




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (12): 90-100.DOI: 10.13304/j.nykjdb.2022.1031
• INNOVATIVE TECHNOLOGY • Previous Articles Next Articles
Weijun GUO( ), Dongwei LI(
), Dongwei LI( ), Shang XIE(
), Shang XIE( ), Liwen YANG, Cong LI, Jian TIAN, Li PU, Xiaofeng GU(
), Liwen YANG, Cong LI, Jian TIAN, Li PU, Xiaofeng GU( )
)
Received:2022-11-15
Accepted:2022-12-02
Online:2022-12-15
Published:2023-02-06
Contact:
Xiaofeng GU
郭位军( ), 李东维(
), 李东维( ), 谢上(
), 谢上( ), 杨立文, 李聪, 田健, 普莉, 谷晓峰(
), 杨立文, 李聪, 田健, 普莉, 谷晓峰( )
)
通讯作者:
谷晓峰
作者简介:郭位军、郭位军E-mail:guoweijun01@163.com基金资助:CLC Number:
Weijun GUO, Dongwei LI, Shang XIE, Liwen YANG, Cong LI, Jian TIAN, Li PU, Xiaofeng GU. Artificial Intelligence Accelerates Epigenetics and Plant Breeding[J]. Journal of Agricultural Science and Technology, 2022, 24(12): 90-100.
郭位军, 李东维, 谢上, 杨立文, 李聪, 田健, 普莉, 谷晓峰. 人工智能加速作物表观遗传设计育种[J]. 中国农业科技导报, 2022, 24(12): 90-100.
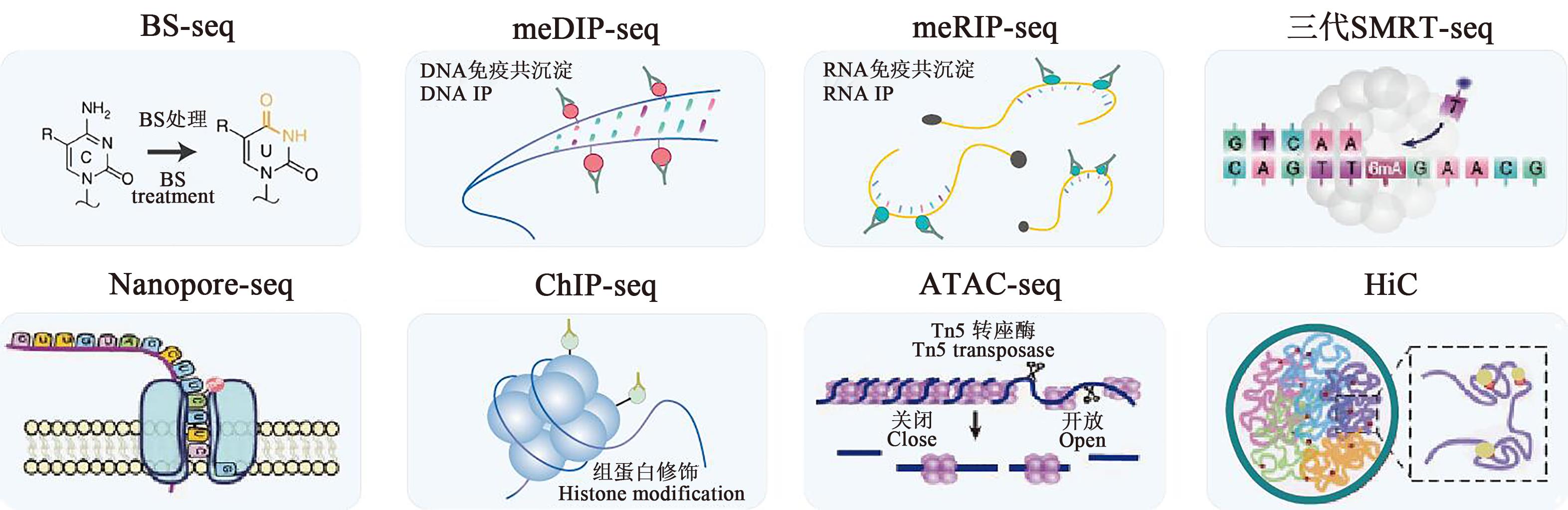
Fig. 2 Technologies relevant to epigenomeBS-seq:Bisulfite sequencing; MeDIP-seq: Methylated DNA immunoprecipitation sequencing; MeRIP-seq: Methylated RNA immunoprecipitation sequencing; ChIP-seq: Chromatin immunoprecipitation sequencing; SMRT-seq: Single-molecule real-time sequencing;Nanopore-seq: Nanopore sequencing; ATAC-seq: Assay for transposase-accessible chromatin with high-throughput sequencing (HTS); HiC: High-throughput chromosome conformation capture technology
| 1 | HOLLIDAY R. Epigenetics: a historical overview [J]. Epigenetics, 2006,1(2):76-80. |
| 2 | LIANG Z, RIAZ A, CHACHAR S, et al.. Epigenetic modifications of mRNA and DNA in plants [J]. Mol. Plant, 2020, 13(1):14-30. |
| 3 | BAYLIN S B, OHM J E. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? [J]. Nat. Rev. Cancer, 2006, 6(2):107-116. |
| 4 | KOUZARIDES T, Chromatin modifications and their function [J]. Cell, 2007, 128(4):693-705. |
| 5 | EGGER G, LIANG G, APARICIO A, et al.. Epigenetics in human disease and prospects for epigenetic therapy [J]. Nature, 2004, 429(6990):457-463. |
| 6 | ZHANG Q, LIANG Z, CUI X, et al.. N 6-methyladenine DNA methylation in Japonica and Indica rice genomes and its association with gene expression, plant development, and stress responses [J]. Mol. Plant, 2018, 11(12):1492-1508. |
| 7 | LIANG Z, GENG Y, GU X, et al.. Adenine methylation: new epigenetic marker of DNA and mRNA [J]. Mol. Plant, 2018, 11(10):1219-1221. |
| 8 | FROMMER M, MCDONALD L E, MILLAR D S, et al.. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands [J]. Proc. Natl. Acad. Sci. USA, 1992, 89(5):1827-1831. |
| 9 | MELNIKOV A A, GARTENHAUS R B, LEVENSON A S, et al.. MSRE-PCR for analysis of gene-specific DNA methylation [J/OL]. Nucleic Acids Res., 2005, 33(10): e93 [2022-11-30].. |
| 10 | BANNISTER A J, KOUZARIDES T. Regulation of chromatin by histone modifications [J]. Cell Res., 2011, 21(3):381-395. |
| 11 | GADE P, KALVAKOLANU D V. Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity [J]. Methods Mol. Biol., 2012, 809:85-104. |
| 12 | DAVIES J O, OUDELAAR A M, HIGGS D R, et al.. How best to identify chromosomal interactions: a comparison of approaches [J]. Nat. Methods, 2017, 14(2):125-134. |
| 13 | DI PIERRO M, CHENG R R, AIDEN L E, et al.. De novo prediction of human chromosome structures: epigenetic marking patterns encode genome architecture [J]. Proc. Natl. Acad. Sci. USA, 2017, 114(46):12126-12131. |
| 14 | YE G, ZHANG H, CHEN B, et al.. De novo genome assembly of the stress tolerant forest species Casuarina equisetifolia provides insight into secondary growth [J]. Plant J., 2019, 97(4):779-794. |
| 15 | YUAN D H, XING J F, LUAN M W, et al.. DNA N 6-methyladenine modification in wild and cultivated soybeans reveals different patterns in nucleus and cytoplasm [J/OL]. Front. Genet., 2020, 11:736 [2022-11-30]. . |
| 16 | ZHOU C, WANG C, LIU H, et al.. Identification and analysis of adenine N 6-methylation sites in the rice genome [J]. Nat. Plants, 2018, 4(8):554-563. |
| 17 | LIANG Z, ZHANG Q, JI C, et al.. Reorganization of the 3D chromatin architecture of rice genomes during heat stress [J/OL]. BMC Biol., 2021, 19(1):53 [2022-11-30]. . |
| 18 | ROUNDTREE I A, EVANS M E, PAN T, et al.. Dynamic RNA modifications in gene expression regulation [J]. Cell, 2017,169(7):1187-1200. |
| 19 | SHEN H, ONTIVEROS R J, OWENS M C, et al.. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation [J/OL]. J. Biol. Chem., 2021,296:100087 [2022-11-30]. . |
| 20 | HOU Y, SUN J, WU B, et al.. CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis [J]. Mol. Plant, 2021, 14(4):688-699. |
| 21 | SHAO Y, WONG C E, SHEN L, et al.. N 6-methyladenosine modification underlies messenger RNA metabolism and plant development [J/OL]. Curr. Opin. Plant Biol., 2021, 63:102047 [2022-11-30]. . |
| 22 | SONG P, YANG J, WANG C, et al.. Arabidopsis N 6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies [J]. Mol. Plant, 2021, 14(4):571-587. |
| 23 | XU T, WU X, WONG C E, et al.. FIONA1-mediated m6A modification regulates the floral transition in Arabidopsis [J/OL]. Adv. Sci., 2022, 9(6):e2103628 [2022-11-30]. . |
| 24 | JIA G, FU Y, ZHAO X, et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO [J]. Nat. Chem. Biol., 2011, 7(12):885-887. |
| 25 | DUAN H C, WEI L H, ZHANG C, et al.. ALKBH10B is an RNA N 6-methyladenosine demethylase affecting Arabidopsis floral transition [J]. Plant Cell, 2017, 29(12):2995-3011. |
| 26 | MARTÍNEZ-PÉREZ M, APARICIO F, LÓPEZ-GRESA M P, et al.. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs [J]. Proc. Natl. Acad. Sci. USA, 2017, 114(40):10755-10760. |
| 27 | ZHENG G, DAHL J A, NIU Y, et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility [J]. Mol. Cell, 2013,49(1):18-29. |
| 28 | SHEN L, LIANG Z, GU X, et al.. N 6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis [J]. Dev. Cell, 2016, 38(2):186-200. |
| 29 | ZHANG F, ZHANG Y C, LIAO J Y, et al.. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice [J/OL]. PLoS Genet., 2019, 15(5):e1008120 [2022-11-30]. . |
| 30 | ARRIBAS-HERNáNDEZ L, BRESSENDORFF S, HANSEN M H, et al.. An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis [J]. Plant Cell, 2018, 30(5):952-967. |
| 31 | WEI L H, SONG P, WANG Y, et al.. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis [J]. Plant Cell, 2018, 30(5):968-985. |
| 32 | CHENG P, BAO S, LI C, et al.. RNA N 6-methyladenosine modification promotes auxin biosynthesis required for male meiosis in rice [J]. Dev. Cell, 2022, 57(2):246-259. |
| 33 | WANG C, YANG J, SONG P, et al.. FIONA1 is an RNA N 6-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering [J]. Genome Biol., 2022, 23(1):40 [2022-11-30]. . |
| 34 | HU J, CAI J, PARK S J, et al.. N 6-methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis [J]. Plant J., 2021, 106(6):1759-1775. |
| 35 | ZHANG T Q, CHEN Y, LIU Y, et al.. Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root [J/OL]. Nat. Commun., 2021, 12(1):2053 [2022-11-30]. . |
| 36 | CUI X, LIANG Z, SHEN L, et al.. 5-methylcytosine RNA methylation in Arabidopsis thaliana [J]. Mol. Plant, 2017, 10(11):1387-1399. |
| 37 | DAVID R, BURGESS A, PARKER B, et al.. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs [J]. Plant Cell, 2017, 29(3):445-460. |
| 38 | TANG Y, GAO C C, GAO Y, et al.. OsNSUN2-mediated 5-methylcytosine mRNA modification enhances rice adaptation to high temperature [J]. Dev. Cell, 2020, 53(3):272-286. |
| 39 | PU L, SUNG Z R. PcG and trxG in plants-friends or foes [J]. Trends Genet., 2015, 31(5):252-262. |
| 40 | KOLASINSKA-ZWIERZ P, DOWN T, LATORRE I, et al.. Differential chromatin marking of introns and expressed exons by H3K36me3 [J]. Nat. Genet., 2009, 41(3):376-381. |
| 41 | PIEN S, FLEURY D, MYLNE J S, et al.. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation [J]. Plant Cell, 2008, 20(3):580-588. |
| 42 | LIU C, LU F, CUI X, et al.. Histone methylation in higher plants [J]. Annu. Rev. Plant Biol., 2010, 61:395-420. |
| 43 | ZHANG X, CLARENZ O, COKUS S, et al.. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis [J/OL]. PLoS Biol., 2007, 5(5):e129 [2022-11-30]. . |
| 44 | PARK M, PATEL N, KEUNG A J, et al.. Engineering epigenetic regulation using synthetic read-write modules [J]. Cell, 2019, 176(1-2):227-238. |
| 45 | LIU X M, ZHOU J, MAO Y, et al.. Programmable RNA N 6-methyladenosine editing by CRISPR-Cas9 conjugates [J]. Nat. Chem. Biol., 2019, 15(9):865-871. |
| 46 | NUÑEZ J K, CHEN J, POMMIER G C, et al.. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing [J]. Cell, 2021, 184(9):2503-2519. |
| 47 | JOHNSON L M, DU J, HALE C J, et al.. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation [J]. Nature, 2014, 507(7490):124-128. |
| 48 | PAPIKIAN A, LIU W, GALLEGO-BARTOLOMÉ J, et al.. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems [J/OL]. Nat. Commun., 2019, 10(1):729 [2022-11-30]. . |
| 49 | GALLEGO-BARTOLOMé J, GARDINER J, LIU W, et al.. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain [J]. Proc. Natl. Acad. Sci. USA, 2018, 115(9):E2125-e2134. |
| 50 | WANG H, CIMEN E, SINGH N, et al.. Deep learning for plant genomics and crop improvement [J]. Curr. Opin. Plant Biol., 2020,54:34-41. |
| 51 | PADOVANI DE SOUZA K, SETUBAL J C, PONCE DE LEON F D C A C, et al.. Machine learning meets genome assembly [J]. Brief Bioinform., 2019. 20(6):2116-2129. |
| 52 | CHAMPIGNY M J, UNDA F, SKYBA O, et al.. Learning from methylomes: epigenomic correlates of Populus balsamifera traits based on deep learning models of natural DNA methylation [J]. Plant Biotechnol. J., 2020, 18(6):1361-1375. |
| 53 | CHEN W, LV H, NIE F, et al.. i6mA-Pred: identifying DNA N6-methyladenine sites in the rice genome [J]. Bioinformatics, 2019, 35(16):2796-2800. |
| 54 | LV H, DAO F Y, GUAN Z X, et al.. iDNA6mA-rice: a computational tool for fetecting N6-methyladenine sites in rice [J/OL]. Front. Genet., 2019, 10:793 [2022-11-30]. . |
| 55 | PIAN C, ZHANG G, LI F, et al.. MM-6mAPred: identifying DNA N6-methyladenine sites based on Markov model [J]. Bioinformatics, 2020. 36(2):388-392. |
| 56 | YU H, DAI Z. SNNRice6mA: a eeep learning method for predicting DNA N6-methyladenine sites in rice genome [J]. Front. Genet., 2019, 10:107110 [2022-11-30]. . |
| 57 | ZHANG P, WANG Y, CHACHAR S, et al.. eRice: a refined epigenomic platform for japonica and indica rice [J]. Plant Biotechnol. J., 2020, 18(8):1642-1644. |
| 58 | WANG Y, ZHANG P, GUO W, et al.. A deep learning approach to automate whole-genome prediction of diverse epigenomic modifications in plants [J]. New Phytol., 2021, 232(2):880-897. |
| 59 | MA C, XIN M, FELDMANN K A, et al.. Machine learning-based differential network analysis: a study of stress-responsive transcriptomes in Arabidopsis [J]. Plant Cell, 2014, 26(2):520-537. |
| 60 | YE W, LIAN Q, YE C, et al.. A survey on methods for predicting polyadenylation sites from DNA sequences, bulk RNA-seq, and single-cell RNA-seq [J]. Genomics Proteomics Bioinformatics, 2022. |
| 61 | ZHAI J, SONG J, ZHANG T, et al.. deepEA: a containerized web server for interactive analysis of epitranscriptome sequencing data [J]. Plant Physiol., 2021, 185(1):29-33. |
| 62 | WU X, FENG H, WU D, et al.. Using high-throughput multiple optical phenotyping to decipher the genetic architecture of maize drought tolerance [J/OL]. Genome Biol., 2021, 22(1):185 [2022-11-30]. . |
| 63 | GUO W, LIU H, WANG Y, et al.. SMOC: a smart model for open chromatin region prediction in rice genomes [J]. J. Genet. Genomics, 2022, 49(5):514-517. |
| 64 | OBUDULU O, MäHLER N, SKOTARE T, et al.. A multi-omics approach reveals function of secretory carrier-associated membrane proteins in wood formation of Populus trees [J/OL]. BMC Genomics, 2018, 19(1):11 [2022-11-30]. . |
| 65 | MESNAGE R, AGAPITO-TENFEN S Z, VILPERTE V, et al.. An integrated multi-omics analysis of the NK603 Roundup-tolerant GM maize reveals metabolism disturbances caused by the transformation process [J/OL]. Sci. Rep., 2016,6:37855 [2022-11-30]. . |
| 66 | DE ABREU E L F, LI K, WEN W, et al.. Unraveling lipid metabolism in maize with time-resolved multi-omics data [J]. Plant J., 2018, 93(6):1102-1115. |
| 67 | GAVICHO UARROTA V, FUENTEALBA C, HERNáNDEZ I, et al.. Integration of proteomics and metabolomics data of early and middle season Hass avocados under heat treatment [J]. Food Chem., 2019, 289:512-521. |
| 68 | ZHOU J, THEESFELD C L, YAO K, et al.. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk [J]. Nat. Genet., 2018, 50(8):1171-1179. |
| 69 | AVSEC Ž, AGARWAL V, VISENTIN D, et al.. Effective gene expression prediction from sequence by integrating long-range interactions [J]. Nat. Methods, 2021, 18(10):1196-1203. |
| 70 | ZHOU J, TROYANSKAYA O G. Predicting effects of noncoding variants with deep learning-based sequence model [J]. Nat. Methods, 2015, 12(10):931-934. |
| 71 | ALIPANAHI B, DELONG A, WEIRAUCH M T, et al.. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning [J]. Nat. Biotechnol., 2015, 33(8):831-838. |
| 72 | AVSEC Ž, WEILERT M, SHRIKUMAR A, et al.. Base-resolution models of transcription-factor binding reveal soft motif syntax [J]. Nat. Genet., 2021, 53(3):354-366. |
| 73 | MOORE J E, PURCARO M J, PRATT H E, et al.. Expanded encyclopaedias of DNA elements in the human and mouse genomes [J]. Nature, 2020. 583(7818):699-710. |
| 74 | KELLEY D R, SNOEK J, RINN J L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks [J]. Genome Res., 2016, 26(7):990-999. |
| 75 | ZHAO H, LI J, YANG L, et al.. An inferred functional impact map of genetic variants in rice [J]. Mol. Plant, 2021, 14(9):1584-1599. |
| 76 | KELLEY D R, RESHEF Y A, BILESCHI M, et al.. Sequential regulatory activity prediction across chromosomes with convolutional neural networks [J]. Genome Res., 2018, 28(5):739-750. |
| 77 | ZHOU J, CHEN Q, BRAUN P R, et al.. Deep learning predicts DNA methylation regulatory variants in the human brain and elucidates the genetics of psychiatric disorders [J/OL]. Proc. Natl. Acad. Sci. USA, 2022, 119(34):e2206069119 [2022-11-30]. . |
| 78 | FUDENBERG G, KELLEY D R, POLLARD K S. Predicting 3D genome folding from DNA sequence with Akita [J]. Nat. Methods, 2020, 17(11):1111-1117. |
| 79 | ZHOU J. Sequence-based modeling of three-dimensional genome architecture from kilobase to chromosome scale [J]. Nat. Genet., 2022, 54(5):725-734. |
| 80 | Meuwissen T H E, Hayes B J, Goddard M E. Prediction of total genetic value using genome-wide dense marker maps [J]. Genetics, 2001, 157: 1819-1829. |
| 81 | ENDELMAN J B. Ridge regression and other kernels for genomic selection with R package rrBLUP [J]. Plant Genome, 2011, 4(3):250-255. |
| 82 | PéREZ P, DE LOS CAMPOS G. Genome-wide regression and prediction with the BGLR statistical package [J]. Genetics, 2014, 198(2):483-495. |
| 83 | YAN J, XU Y, CHENG Q, et al.. LightGBM: accelerated genomically designed crop breeding through ensemble learning [J/OL]. Genome Biol., 2021, 22(1):271 [2022-11-30]. . |
| 84 | MA W, QIU Z, SONG J, et al.. A deep convolutional neural network approach for predicting phenotypes from genotypes [J]. Planta, 2018,248(5):1307-1318. |
| 85 | LIU Y, WANG D, HE F, et al.. Phenotype prediction and genome-wide association study using deep convolutional neural network of soybean [J/OL]. Front. Genet., 2019, 10:1091 [2022-11-30]. . |
| 86 | WANG K, ABID M A, RASHEED A, et al.. DNNGP, a deep neural network-based method for genomic prediction using multi-omics data in plants [J/OL]. Mol. Plant, 2023,16:1-15[2022-11-30]. . |
| 87 | CHENG Q, JIANG S, XU F, et al.. Genome optimization via virtual simulation to accelerate maize hybrid breeding [J/OL]. Brief. Bioinform., 2022, 23(1):bbab447 [2022-11-30]. . |
| 88 | JIANG S, CHENG Q, YAN J, et al.. Genome optimization for improvement of maize breeding [J]. Theor. Appl. Genet., 2020, 133(5):1491-1502. |
| 89 | YANG L, ZHANG P, WANG Y, et al.. Plant synthetic epigenomic engineering for crop improvement [J]. Sci. China Life Sci., 2022,65(11):2191-220. |
| 90 | RODRíGUEZ-LEAL D, LEMMON Z H, MAN J, et al.. Engineering quantitative trait variation for crop improvement by genome editing [J]. Cell, 2017, 171(2):470-480. |
| 91 | YU Q, LIU S, YU L, et al.. RNA demethylation increases the yield and biomass of rice and potato plants in field trials [J]. Nat. Biotechnol., 2021, 39(12):1581-1588. |
| [1] | Hai WANG, Jinsheng LAI, Haiyang WANG, Xinhai LI. Bipartite Intelligent Design of Crops—Intelligent Combination of Natural Variation and Intelligent Creation of Artificial Variation [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 1-8. |
| [2] | YUAN Fang, GUO Pei-yuan, WU Hao, SUN Mei. Study on the Development of Pork Freshness Detection Techniques [J]. , 2009, 11(S1): 72-74. |
| [3] | WEI Hua-li, YANG Wen-hua, HAN Su-ying, QI Li-wang. Research Strategy of Epigenetics and its Utilization in Wooden Plants [J]. , 2009, 11(2): 10-16. |
| [4] | ZHENG Xiao-mei, WU Ning-feng. Biological Function of DNA Methylation [J]. , 2009, 11(1): 33-39. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号