and prediction [J]. Genom. Proteom. Bioinform., 2021,19(4):629-640. |
| 19 | HUANG M, LIU X L, ZHOU Y, et al.. BLINK:a package for the next level of genome-wide association studies with both individuals and markers in the millions [J]. Gigascience, 2019,8(2):giy154 [2024-11-06]. . |
| 20 | VOSS-FELS K, FRISCH M, QIAN L W,et al.. Subgenomic diversity patterns caused by directional selection in bread wheat gene pools [J/OL]. Plant Genome,2015,8(2):03.0013 [2024-11-06]. . |
| 21 | PONT C, LEROY T, SEIDEL M, et al.. Tracing the ancestry of modern bread wheats [J]. Nat. Genet., 2019,51(5):905-911. |
| 22 | 陈华斌.小麦粒形、品质和农艺相关性状的全基因组关联分析[D].杭州:浙江大学, 2021. |
| CHEN H B. Genome-wide association study of grain shape, quality and agronomic related traits in wheat [D]. Hangzhou: Zhejiang University, 2021. | |
| 23 | 彭小爱,卢茂昂,张玲,等.基于55K SNP芯片的小麦籽粒主要品质性状的全基因组关联分析[J].作物学报, 2024, 50(8): 1948-1960. |
| PENG X A, LU M A, ZHANG L, et al.. Genome-wide association study of major grain quality traits in wheat based on 55K SNP arrays [J]. Acta Agron. Sin., 2024, 50(8):1948-1960. | |
| 24 | 任生林,吴才文,经艳芬,等.全基因组关联分析在作物中的研究进展[J].分子植物育种,2024,22(11):3594-3602. |
| REN S L, WU C W, JING Y F, et al.. Research progress of genome-wide association analysis in crops [J]. Mol. Plant Breeding, 2024, 22(11): 3594-3602. | |
| 25 | 吴宁宁.玉米种质粒型及粒重性状的鉴定和全基因组关联分析(GWAS)[D].杭州: 浙江农林大学, 2023. |
| WU N N. Identification of grain type and grain weight traits in maize germplasms and genome-wide association analysis (GWAS) [D]. Hangzhou: Zhejiang A&F University, 2023. | |
| 26 | WEN T W, DAI B S, WANG T, et al.. Genetic variations in plant architecture traits in cotton (Gossypium hirsutum) revealed by a genome-wide association study [J]. Crop J., 2019,7(2):209-216. |
| 27 | SETTER T L, YAN J B, WARBURTON M, et al.. Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought [J]. J. Exp. Bot., 2011,62(2):701-716. |
| 28 | TANG S, ZHAO H, LU S P, et al.. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus [J]. Mol. Plant, 2021,14(3):470-487. |
| 29 | ANACLETO R, BADONI S, PARWEEN S,et al.. Integrating a genome-wide association study with a large-scale transcriptome analysis to predict genetic regions influencing the glycaemic index and texture in rice [J]. Plant Biotechnol. J., 2019,17(7):1261-1275. |
| 30 | ZHUANG X L, XU Y, CHONG K, et al.. OsAGAP,an ARF-GAP from rice,regulates root development mediated by auxin in Arabidopsis [J]. Plant Cell Environ., 2005,28(2):147-156. |
| 31 | ZHUANG X L, JIANG J F, LI J H, et al.. Over-expression of OsAGAP,an ARF-GAP,interferes with auxin influx,vesicle trafficking and root development [J]. Plant J., 2006,48(4):581-591. |
| 32 | UAUY C, DISTELFELD A, FAHIMA T, et al.. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat [J]. Science, 2006,314(5803):1298-1301. |
| 33 | COLLARD B C, MACKILL D J. Marker-assisted selection:an approach for precision plant breeding in the twenty-first century [J]. Philos. Trans. R. Soc. Lond. Ser. Biol. Sci., 2008,363(1491):557-572. |
| 34 | 杨青青,唐家琪,张昌泉,等. KASP标记技术在主要农作物中的应用及展望[J]. 生物技术通报,2022,38(4):58-71. |
| YANG Q Q, TANG J Q, ZHANG C Q, et al.. Application and prospect of KASP marker technology in main crops [J]. Biotechnol. Bull., 2022,38(4):58-71. | |
| 35 | 魏广辉,李执,陈强,等. 人工合成小麦SHW-L1高硒含量KASP分子标记开发及其应用[J].中国农业科学,2020,53(20):4103-4112. |
| WEI G H, LI Z, CHEN Q,et al.. Development and utilization of KASP marker for Se concentration in synthetic wheat SHW-L1 [J]. Sci. Agric. Sin., 2020,53(20):4103-4112. | |
| 36 | LIU D, YANG H, ZHANG Z, et al.. An elite γ-gliadin allele improves end-use quality in wheat [J]. New Phytol., 2023,239(1):87-101. |
| 37 | 毕俊鸽,曾占奎,李琼,等. 两个RIL群体中小麦籽粒品质相关性状QTL定位及KASP标记开发[J].作物学报, 2024, 50(7): 1669-1683. |
| BI J G, ZENG Z K, LI Q, et al.. QTL mapping and KASP marker development of grain quality-relating traits in two wheat RIL populations [J]. Acta Agron. Sin., 2024, 50(7):1669-1683. |
| [1] | Yifan SUN, Zhilei HUANG, Baochun LI, Lirong YAO, Juncheng WANG, Erjing SI, Ke YANG, Yaxiong MENG, Xiaole MA, Huajun WANG. Evaluation of Resistance to Spot Blotch in Core Germplasm Resources [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 43-54. |
| [2] | GUO Guangyan§, YANG Yaling§, CAO Lu, LIU Wei, BI Caili*. RF2 Basic Leucine Zipper Transcription Factor TabZIP3 Involved in Salt Stress Response in Wheat [J]. Journal of Agricultural Science and Technology, 2019, 21(6): 20-27. |
| [3] | LI Yang1,2, ZOU Junjie2, YU Jia2, XU Yu2,3, XU Miaoyun2, LUO Hongfa1*, WANG Lei2*. Construction of Maize Near-Isogenic Lines and Its Application [J]. Journal of Agricultural Science and Technology, 2019, 21(12): 14-22. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号