




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (8): 239-249.DOI: 10.13304/j.nykjdb.2025.0124
• 方法与技术创新 • 上一篇
马燕勤1,2( ), 周玉洁1,2, 龙海成1,2, 李菊1,2, 王海娥1,2, 常伟3, 李志1,2, 钟建1,2, 苗明军1,2, 杨亮1,2(
), 周玉洁1,2, 龙海成1,2, 李菊1,2, 王海娥1,2, 常伟3, 李志1,2, 钟建1,2, 苗明军1,2, 杨亮1,2( )
)
收稿日期:2025-02-26
接受日期:2025-06-23
出版日期:2025-08-15
发布日期:2025-08-26
通讯作者:
杨亮
作者简介:马燕勤 E-mail:dora0514@sina.cn;
基金资助:
Yanqin MA1,2( ), Yujie ZHOU1,2, Haicheng LONG1,2, Ju LI1,2, Haie WANG1,2, Wei CHANG3, Zhi LI1,2, Jian ZHONG1,2, Mingjun MIAO1,2, Liang YANG1,2(
), Yujie ZHOU1,2, Haicheng LONG1,2, Ju LI1,2, Haie WANG1,2, Wei CHANG3, Zhi LI1,2, Jian ZHONG1,2, Mingjun MIAO1,2, Liang YANG1,2( )
)
Received:2025-02-26
Accepted:2025-06-23
Online:2025-08-15
Published:2025-08-26
Contact:
Liang YANG
摘要:
为构建烟草脆裂病毒(tobacco rattle virus,TRV)介导的十字花科植物的病毒诱导的基因沉默(virus-induced gene silencing,VIGS)体系,以十字花科蔬菜上海青和芥菜为研究对象,以内源八氢番茄红素脱氢酶(phytoene desaturase,PDS)基因为标记基因,构建pTRV2-BrPDS、pTRV2-BjuPDS-g和pTRV2-BjuPDS-c载体,并进一步对农杆菌的侵染水平进行优化,构建上海青和芥菜的高效VIGS系统。结果显示,pTRV2-BrPDS、pTRV2-BjuPDS-g和pTRV2-BjuPDS-c载体构建成功。当农杆菌OD600=1.0时,上海青植株白化表型最明显,当农杆菌OD600= 0.8时,芥菜的沉默效果最佳。通过qRT-PCR证实,白化表型是由TRV重组病毒侵染植株后,内源性PDS基因发生沉默而引起的。以上研究结果为利用VIGS技术研究十字花科植物的基因功能提供了理论依据。
中图分类号:
马燕勤, 周玉洁, 龙海成, 李菊, 王海娥, 常伟, 李志, 钟建, 苗明军, 杨亮. TRV介导的上海青和芥菜VIGS体系的构建[J]. 中国农业科技导报, 2025, 27(8): 239-249.
Yanqin MA, Yujie ZHOU, Haicheng LONG, Ju LI, Haie WANG, Wei CHANG, Zhi LI, Jian ZHONG, Mingjun MIAO, Liang YANG. Construction of TRV-mediated VIGS System in Brassica rapa subsp. chinensis and Brassica juncea[J]. Journal of Agricultural Science and Technology, 2025, 27(8): 239-249.

图1 基于TRV的VIGS载体结构注:Replicase—依赖于RNA的RNA聚合酶;16 KD—16 kD富半胱氨酸蛋白;MP—移动蛋白;CP—外壳蛋白;LB 和RB—T-DNA左、右边界;Rz—自剪切核酸酶;MCS—多克隆位点。
Fig. 1 Structure of TRV-based VIGS vectorNote: Replicase—RNA-dependent RNA polymerase; 16 KD—16 kD cysteine-rich protein; MP—Move protein; CP—Coat protein; LB and RB—left and right border of T-DNA; Rz—Ribozyme; MCS—Multiple clone site.
引物名称 Primer name | 序列 Sequence (5’-3’) | 备注 Reference |
|---|---|---|
| BrPDS-F | ATGGTTGTGTTTGGGAATGT | 扩增BrPDS开放阅读框序列 |
| BrPDS-R | TCATGTTGATACAGTTGTCTC | Amplify BrPDS open reading frame |
| BjuPDS-g-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-g 开放阅读框序列 |
| BjuPDS-g-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BjuPDS-c-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-c 开放阅读框序列 |
| BjuPDS-c-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BrPDS-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BrPDS沉默表达载体 Construct TRV2-BrPDS vector |
| BrPDS-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-g-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BjuPDS-g沉默表达载体 Construct TRV2-BjuPDS-g vector |
| BjuPDS-g-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-c-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2- BjuPDS-c沉默表达载体 Construct TRV2-BjuPDS-c vector |
| BjuPDS-c-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| YL192-F | CTTGAAGAAGAAGACTTTCGAAGTCTC | 鉴定TRV1,约900 bp Identify TRV1, 900 bp |
| YL192-R | GTAAAATCATTGATAACAACACAGACAAAC | |
| YL156-F | GGTCAAGGTACGTAGTAGAG | 鉴定TRV2,约390 bp Identify TRV1, 390 bp |
| YL156-R | CGAGAATGTCAATCTCGTAGG | |
| BrPDS-F2 | CCTGATCGCGTGACTGATG | 内源BrPDS表达检测 Detect expression of BrPDS |
| BrPDS-R2 | TGTTCAACAATCGGCATGCA | |
| BrActin-F | GTCTCCATCTCCTGCTCATAGT | 上海青内参基因 |
| BrActin-R | GCTGACCGTATGAGCAAAGA | actin gene of B. rapa subsp. chinensis |
| BjuPDS-g-F2 | CTGATCGCGTGACTGATGAG | 内源BjuPDS-g表达检测 |
| BjuPDS-g-R2 | CCATGTTTCTCCTGAAGAAACC | Detect expression of BjuPDS-g |
| BjuPDS-c-F2 | TATAGCCATGTCAAAGGCGC | 内源BjuPDS-c表达检测 |
| BjuPDS-c-R2 | GCTCAATCTTCCTTATCCTTG | Detect expression of BjuPDS-c |
| BjuActin-R | GCTGACCGTATGAGCAAAGA | 芥菜内参基因 |
| BjuActin-R | GTTGGAAAGTGCTGAGGGAT | actin gene of B. juncea |
表1 载体构建引物
Table 1 Primers for vector construction
引物名称 Primer name | 序列 Sequence (5’-3’) | 备注 Reference |
|---|---|---|
| BrPDS-F | ATGGTTGTGTTTGGGAATGT | 扩增BrPDS开放阅读框序列 |
| BrPDS-R | TCATGTTGATACAGTTGTCTC | Amplify BrPDS open reading frame |
| BjuPDS-g-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-g 开放阅读框序列 |
| BjuPDS-g-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BjuPDS-c-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-c 开放阅读框序列 |
| BjuPDS-c-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BrPDS-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BrPDS沉默表达载体 Construct TRV2-BrPDS vector |
| BrPDS-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-g-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BjuPDS-g沉默表达载体 Construct TRV2-BjuPDS-g vector |
| BjuPDS-g-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-c-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2- BjuPDS-c沉默表达载体 Construct TRV2-BjuPDS-c vector |
| BjuPDS-c-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| YL192-F | CTTGAAGAAGAAGACTTTCGAAGTCTC | 鉴定TRV1,约900 bp Identify TRV1, 900 bp |
| YL192-R | GTAAAATCATTGATAACAACACAGACAAAC | |
| YL156-F | GGTCAAGGTACGTAGTAGAG | 鉴定TRV2,约390 bp Identify TRV1, 390 bp |
| YL156-R | CGAGAATGTCAATCTCGTAGG | |
| BrPDS-F2 | CCTGATCGCGTGACTGATG | 内源BrPDS表达检测 Detect expression of BrPDS |
| BrPDS-R2 | TGTTCAACAATCGGCATGCA | |
| BrActin-F | GTCTCCATCTCCTGCTCATAGT | 上海青内参基因 |
| BrActin-R | GCTGACCGTATGAGCAAAGA | actin gene of B. rapa subsp. chinensis |
| BjuPDS-g-F2 | CTGATCGCGTGACTGATGAG | 内源BjuPDS-g表达检测 |
| BjuPDS-g-R2 | CCATGTTTCTCCTGAAGAAACC | Detect expression of BjuPDS-g |
| BjuPDS-c-F2 | TATAGCCATGTCAAAGGCGC | 内源BjuPDS-c表达检测 |
| BjuPDS-c-R2 | GCTCAATCTTCCTTATCCTTG | Detect expression of BjuPDS-c |
| BjuActin-R | GCTGACCGTATGAGCAAAGA | 芥菜内参基因 |
| BjuActin-R | GTTGGAAAGTGCTGAGGGAT | actin gene of B. juncea |
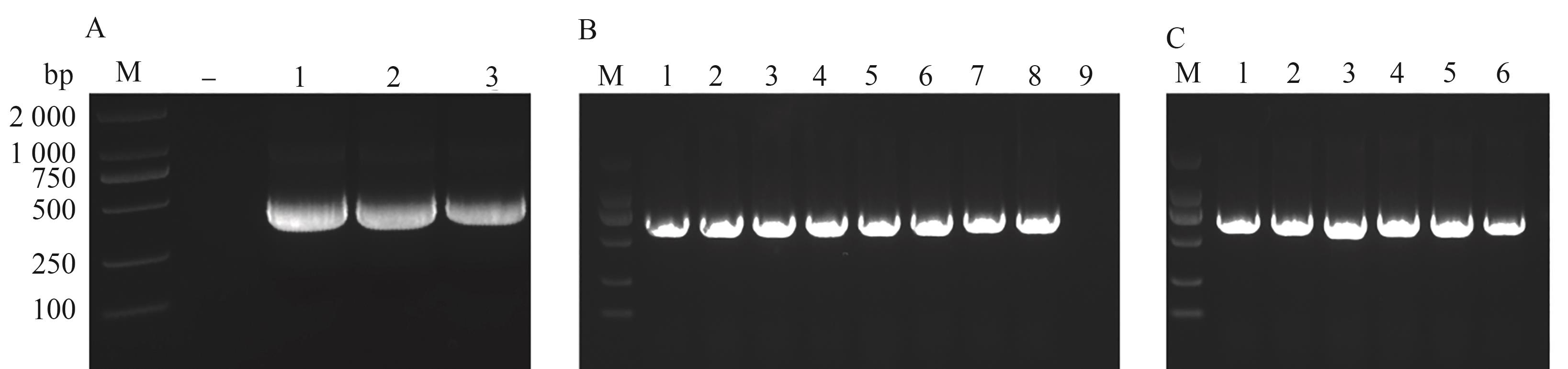
图3 VIGS载体的构建A:内源PDS基因扩增片段,-表示阴性对照,1~3分别为BrPDS、BjuPDS-g和BjuPDS-c基因片段;B:重组载体转化大肠杆菌DH5α菌落PCR检测,1~3为pTRV2-BrPDS DH5α菌落PCR 检测,4~6为pTRV2-BjuPDS-g DH5α菌落PCR 检测,7~9为pTRV2-BjuPDS-c DH5α菌落PCR检测;C:重组载体转化根癌农杆菌GV3101菌株菌落PCR检测,1~2为pTRV2-BrPDS GV3101菌落PCR 检测,3~4为pTRV2-BjuPDS-g大GV3101菌落PCR 检测,5~6为pTRV2-BjuPDS-c GV3101菌落PCR 检测。M—DL2000 DNA Marker。
Fig. 3 Construction of VIGS vectorsA: Amplification fragments of Endogenous PDS gene, - indicates negative control; 1~3 are BrPDS, BjuPDS-g and BjuPDS-c gene fragments, respectively; B: PCR detection of recombinant vector-transformed Escherichia coli DH5α colonies, 1~3 are pTRV2-BrPDS DH5αcolonies PCR detection, respectively; 4~6 are pTRV2-BjuPDS-g DH5αcolonies PCR detection, respectively; 7~9 are pTRV2-BjuPDS-c DH5αcolonies PCR detection, respectively; C: PCR detection of recombinant vector-transformed Agrobacterium tumefaciens GV3101 strain colonies, 1~2 are pTRV2-BrPDS GV3101 colonies PCR detection, respectively; 3~4 are pTRV2-BjuPDS-g GV3101 colonies PCR detection, respectively; 5~6 are pTRV2-BjuPDS-c GV3101 colonies PCR detection. M—DL2000 DNA Marker

图4 不同农杆菌水平下植株的白化率注:不同小写字母表示不同处理间在P<0.05水平差异显著。
Fig. 4 Albinism rate of plant under different Agrobacterium levelsNote: Different lowercase letters indicate significant differences between different treatments at P<0.05 level.

图5 上海青BrPDS沉默植株的白化表型A:OD600=0.3;B:OD600=0.5;C:OD600=0.8;D:OD600=1.0。标尺=1 cm
Fig. 5 Albinism phenotype of Chinese cabbage plant with silenced BrPDSA: OD600=0.3; B: OD600=0.5; C: OD600=0.8; D: OD600=1.0. Scale bar=1 cm

图6 芥菜PDS基因下调导致植株叶片白化现象A:抱子芥OD600=0.3;B:抱子芥OD600=0.5;C:抱子芥OD600=0.8;D:抱子芥OD600=1.0; E:结球芥OD600=0.3;F:结球芥OD600=0.5;G:结球芥OD600=0.8;H:结球芥OD600=1.0。标尺=1 cm
Fig. 6 Leaf albinismof B. juncea plant with downregulation of PDSA: B. juncea var. gemmifera with OD600=0.3; B: B. juncea var. gemmifera with OD600=0.5; C: B. juncea var. gemmifera with OD600=0.8; D: B. juncea var. gemmifera with OD600=1.0;E: B. juncea var. capitate with OD600=0.3; F: B. juncea var. capitate with OD600=0.5; G: B. juncea var. capitate with OD600=0.8; H: B. juncea var. capitate with OD600=1.0. Scale bar =1 cm

图7 芥菜内源PDS基因下调导致植株花序白化现象A:抱子芥;B:结球芥。标尺=1 mm
Fig. 7 Inflorescence albinism of B. juncea plant with downregulation of PDSA: B. juncea var. gemmifera; B: B.juncea var. capitata. Scale bar =1 mm

图8 芥菜内源PDS基因下调导致植株花冠白化现象A:抱子芥;B:结球芥。标尺=1 mm
Fig. 8 Corolla albinism of B. juncea plant with downregulation of PDSA: B. juncea var. gemmifera; B: B.juncea var. capitata. Scale bar =1 mm.

图9 叶片中TRV RNA的表达分析A:上海青;B:抱子芥;C:结球芥。TRV1为叶片中TRV1的表达量检测,TRV2为叶片中TRV2的表达量检测
Fig. 9 Analysis of TRV RNA expression in leafA: B. rapa subsp. Chinensis; B: B. juncea var. gemmifera; C: B. juncea var. capitata. TRV1 is the detection of TRV1 expression in leaves, TRV2 is the detection of TRV1 expression in leaves

图10 内源PDS基因的相对表达量A~B:上海青;C~D:抱子芥;E~F:结球芥
Fig. 10 Relative expression of endogenous PDS geneA~B: B. rapa subsp. Chinensis; C~D: B. juncea var. gemmifera; E~F: B. juncea var. capitata
| [1] | WATERHOUSE P M, WANG M B, LOUGH T. Gene silencing as an adaptive defence against viruses [J]. Nature, 2001, 411(6839): 834-842. |
| [2] | LIU Y, SUN W, ZENG S, et al.. Virus-induced gene silencing in two novel functional plants, Lycium barbarum L. and Lycium ruthenicum Murr [J]. Sci. Hortic., 2014, 170: 267-274. |
| [3] | ARCE-RODRÍGUEZ M L, OCHOA-ALEJO N. Virus-induced gene silencing (VIGS) in chili pepper (Capsicum spp.) [J]. Methods Mol. Biol., 2020, 2172: 27-38. |
| [4] | BACHAN S, DINESH-KUMAR S P. Tobacco rattle virus (TRV)-based virus-induced gene silencing [J]. Methods Mol. Biol., 2012, 894: 83-92. |
| [5] | RATCLIFF F, MARTIN-HERNANDEZ A M, BAULCOMBE D C. Technical advance. tobacco rattle virus as a vector for analysis of gene function by silencing [J]. Plant J., 2001, 25(2): 237-245. |
| [6] | SINGH A K, GHOSH D, CHAKRABORTY S. Optimization of tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) in tomato [J]. Methods Mol. Biol., 2022, 2408: 133-145. |
| [7] | GOULD B, KRAMER E M. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (Columbine, Ranunculaceae) [J/OL]. Plant Methods, 2007, 3: 6 [2025-01-20]. . |
| [8] | ZHANG J, WANG F, ZHANG C, et al.. A novel VIGS method by agroinoculation of cotton seeds and application for elucidating functions of GhBI-1 in salt-stress response [J]. Plant Cell Rep., 2018, 37(8): 1091-1100. |
| [9] | CAI C, WANG X, ZHANG B, et al.. Tobacco rattle virus-induced gene silencing in cotton [J]. Methods Mol. Biol., 2019, 1902: 105-119. |
| [10] | YAN H, SHI S, MA N, et al.. Graft-accelerated virus-induced gene silencing facilitates functional genomics in rose flowers [J]. J. Integr. Plant Biol., 2018, 60(1): 34-44. |
| [11] | CHEN R, CHEN X, HAGEL J M, et al.. Virus-induced gene silencing to investigate alkaloid biosynthesis in opium poppy [J]. Methods Mol. Biol., 2020, 2172: 75-92. |
| [12] | MA Y Q, LI Q, CHENG H, et al.. Alternative splicing variants of IiSEP3 in Isatis indigotica are involved in floral transition and flower development [J/OL]. Plant Physiol. Biochem., 2024, 216: 109153 [2025-01-20]. . |
| [13] | SHEN Z, SUN J, YAO J, et al.. High rates of virus-induced gene silencing by tobacco rattle virus in Populus [J]. Tree Physiol., 2015, 35(9): 1016-1029. |
| [14] | LI H L, GUO D, WANG Y, et al.. Tobacco rattle virus-induced gene silencing in Hevea brasiliensis Free [J]. Biosci. Biotechnol. Biochem., 2021, 85(3): 562-567. |
| [15] | KOUDOUNAS K, THOMOPOULOU M, ANGELI E, et al.. Virus-induced gene silencing in olive tree (Oleaceae) [J]. Methods Mol. Biol., 2020, 2172: 165-182. |
| [16] | ZHANG Y, NIU N, LI S, et al.. Virus-induced gene silencing (VIGS) in Chinese jujube [J/OL]. Plants (Basel), 2023, 12(11): 2115 [2025-01-20]. . |
| [17] | YU J, YANG X D, WANG Q, et al.. Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector [J]. Biol. Plant., 2018, 62(4): 826-834. |
| [18] | WANG L, WU Y, DU W, et al.. Virus-induced gene silencing (VIGS) analysis shows involvement of the LsSTPK gene in lettuce (Lactuca sativa L.) in high temperature-induced bolting [J/OL]. Plant Signal. Behav., 2021, 16(7): 1913845 [2025-01-20]. . |
| [19] | LI G, LI Y, YAO X, et al.. Establishment of a virus-induced gene-silencing (VIGS) system in tea plant and its use in the functional analysis of CsTCS1 [J/OL]. Int. J. Mol. Sci., 2022, 24(1): 392 [2025-01-20]. . |
| [20] | PARK H, KREUNEN S S, CUTTRISS A J, et al.. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis [J]. Plant Cell, 2002, 14(2): 321-332. |
| [21] | ISAACSON T, RONEN G, ZAMIR D, et al.. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants [J]. Plant Cell, 2002, 14(2): 333-342. |
| [22] | KANT R, DASGUPTA I. Phenotyping of VIGS-mediated gene silencing in rice using a vector derived from a DNA virus [J]. Plant Cell Rep., 2017, 36(7): 1159-1170. |
| [23] | ZHANG J, YU D, ZHANG Y, et al.. Vacuum and co-cultivation agroinfiltration of (germinated) seeds results in tobacco rattle virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize [J/OL]. Front. Plant Sci., 2017, 8: 393 [2025-01-20]. . |
| [24] | YAMAGISHI N, YOSHIKAWA N. Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with apple latent spherical virus vectors [J]. Plant Mol. Biol., 2009, 71(1-2): 15-24. |
| [25] | ROMERO I, TIKUNOV Y, BOVY A. Virus-induced gene silencing in detached tomatoes and biochemical effects of phytoene desaturase gene silencing [J]. J. Plant Physiol., 2011, 168(10): 1129-1135. |
| [26] | C-MRYU, ANAND A, KANG L, et al.. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species [J]. Plant J., 2004, 40(2): 322-331. |
| [27] | SENTHIL-KUMAR M, MYSORE K S. Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato [J]. Plant Biotechnol. J., 2011, 9(7): 797-806. |
| [28] | BURCH-SMITH T M, ANDERSON J C, MARTIN G B, et al.. Applications and advantages of virus-induced gene silencing for gene function studies in plants [J]. Plant J., 2004, 39(5): 734-746. |
| [29] | BECKER A, LANGE M. VIGS: genomics goes functional [J]. Trends Plant Sci., 2010, 15(1): 1-4. |
| [30] | ZHANG F, WEN Y, GUO X. CRISPR/Cas9 for genome editing: progress, implications and challenges [J]. Huuman Mol. Genet., 2014, 23(r1): R40-R46. |
| [1] | 翁慧婷, 刘海洋, 郭惠明, 程红梅, 李君, 张超, 苏晓峰. 棉花抗黄萎病相关基因GhERF020功能的初步分析[J]. 中国农业科技导报, 2024, 26(9): 112-121. |
| [2] | 王琴琴, 陈修贵, 陆许可, 王帅, 张悦新, 范亚朋, 陈全家, 叶武威. 陆地棉GhPKE1的生物信息学分析及功能验证[J]. 中国农业科技导报, 2022, 24(1): 38-45. |
| [3] | 王瑞霞,李小玉,田宏先*. 晋北区芥菜型油菜抗旱性鉴定及综合抗旱指标筛选[J]. 中国农业科技导报, 2020, 22(11): 42-51. |
| [4] | 杨笑敏,芮存,张悦新,王德龙,王俊娟,陆许可,陈修贵,郭丽雪,王帅,陈超,叶武威*. 棉花DNA甲基转移酶GhDMT9抗逆性分析[J]. 中国农业科技导报, 2019, 21(10): 12-19. |
| [5] | 靳巧春1,于放2,于宗霞1*,冯宝民1*. 利用VIGS技术研究广藿香醇合酶基因PatPTS的功能[J]. 中国农业科技导报, 2018, 20(3): 39-45. |
| [6] | 任向辉,崔建新,杨萌,李鹏,牛新月,李文杰. 上海青菜田黄曲条跳甲发生环境因子的随机森林建模分析[J]. 中国农业科技导报, 2017, 19(7): 117-123. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号