




















中国农业科技导报 ›› 2024, Vol. 26 ›› Issue (5): 30-43.DOI: 10.13304/j.nykjdb.2024.0186
徐佳睿1,2( ), 王逸茹2, 赵绍赓1,2, 李坤2(
), 王逸茹2, 赵绍赓1,2, 李坤2( ), 郑军1,2(
), 郑军1,2( )
)
收稿日期:2024-03-12
接受日期:2024-03-28
出版日期:2024-05-15
发布日期:2024-05-14
通讯作者:
李坤,郑军
作者简介:徐佳睿 E-mail :xjr5299@163.com
基金资助:
Jiarui XU1,2( ), Yiru WANG2, Shaogeng ZHAO1,2, Kun LI2(
), Yiru WANG2, Shaogeng ZHAO1,2, Kun LI2( ), Jun ZHENG1,2(
), Jun ZHENG1,2( )
)
Received:2024-03-12
Accepted:2024-03-28
Online:2024-05-15
Published:2024-05-14
Contact:
Kun LI,Jun ZHENG
摘要:
青贮玉米是一种优良的饲料作物,是畜牧业的优质粗饲料来源。咖啡酰辅酶A-O-甲基转移酶(caffeoyl-CoA-O-methyltransferase, CCoAOMT)是木质素合成途径中的关键甲基转移酶。从玉米自交系B73的EMS突变体库获得了ZmCCoAOMT1基因的单碱基突变体,通过表型分析、生理生化分析、基因表达分析验证了ZmCCoAOMT1的功能。研究发现,ZmCCoAOMT1突变造成茎秆木质化程度降低、木质素含量显著减少、G型木质素(guaiacyl lignin)和S型木质素(syringa lignin)含量下降、体外干物质消化率提高。此外,突变体茎秆中ZmCCoAOMT1的表达量也显著低于野生型。转录组分析表明,ZmCCoAOMT1基因的突变可能调控苯丙烷代谢途径中相关基因的表达,进而引起木质素含量和单体组成发生变化。因此,ZmCCoAOMT1基因是培育高消化率优质青贮玉米品种的重要基因资源,研究结果为木质素合成代谢途径遗传机理研究提供了重要理论依据。
中图分类号:
徐佳睿, 王逸茹, 赵绍赓, 李坤, 郑军. 玉米木质素合成途径基因ZmCCoAOMT1功能研究及转录组分析[J]. 中国农业科技导报, 2024, 26(5): 30-43.
Jiarui XU, Yiru WANG, Shaogeng ZHAO, Kun LI, Jun ZHENG. Functional Study and Transcriptome Analysis of Corn Gene ZmCCoAOMT1 Involved in Lignin Synthesis Pathway[J]. Journal of Agricultural Science and Technology, 2024, 26(5): 30-43.
| 引物名称Primer name | 正向序列Forward sequence (5’-3’) | 反向序列Reverse sequence (5’-3’) |
|---|---|---|
| ZmCCoAOMT1-EMS-F1/R1 | ATCAACCGCGAGAACTACGA | GCTGCGGGTCGTCTATTATG |
| GAPDH-F/R | AGGATATCAAGAAAGCTATTAAGGC | GTAGCCCCACTCGTTGTCG |
| ZmCCoAOMT1-Q-F1/R1 | ACTTCGTGCTCGTCCTCAAC | GCTGCGGGTCGTCTATTATG |
| ZmCCoAOMT2-Q-F1/R1 | GGCCACAAGATCGACTTCC | GTGTTGTCGTAGCCGATGAG |
| ZmCCoAOMT3-Q-F1/R1 | CTTCGACTTCGCCTTCGTC | CGAGAACCTCCTGTCGATGT |
| ZmCCoAOMT4-Q-F1/R1 | CGTCAGCGAAGAAGAGGTG | CTCTCGAGGTCAGCCTCAAC |
| Zm20961-F/R | GCCTAATCGGATCAGCACAAAC | TAACCCTCTCCATCACGCAAAA |
| Zm02901-F/R | GCCTTCGACAACGTCTACTACA | GCTCATCTTGGCGTTATTGGTC |
| Zm14603-F/R | CAGAGAATCCGTCTACCAGAGC | TGTACTGGAAGAAAGGGCAGTC |
| Zm52336-F/R | CGACCGTCTTCGAGAACAACTA | AGTTAACGGAGTGATGTCACCC |
| Zm32405-F/R | GTCATCGACGACATCAAGGAGG | TGAGAGAACAGCGAGATCCTTG |
| Zm40581-F/R | CCTGGACAACTCCTTCTACCAC | TTAGTATAGGCGGTGGTTGACG |
| Zm37359-F/R | GGAAGACTGGACTCCAAGAAGG | GGGTTGTACTTGGACGTCATCT |
| Zm32854-F/R | TTAGTGGAGGGCCTTACTACGA | GGCGAACTTGTGAATGATGGAG |
| Zm32406-F/R | TTCGCGAAGTCGATGGTGAG | GTTAAACCTTCGGCAGTTGAGC |
| Zm46184-F/R | CTCTCTGCTCAGGCTCTTCTTC | CTTGATGGTGTCGATGACCTCA |
| Zm04387-F/R | AATGACCAGCTGTGCTTGGATA | TGACGGCACAATACCCACTTTA |
| Zm07828-F/R | TCTGAACTCCTCATCCACAAGC | TTCCTCATCGGGATGGAGTAGT |
| Zm48354-F/R | TCGTCAAGATGCAGAAGGAACA | TGTTCCTTCTGCATCTTGACGA |
| Zm13212-F/R | GCCTTCGACAACGTCTACTACA | GCTCATCTTGGCGTTATTGGTC |
| Zm50572-F/R | TGAGAAAGGAAGTGGAGAAGGC | AACCATAGACCCAGCAGTCATG |
表 1 研究所用的引物
Table 1 Primers used in this study
| 引物名称Primer name | 正向序列Forward sequence (5’-3’) | 反向序列Reverse sequence (5’-3’) |
|---|---|---|
| ZmCCoAOMT1-EMS-F1/R1 | ATCAACCGCGAGAACTACGA | GCTGCGGGTCGTCTATTATG |
| GAPDH-F/R | AGGATATCAAGAAAGCTATTAAGGC | GTAGCCCCACTCGTTGTCG |
| ZmCCoAOMT1-Q-F1/R1 | ACTTCGTGCTCGTCCTCAAC | GCTGCGGGTCGTCTATTATG |
| ZmCCoAOMT2-Q-F1/R1 | GGCCACAAGATCGACTTCC | GTGTTGTCGTAGCCGATGAG |
| ZmCCoAOMT3-Q-F1/R1 | CTTCGACTTCGCCTTCGTC | CGAGAACCTCCTGTCGATGT |
| ZmCCoAOMT4-Q-F1/R1 | CGTCAGCGAAGAAGAGGTG | CTCTCGAGGTCAGCCTCAAC |
| Zm20961-F/R | GCCTAATCGGATCAGCACAAAC | TAACCCTCTCCATCACGCAAAA |
| Zm02901-F/R | GCCTTCGACAACGTCTACTACA | GCTCATCTTGGCGTTATTGGTC |
| Zm14603-F/R | CAGAGAATCCGTCTACCAGAGC | TGTACTGGAAGAAAGGGCAGTC |
| Zm52336-F/R | CGACCGTCTTCGAGAACAACTA | AGTTAACGGAGTGATGTCACCC |
| Zm32405-F/R | GTCATCGACGACATCAAGGAGG | TGAGAGAACAGCGAGATCCTTG |
| Zm40581-F/R | CCTGGACAACTCCTTCTACCAC | TTAGTATAGGCGGTGGTTGACG |
| Zm37359-F/R | GGAAGACTGGACTCCAAGAAGG | GGGTTGTACTTGGACGTCATCT |
| Zm32854-F/R | TTAGTGGAGGGCCTTACTACGA | GGCGAACTTGTGAATGATGGAG |
| Zm32406-F/R | TTCGCGAAGTCGATGGTGAG | GTTAAACCTTCGGCAGTTGAGC |
| Zm46184-F/R | CTCTCTGCTCAGGCTCTTCTTC | CTTGATGGTGTCGATGACCTCA |
| Zm04387-F/R | AATGACCAGCTGTGCTTGGATA | TGACGGCACAATACCCACTTTA |
| Zm07828-F/R | TCTGAACTCCTCATCCACAAGC | TTCCTCATCGGGATGGAGTAGT |
| Zm48354-F/R | TCGTCAAGATGCAGAAGGAACA | TGTTCCTTCTGCATCTTGACGA |
| Zm13212-F/R | GCCTTCGACAACGTCTACTACA | GCTCATCTTGGCGTTATTGGTC |
| Zm50572-F/R | TGAGAAAGGAAGTGGAGAAGGC | AACCATAGACCCAGCAGTCATG |

图 1 ZmCCoAOMT1突变体的鉴定A:ZmCCoAOMT1突变位置。B:野生型和ccoaomt1突变体测序结果,红框为突变位点;C:氨基酸序列比对;D: ZmCCoAOMT1基因在野生型和ccoaomt1突变体中的表达量,**表示在P<0.01水平与野生型差异显著(t检验)
Fig. 1 Identification of ZmCCoAOMT1 mutantA: Mutation location of the ZmCCoAOMT1; B: Sequencing of WT and ccoaomt1 mutant, red box indicates mutation location; C: Amino acid sequence alignment; D: Expression levels of ZmCCoAOMT1 in WT and ccoaomt1 mutant, ** represents significant difference compared with WT at P<0.05 level (t-test)
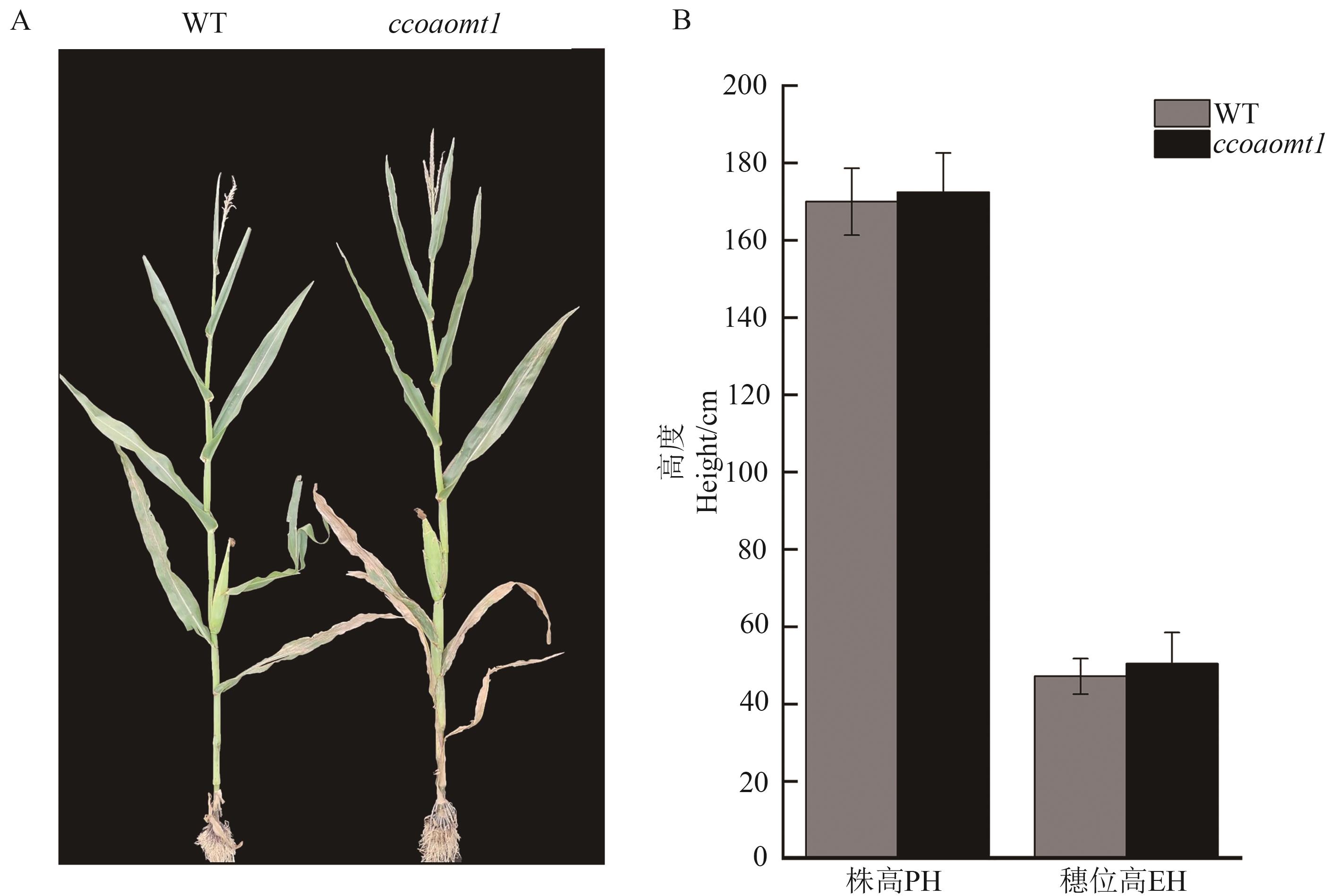
图 3 ZmCCoAOMT1对玉米株型的影响A:野生型和ccoaomt1突变体在开花后20 d的表型;B:野生型和ccoaomt1突变体开花后20 d的株高(PH)和穗位高(EH)比较
Fig. 3 Effect of ZmCCoAOMT1 on corn plant architectureA: Phenotype of WT and ccoaomt1 mutant at 20 d after flowering. B: Comparison of plant height (PH) and ear height (EH) at 20 d after flowering between wild-type and ccoaomt1 mutant
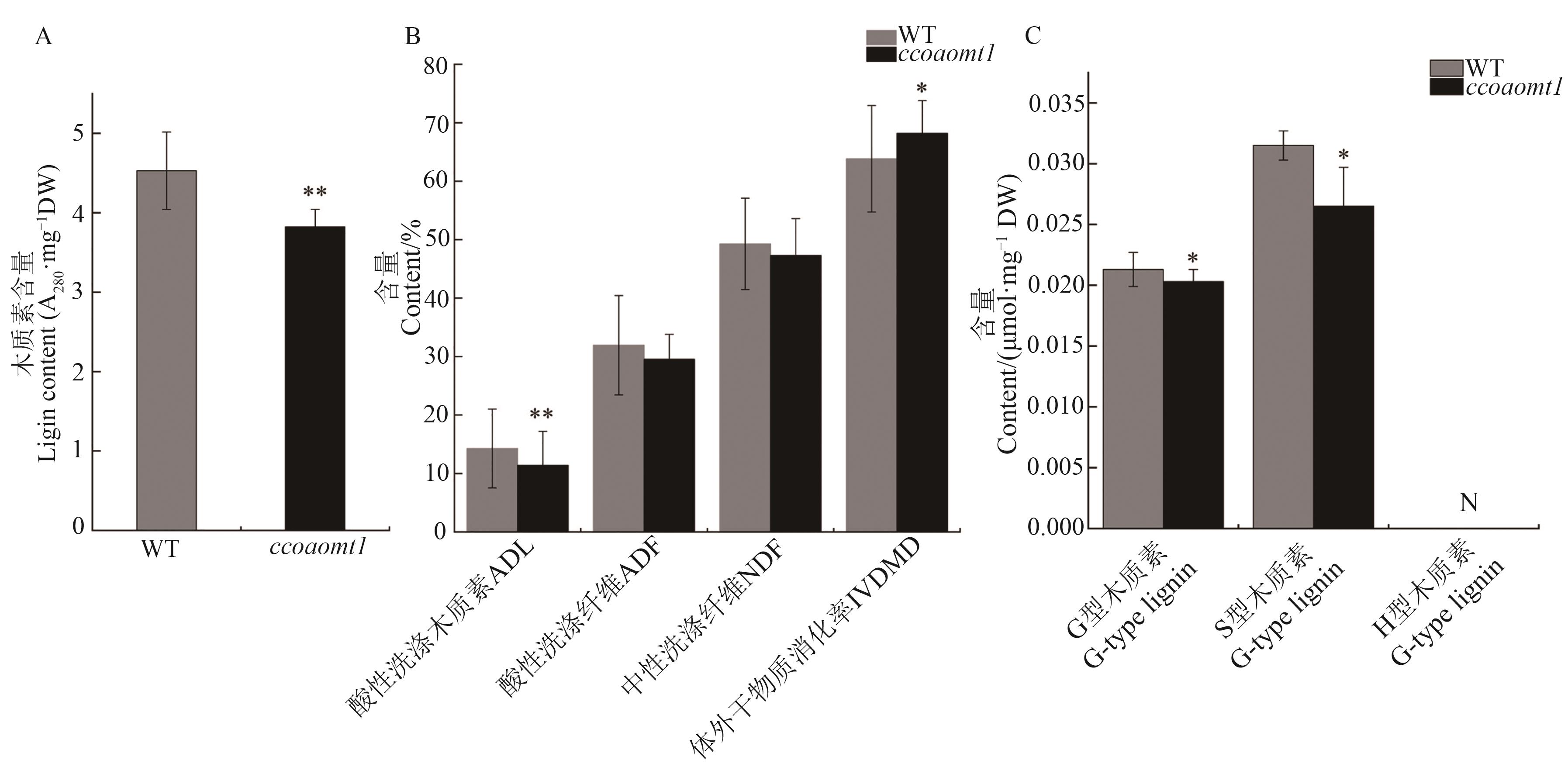
图5 ccoaomt1对木质素含量、组成及消化品质的影响A:成熟期野生型和ccoaomt1突变体地上部3~4茎节的木质素含量;B: 成熟期野生型和ccoaomt1突变体地上部3~4茎节的酸性洗涤木质素(ADL)、酸性洗涤纤维(ADF)、中性洗涤纤维(NDF)、体外干物质消化率(IVDMD)差异; C:成熟期野生型和ccoaomt1突变体地上部3~4茎节的木质素单体(G型、S型、H型)差异。*和**分别表示与野生型相比在P<0.05和P<0.01水平差异显著(t检验)
Fig. 5 Effects of ccoaomt1 on lignin content, composition and digestive qualityA: Differences of lignin content in the shoots of three or four stalk nodes between wild type and ccoaomt1 mutant at corn maturity stage; B: Differences of acid detergent lignin(ADL), acid detergent fiber ( ADF ), neutral detergent fiber (NDF ) and in vitro digestibility (IVDMD ) in 3~4 shoots of corn between wild type and ccoaomt1 mutants; C: Difference of lignin monomer (G-type, S-type, H-type) in the shoots of wild-type and ccoaomt1 mutants at maturity stage. * and ** represent significant differences compared with WT at P<0.05 and P<0.01 levels (t-test) , respectively
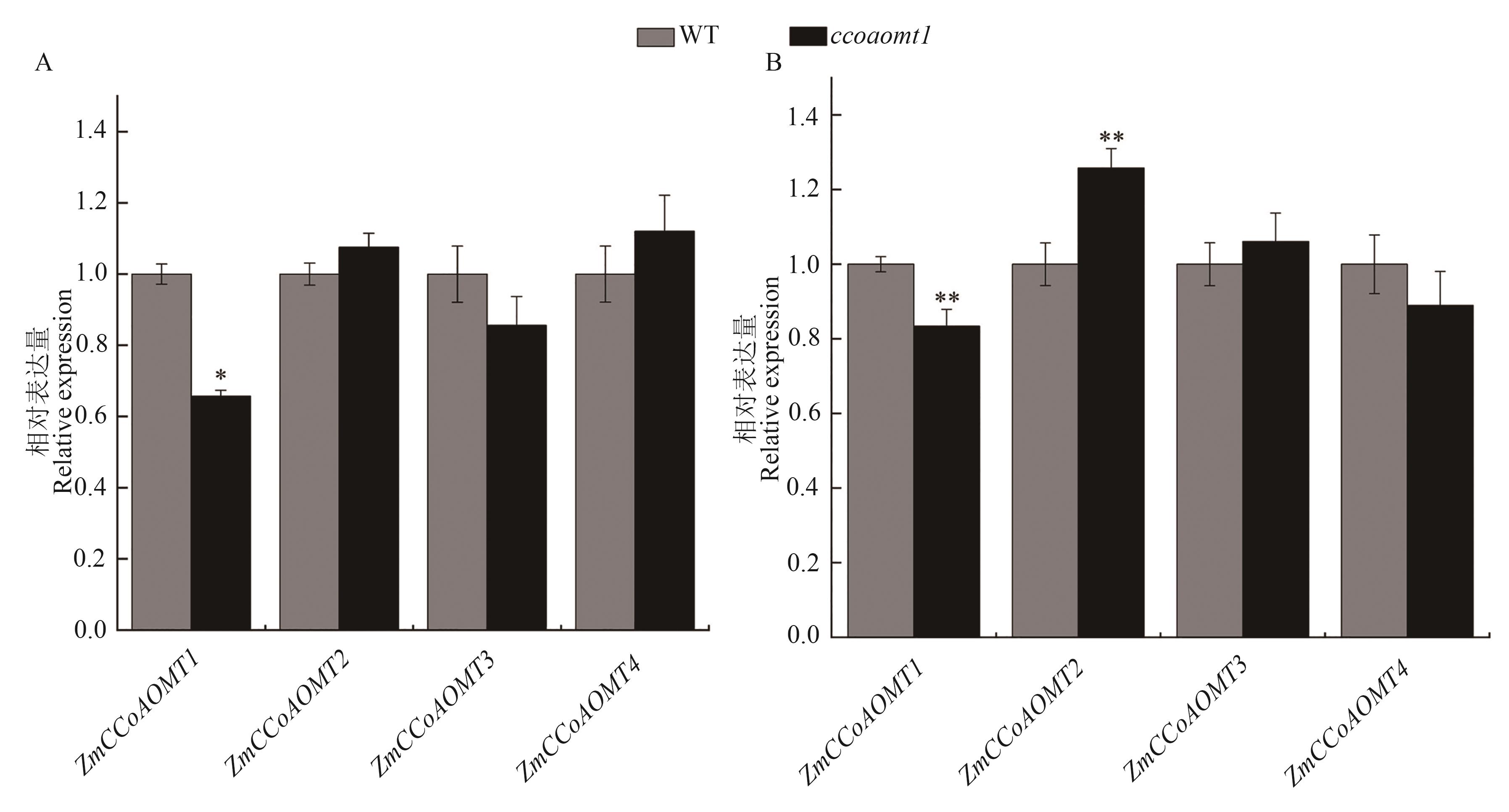
图 6 野生型和突变体 ZmCCoAOMT基因不同时期表达量A: V9期;B吐丝期。*和**分别表示与野生型相比在P<0.05和P<0.01水平差异显著(t检验)
Fig. 6 Expression levels of ZmCCoAOMT genes at different times in wild type and mutantA: V9 stage; B: Silking stage. * and ** represent significant differences compared with WT at P<0.05 and P<0.01 levels (t-test) , respectively
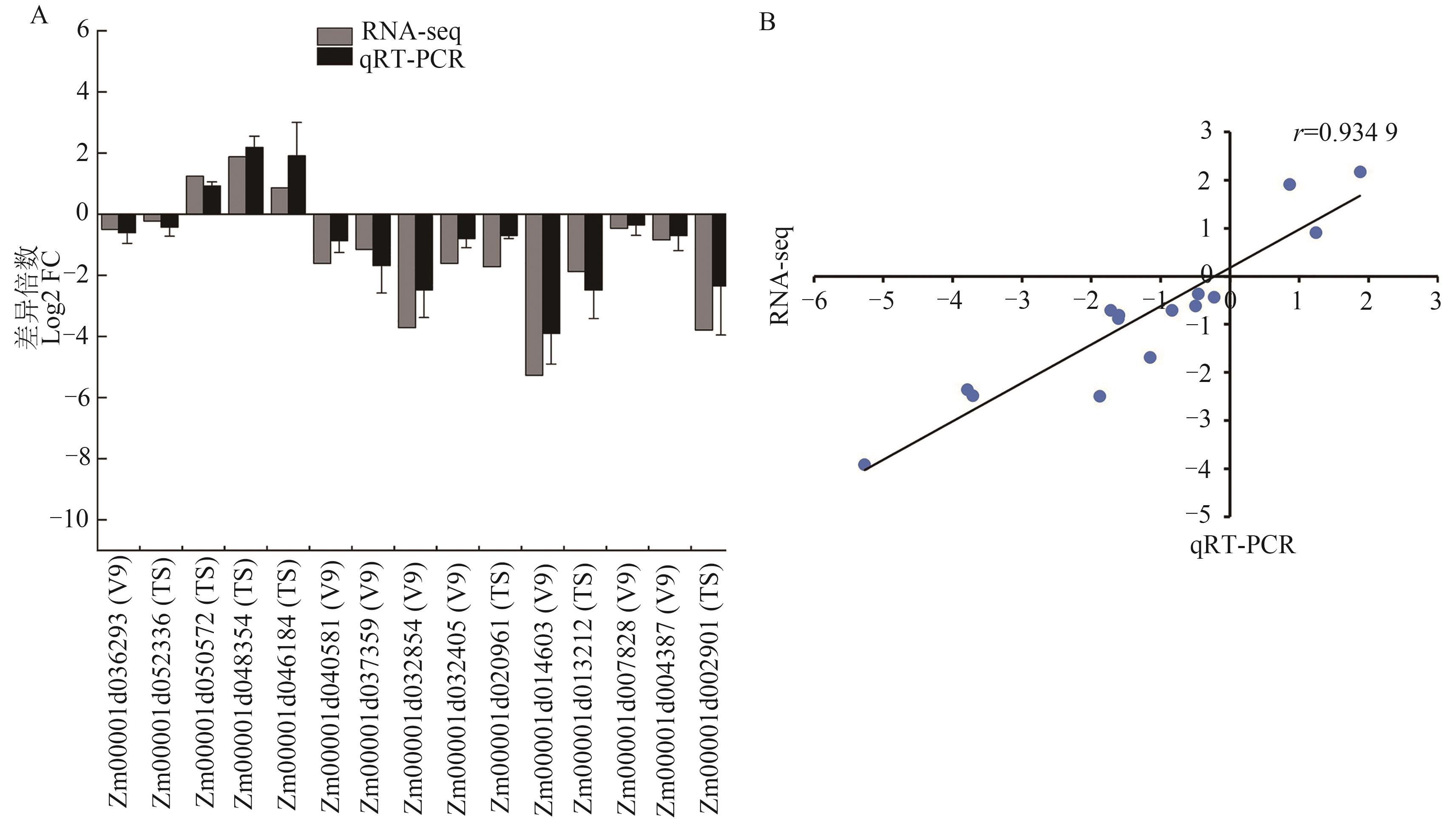
图 7 差异表达基因的qRT-PCR验证A: RNA-seq数据的qRT-PCR验证结果;B: RNA-Seq和qRT-PCR结果相关性分析
Fig. 7 qRT-PCR verification of differentially expressed genesA: qRT-PCR verification of RNA-seq data, ; B: Correlation analysis of RNA-Seq and qRT-PCR data
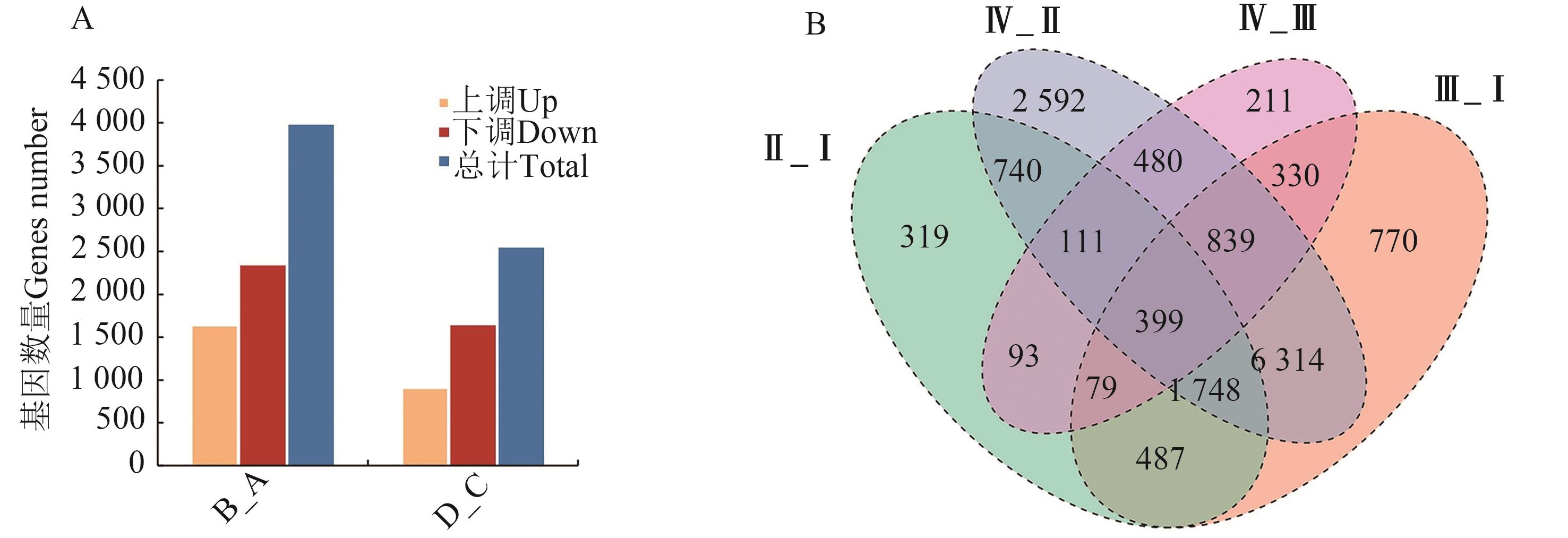
图 8 不同时期野生型和突变体的差异表达基因分析A:差异表达基因数量;B:差异表达基因韦恩图;4个组分别是Ⅰ(V9-野生型)、Ⅱ(V9-ccoaomt1突变体)、Ⅲ(吐丝期-野生型)、Ⅳ(吐丝期-ccoaomt1突变体)。
Fig. 8 Analysis of differentially expressed genes in wild type and mutant at different stagesA: Number of differentially expressed genes; B: Venn of differentially expressed genes; the 4 groups were Ⅰ (V9-WT), Ⅱ (V9-ccoaomt1 mutant), Ⅲ (silking-WT) and Ⅳ (silking-ccoaomt1 mutant)
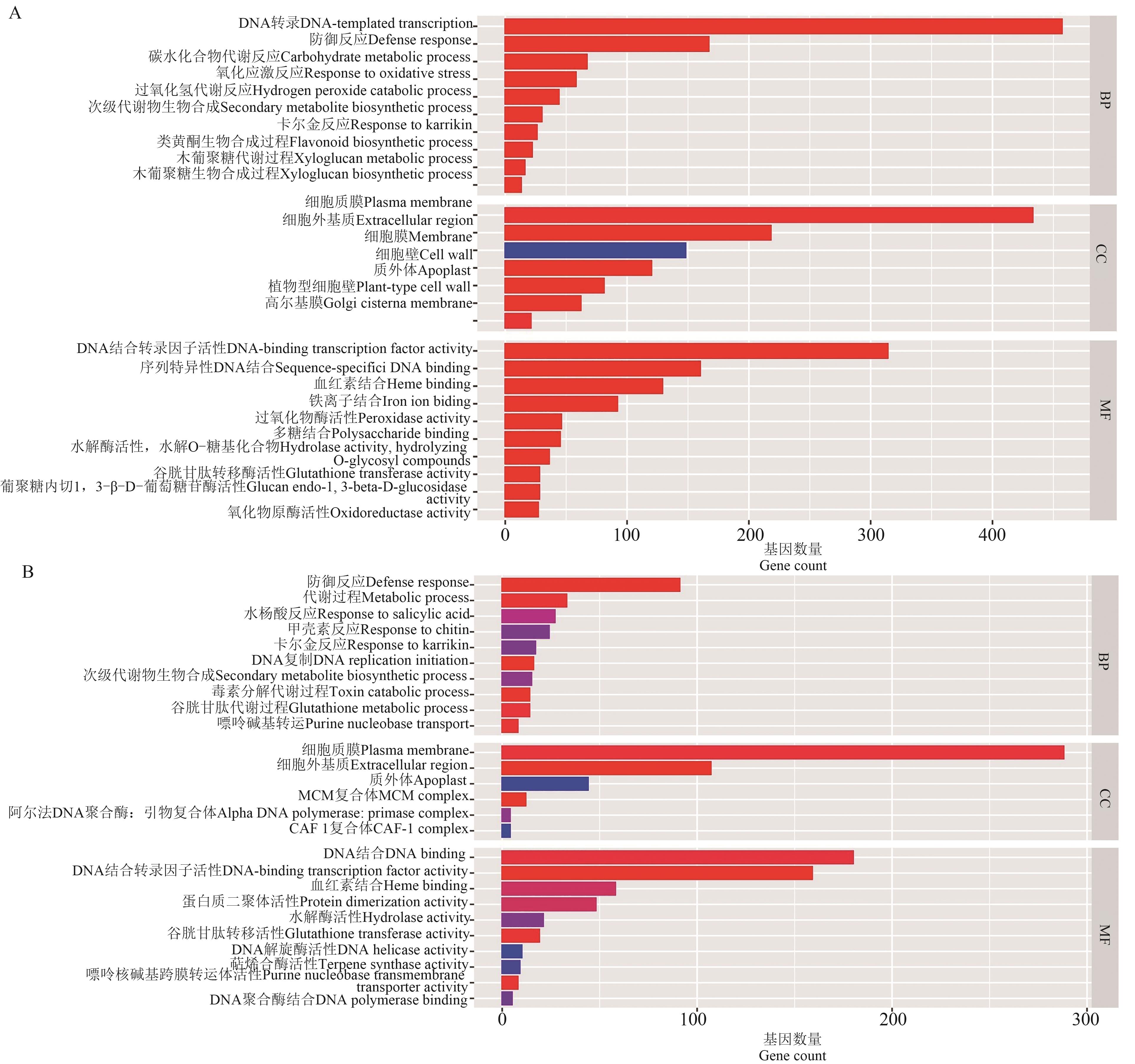
图9 V9时期和吐丝期野生型和突变体差异表达基因的GO富集分析A:V9期;B:吐丝期
Fig. 9 GO enrichment analysis of differentially expressed genes in wild type and mutant at V9 and silking stagesA: V9 stage; B:Silking stage
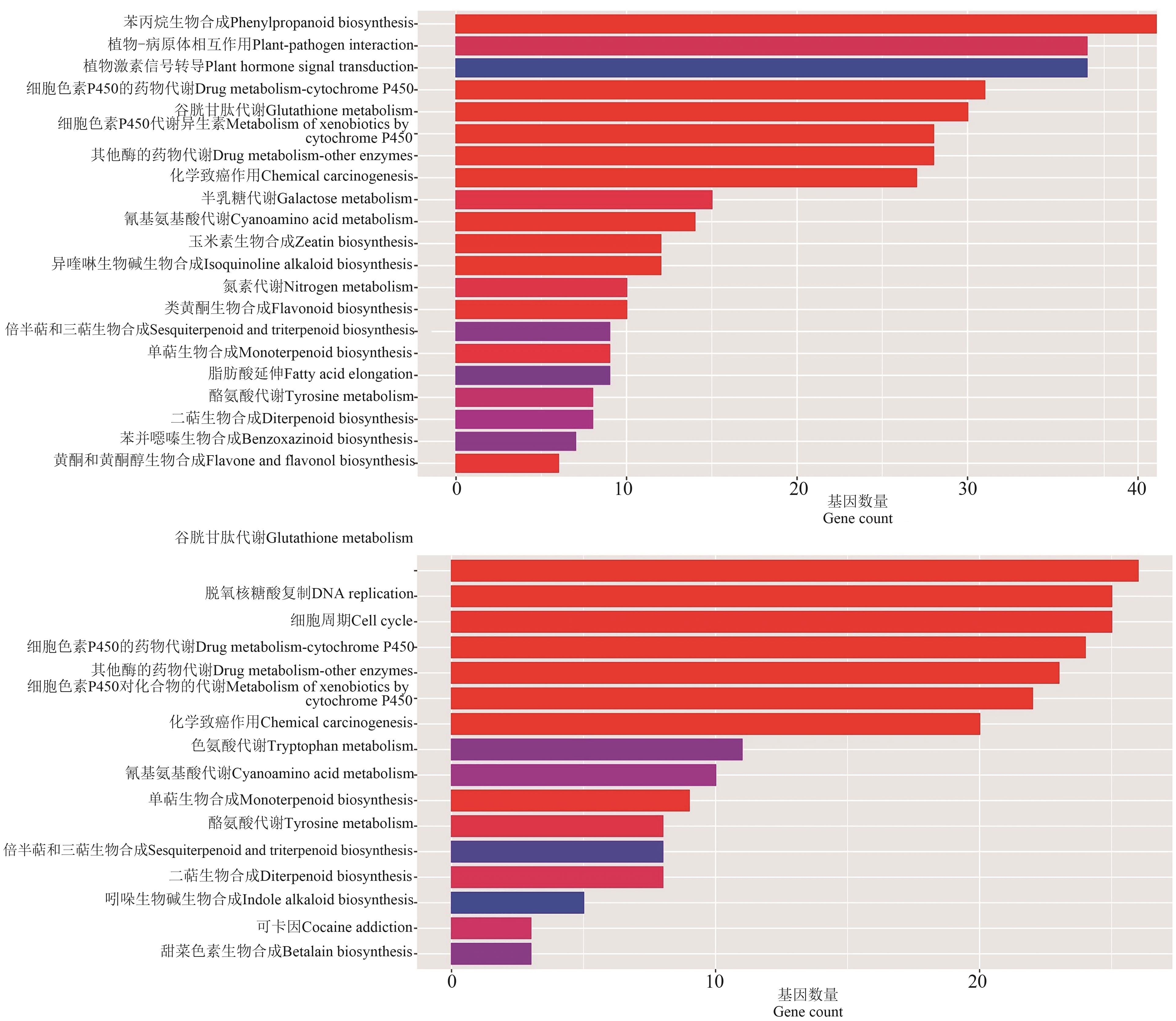
图 10 V9时期和吐丝期野生型和突变体差异表达基因的KEGG富集分析A:V9时期;B:吐丝期
Fig. 10 KEGG analysis of differentially expressed genes of wild-type and mutant at V9 and silking stagesA: V9 stage; B:Silking stage
基因ID Gene ID | 差异倍数 log2FC | 假定值 Postulate value | 调节 Regulate | 蛋白 Protein |
|---|---|---|---|---|
| Zm00001d002898 | -1.311 629 353 | 0.016 167 | 下调 Down | 过氧化物酶12 Peroxidase 12 |
| Zm00001d004443 | -2.003 251 187 | 0.195 030 | 下调 Down | 肉桂醇脱氢酶6 CAD 6 |
| Zm00001d005279 | -1.150 411 362 | 0.419 242 | 下调Down | 过氧化物酶51 Peroxidase 51 |
| Zm00001d007161 | -1.111 930 889 | 0.335 002 | 下调 Down | 过氧化物酶42 Peroxidase 42 |
| Zm00001d008173 | -2.638 001 029 | 0.009 849 | 下调 Down | 过氧化物酶2 Peroxidase 2 |
| Zm00001d008898 | 1.233 030 247 | 0.002 509 | 上调 Up | 过氧化物酶4 Peroxidase 4 |
| Zm00001d009373 | 1.494 820 928 | 0.001 897 | 上调 Up | 过氧化物酶72 Peroxidase 72 |
| Zm00001d014606 | -4.768 298 025 | 0.055 246 | 下调 Down | 过氧化物酶45 Peroxidase 45 |
| Zm00001d018620 | 1.666 833 782 | 0.019 950 | 上调 Up | 过氧化物酶39 Peroxidase 39 |
| Zm00001d019534 | -1.111 649 783 | 0.018 069 | 下调 Down | 丝氨酸羧肽酶 SC-PLs |
| Zm00001d022451 | -0.261 406 941 | 0.454 268 | 下调 Down | 胱硫醚β裂解酶29 CBL 29 |
| Zm00001d020401 | -2.7 444 206 | 0.111 547 | 下调 Down | 肉桂醇脱氢酶8 CAD8 |
| Zm00001d024119 | -1.690 949 412 | 0.473 358 | 下调Down | 过氧化物酶40 Peroxidase 40 |
| Zm00001d026268 | -1.179 850 994 | 0.531 814 | 下调 Down | 莽草酸羟基肉桂酰转移酶 HCT |
| Zm00001d026357 | -4.078 721 983 | 0.010 213 | 下调 Down | 过氧化物酶1 Peroxidase 1 |
| Zm00001d028347 | -1.309 639 992 | 0.274 804 | 下调 Down | 过氧化物酶53 Peroxidase 53 |
| Zm00001d030220 | -2.225 592 941 | 0.021 711 | 下调 Down | 咖啡酰莽草酸酯酶 CSE |
| Zm00001d032405 | -1.604 153 903 | 0.019 927 | 下调 Down | 过氧化物酶56 Peroxidase 56 |
| Zm00001d032467 | -1.372 207 308 | 0.152 413 | 下调 Down | 细胞P450氧化酶 Cytochrome P450 84A1 |
| Zm00001d032854 | -3.70 874 872 | 0.00 016 | 下调 Down | 过氧化物酶59 Peroxidase 59 |
| Zm00001d035055 | -2.536 037 556 | 0.013 541 | 下调 Down | 过氧化物酶47 Peroxidase 47 |
| Zm00001d037359 | -1.152 148 355 | 0.715 147 | 下调 Down | 过氧化物酶11 Peroxidase 11 |
| Zm00001d045101 | -1.762 321 261 | 1.41E-07 | 下调 Down | 肉桂酰辅酶A还原酶1 CCR1 |
| Zm00001d047066 | -2.134 983 298 | 0.034 705 | 下调 Down | 过氧化物酶57 Peroxidase 57 |
| Zm00001d053554 | 4.15 134 339 | 0.0 965 01 | 上调 Up | 过氧化物酶70 Peroxidase 70 |
表 3 木质素合成相关的差异表达基因
Table 3 Differentially expressed genes associated with lignin synthesis
基因ID Gene ID | 差异倍数 log2FC | 假定值 Postulate value | 调节 Regulate | 蛋白 Protein |
|---|---|---|---|---|
| Zm00001d002898 | -1.311 629 353 | 0.016 167 | 下调 Down | 过氧化物酶12 Peroxidase 12 |
| Zm00001d004443 | -2.003 251 187 | 0.195 030 | 下调 Down | 肉桂醇脱氢酶6 CAD 6 |
| Zm00001d005279 | -1.150 411 362 | 0.419 242 | 下调Down | 过氧化物酶51 Peroxidase 51 |
| Zm00001d007161 | -1.111 930 889 | 0.335 002 | 下调 Down | 过氧化物酶42 Peroxidase 42 |
| Zm00001d008173 | -2.638 001 029 | 0.009 849 | 下调 Down | 过氧化物酶2 Peroxidase 2 |
| Zm00001d008898 | 1.233 030 247 | 0.002 509 | 上调 Up | 过氧化物酶4 Peroxidase 4 |
| Zm00001d009373 | 1.494 820 928 | 0.001 897 | 上调 Up | 过氧化物酶72 Peroxidase 72 |
| Zm00001d014606 | -4.768 298 025 | 0.055 246 | 下调 Down | 过氧化物酶45 Peroxidase 45 |
| Zm00001d018620 | 1.666 833 782 | 0.019 950 | 上调 Up | 过氧化物酶39 Peroxidase 39 |
| Zm00001d019534 | -1.111 649 783 | 0.018 069 | 下调 Down | 丝氨酸羧肽酶 SC-PLs |
| Zm00001d022451 | -0.261 406 941 | 0.454 268 | 下调 Down | 胱硫醚β裂解酶29 CBL 29 |
| Zm00001d020401 | -2.7 444 206 | 0.111 547 | 下调 Down | 肉桂醇脱氢酶8 CAD8 |
| Zm00001d024119 | -1.690 949 412 | 0.473 358 | 下调Down | 过氧化物酶40 Peroxidase 40 |
| Zm00001d026268 | -1.179 850 994 | 0.531 814 | 下调 Down | 莽草酸羟基肉桂酰转移酶 HCT |
| Zm00001d026357 | -4.078 721 983 | 0.010 213 | 下调 Down | 过氧化物酶1 Peroxidase 1 |
| Zm00001d028347 | -1.309 639 992 | 0.274 804 | 下调 Down | 过氧化物酶53 Peroxidase 53 |
| Zm00001d030220 | -2.225 592 941 | 0.021 711 | 下调 Down | 咖啡酰莽草酸酯酶 CSE |
| Zm00001d032405 | -1.604 153 903 | 0.019 927 | 下调 Down | 过氧化物酶56 Peroxidase 56 |
| Zm00001d032467 | -1.372 207 308 | 0.152 413 | 下调 Down | 细胞P450氧化酶 Cytochrome P450 84A1 |
| Zm00001d032854 | -3.70 874 872 | 0.00 016 | 下调 Down | 过氧化物酶59 Peroxidase 59 |
| Zm00001d035055 | -2.536 037 556 | 0.013 541 | 下调 Down | 过氧化物酶47 Peroxidase 47 |
| Zm00001d037359 | -1.152 148 355 | 0.715 147 | 下调 Down | 过氧化物酶11 Peroxidase 11 |
| Zm00001d045101 | -1.762 321 261 | 1.41E-07 | 下调 Down | 肉桂酰辅酶A还原酶1 CCR1 |
| Zm00001d047066 | -2.134 983 298 | 0.034 705 | 下调 Down | 过氧化物酶57 Peroxidase 57 |
| Zm00001d053554 | 4.15 134 339 | 0.0 965 01 | 上调 Up | 过氧化物酶70 Peroxidase 70 |
| 1 | 李海燕,魏建民,安小虎,等.青贮玉米的发展现状及栽培技术[J].畜牧与饲料科学, 2011, 32(6): 27,43. |
| 2 | 戴忠民,高凤菊,王友平,等.青贮玉米的育种及发展趋势[J].玉米科学,2004, 12(4): 9-11. |
| DAI Z M, GAO F J, WANG Y P, et al.. Silage maize breeding and its development trend [J]. J. Maize Sci., 2004, 12(4): 9-11. | |
| 3 | 王东军.青贮玉米育种目标与育种方法分析[J].新农业,2020, 1(15): 14-15. |
| 4 | ZHONG R, MORRISON W H, HIMMELSBACH D S, et al.. Essential role ofcaffeoyl coenzyme A-O-methyltransferase in lignin biosynthesis in woody poplar plants [J]. Plant Physiol., 2000, 124(2): 563-578. |
| 5 | 薛永常,李金花,卢孟柱,等.木质素单体生物合成途径[J].林业科学, 2003, 39(6): 146-153. |
| XUE Y C, LI J H, LU M Z, et al.. The lignin subunits biosynthesis pathway and its rewriting [J]. Sci. Silvae Sin., 2003, 39(6): 146-153. | |
| 6 | KUMAR M, CAMPBELL L, TURNER S. Secondary cell walls: biosynthesis and manipulation [J]. J. Exp. Bot., 2016, 67(2): 515-531. |
| 7 | SCHMITT D, PAKUSCH A E, MATERN U. Molecular cloning induction and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance [J]. J. Biol. Chem., 1991, 266(26): 17416-17420. |
| 8 | YE Z H, KNEUSEL R E, MATERN U, et al.. An alternative methylation pathway in lignin biosynthesis in Zinnia [J]. Plant Cell, 1994, 6(10): 1427-1439. |
| 9 | YE Z H. Association of caffeoyl coenzyme A 3-O-methyltransferase expression with lignifying tissues in several dicot plans [J]. Plant Physiol., 1997, 115(4): 1341-1350. |
| 10 | 王华美,于延冲,付春祥,等.木质素合成关键酶咖啡酰辅酶 A 氧甲基转移酶的研究进展[J].基因组学与应用生物学, 2014, 33(2): 458-466. |
| WANG H M, YU Y C, FU C X, et al.. Progress of a key Enzyme-Caffeoyl-CoA 3-O-methyltransferase in lignin biosynthesis [J]. Genom. Appl. Biol., 2014,33(2): 458-466. | |
| 11 | CHEN C, MEYERMANS H, BURGGRAEVE B, et al.. Cell-specific and conditional expression of caffeoyl-coenzyme A-3-O-methyltransferase in poplar [J]. Plant Physiol., 2000, 123(3): 853-868. |
| 12 | RAES J, ROHDE A, CHRISTENSEN J H, et al.. Genome-wide characterization of the lignification toolbox in Arabidopsis [J]. Plant Physiol., 2003, 133(3): 1051-1071. |
| 13 | DO C T, POLLET B, THÉVENIN J, et al.. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis [J]. Planta, 2007, 226(5): 1117-1129. |
| 14 | PINÇON G, MAURY S, HOFFMANN L, et al.. Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth [J]. Phytochemistry, 2001, 57(7): 1167-1176. |
| 15 | GUO D, CEN F, INOUE K, et al.. Downregulation of cafeic acid 3-O-methyltransferase and cafeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin [J]. Plant Cell, 2001, 13(1): 73-88. |
| 16 | KWON H, CHO D J, LEE H, et al.. CCOAOMT1, a candidate cargo secreted via VAMP721/722 secretory vesicles in Arabidopsis [J]. Biochem. Biophys. Res. Commun., 2020, 524(4): 977-982. |
| 17 | CHUN H J, LIM L H, CHEONG M S, et al.. Arabidopsis CCoAOMT1 plays a role in drought stress response via ROS-and ABA-dependent manners [J/OL]. Plants, 2021, 10(5): 831 [2024-04-02]. . |
| 18 | XIA Y, LIU J, WANG Y, et al.. Ectopic expression of Vicia sativa Caffeoyl-CoA-O -methyltransferase (VsCCoAOMT) increases the uptake and tolerance of cadmium in Arabidopsis [J]. Environ. Exp. Bot., 2018, 145(1): 47-53. |
| 19 | FORNALÉ S, RENCORET J, GARCÍA C L, et al.. Changes in cell wall polymers and degradability in maize mutants lacking 3’- and 5’-O-methyltransferases involved in lignin biosynthesis [J]. Plant Cell Physiol., 2017, 58(2): 240-255. |
| 20 | BRENNER E A, ZEIN I, CHEN Y, et al.. Polymorphisms in O-methyltransferase genes are associated with stover cell wall digestibility in European maize (Zea mays L.) [J/OL]. BMC Plant Biol., 2010, 10:27 [2024-04-02]. . |
| 21 | YANG Q, HE Y J, KABAHUMA M, et al.. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens [J]. Nat. Genet., 2017, 49(9): 1364-1372. |
| 22 | 刘玄,王梅,李玉龙,等.玉米主要农艺性状相关性分析[J].安徽农学通报, 2021, 27(24): 39-41, 79. |
| 23 | GEIGER H, FUGGERERA H. Über den chemismus der wiesner-reaktion auf lignin on the chemistry of the wiesner reaction on lignin [J]. Zeitschrift Fur Naturforschung B, 1979, 34: 1471-1472. |
| 24 | 马飞前.玉米茎秆纤维性状QTL定位[D].北京:中国农业科学院, 2014. |
| MA F Q. Mapping of quantitative trait loci (QTL) for stalk fiber traits in maize [D]. Beijing:Chinese Academy of Agricultural Sciences, 2014. | |
| 25 | HARMAN-WARE A E, FOSTER C, HAPPS R M, et al.. A thioacidolysis method tailored for higher-throughput quantitative analysis of lignin monomers [J]. Biotechnol. J., 2016, 11(10): 1268-1273. |
| 26 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method [J]. Methods, 2001, 25(4): 402-408. |
| 27 | CHALMEL F, LARDENOIS A, THOMPSON J D, et al.. GOAnno:GO annotation based on multiple alignment [J]. Bioinformatics, 2005, 21(9): 2095-2096. |
| 28 | KANEHISA M, GOTO S. KEGG: kyoto encyclopedia of genes and genomes [J]. Nucleic. Acids. Res., 2000, 28(1): 27-30. |
| 29 | YU G C, WANG L G, HAN Y Y, et al.. ClusterProfiler: an R package comparing biological themes among gene clusters [J]. OMICS, 2012,16(5): 284-287. |
| 30 | ZAKZESKI J, BRUIJNINCX P C A, JONGERIUS A L, et al.. The catalytic valorization of lignin for the production of renewable chemicals [J]. Chem. Rev., 2010, 110(6): 3552-3599. |
| 31 | 龙国辉,武鹏雨,付嘉智,等.过氧化物酶调控木质素合成研究进展[J].现代农业科技, 2021(23): 47-49, 54. |
| LONG G H, WU P Y, FU J Z, et al.. Research progress on regulation of peroxidase on lignin synthesis [J]. Modern Agric. Sci. Technol., 2021(23): 47-49. | |
| 32 | DIXON R A, BARROS J. Lignin biosynthesis: old roads revisited and new roads explored [J/OL]. Open Biol., 2019,9(12):190215 [2024-04-02].. |
| 33 | VANHOLME R, CESARINO I, RATAJ K, et al.. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis [J]. Science, 2013, 341(6150): 1103-1106. |
| [1] | 成广雷, 邱军, 王晓光, 徐田军, 陈传永, 张春原, 夏千千, 吴元奇, 赵久然, 王荣焕. 我国青贮玉米组合(品种)的农艺性状、生物产量和品质变化[J]. 中国农业科技导报, 2022, 24(4): 30-37. |
| [2] | 宋晋辉,瓮巧云,吕爱枝,袁进成,刘颖慧*. 拔节期干旱胁迫对青贮玉米生育与品质的影响[J]. 中国农业科技导报, 2020, 22(6): 161-167. |
| [3] | 张文云1,张建诚2,姚景珍2*. 氮胁迫下小麦叶片转录组分析[J]. 中国农业科技导报, 2020, 22(11): 26-34. |
| [4] | 杨朝元,刘国涛*,李伟雨,李蕾,夏璇,李世博. 堆肥化过程木质素降解和腐殖质形成的研究进展[J]. 中国农业科技导报, 2019, 21(2): 148-154. |
| [5] | 范雅婷,刘胜*. 用多模型方法预测相思树苯醇抽提物含量[J]. 中国农业科技导报, 2017, 19(2): 131-138. |
| [6] | 刘自飞1,2,云鹏2,王盛锋2,陈磊3,高丽丽2,刘荣乐4,汪洪2*. 木质素磺酸铁肥研制及其对花生的施用效果[J]. 中国农业科技导报, 2016, 18(3): 126-133. |
| [7] | 马光路1,吕建波2,曹青2*. 玉米秸秆中木质素、半纤维素和纤维素的组分分离研究[J]. 中国农业科技导报, 2015, 17(6): 70-79. |
| [8] | 张庆芳,于宗莲. 一株高效木质素降解细菌的筛选及产酶条件的优化[J]. , 2014, 16(2): 143-148. |
| [9] | 陈翟微 刘长江. 利用白腐真菌提高秸秆利用率[J]. , 2001, 3(3): 53-56. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号