




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (2): 93-103.DOI: 10.13304/j.nykjdb.2021.0389
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Shaojing MO( ), Zhicheng WANG(
), Zhicheng WANG( ), Xingfen WANG, Zhengwen LIU, Liqiang WU, Guiyin ZHANG, Zhiying MA, Yan ZHANG(
), Xingfen WANG, Zhengwen LIU, Liqiang WU, Guiyin ZHANG, Zhiying MA, Yan ZHANG( ), Huijun DUAN(
), Huijun DUAN( )
)
Received:2021-04-26
Accepted:2021-06-01
Online:2022-02-15
Published:2022-02-22
Contact:
Yan ZHANG,Huijun DUAN
默韶京( ), 王志城(
), 王志城( ), 王省芬, 刘正文, 吴立强, 张桂寅, 马峙英, 张艳(
), 王省芬, 刘正文, 吴立强, 张桂寅, 马峙英, 张艳( ), 段会军(
), 段会军( )
)
通讯作者:
张艳,段会军
作者简介:默韶京和王志城为本文共同第一作者。默韶京 E-mail:msjing1983@163.com基金资助:CLC Number:
Shaojing MO, Zhicheng WANG, Xingfen WANG, Zhengwen LIU, Liqiang WU, Guiyin ZHANG, Zhiying MA, Yan ZHANG, Huijun DUAN. Genome-wide Identification of GELP Family Genes in Cotton and Expression Analysis Under Stress[J]. Journal of Agricultural Science and Technology, 2022, 24(2): 93-103.
默韶京, 王志城, 王省芬, 刘正文, 吴立强, 张桂寅, 马峙英, 张艳, 段会军. 陆地棉GELP家族基因鉴定及其响应胁迫的表达分析[J]. 中国农业科技导报, 2022, 24(2): 93-103.
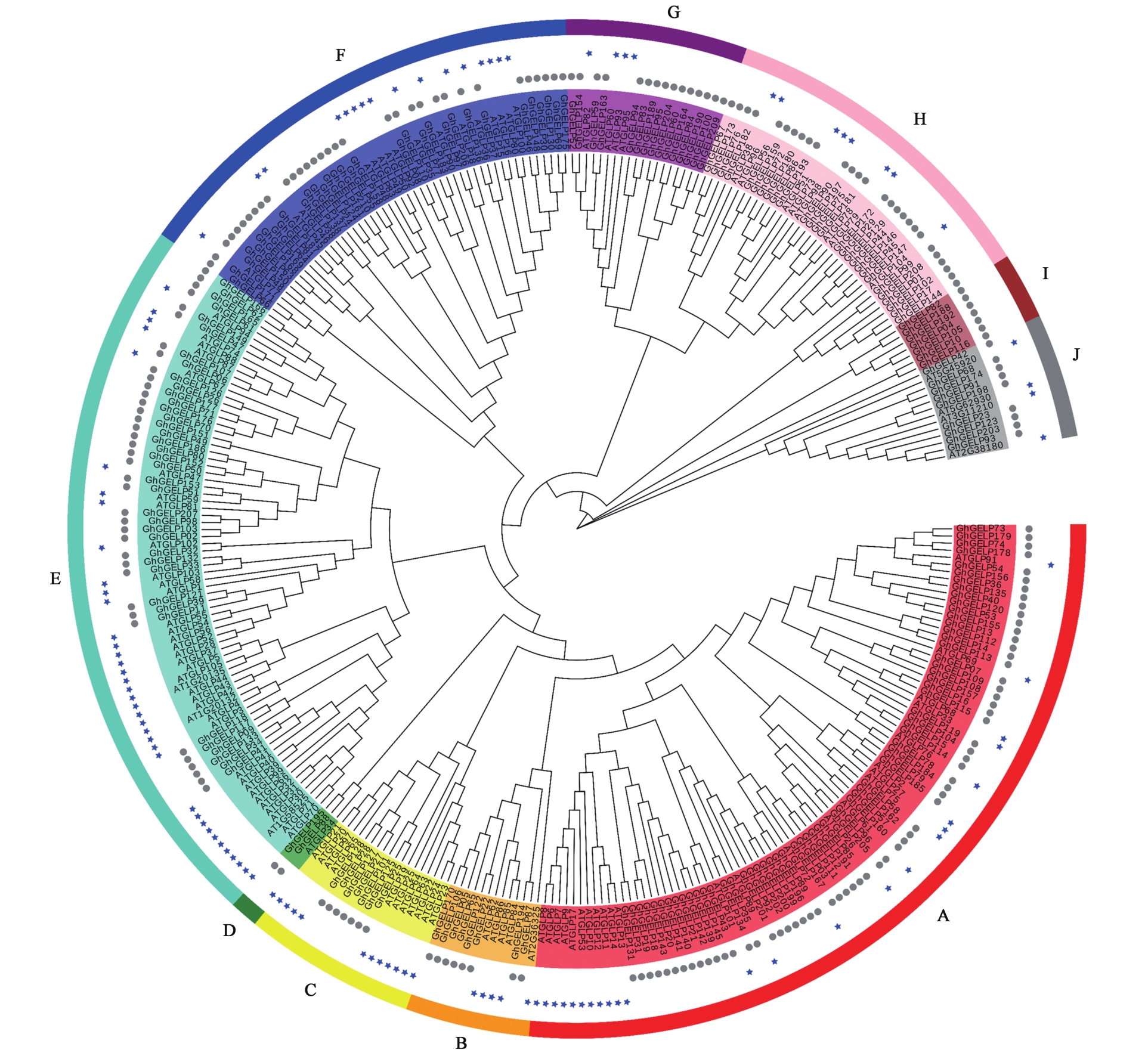
Fig.1 Phylogenetic analysis of GELP proteins in cotton and ArabidopsisNote: Phylogenetic tree of GELP proteins from Arabidopsis and G. hirsutum. The GELP members are divided into 10 subfamilies(A~J). The branches of different subfamilies are marked using different colors.

Fig.3 Motif logos of four conservative blocks detected in GhGELP proteinsNote: Conservative amino acid residue Ser-Gly-Asn-His is marked by red triangle.
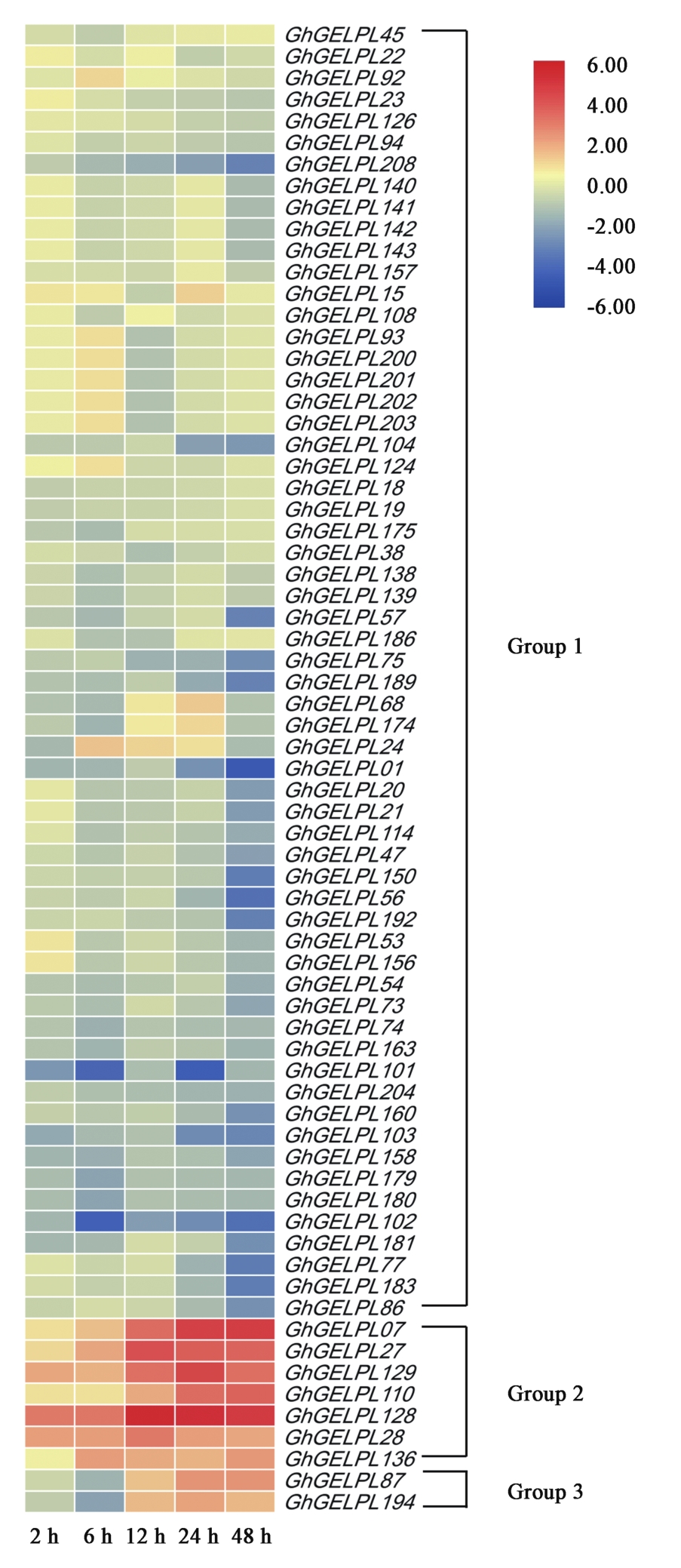
Fig.4 Expression profiles of GhGELPs from upland cotton inoculated with V. dahliaeNote: Red color indicates up-regulation expression. Blue color indicates down-regulation expression. 2 h, 6 h, 12 h, 24 h and 48 h represent the processing time under V.dahliae stress conditions.
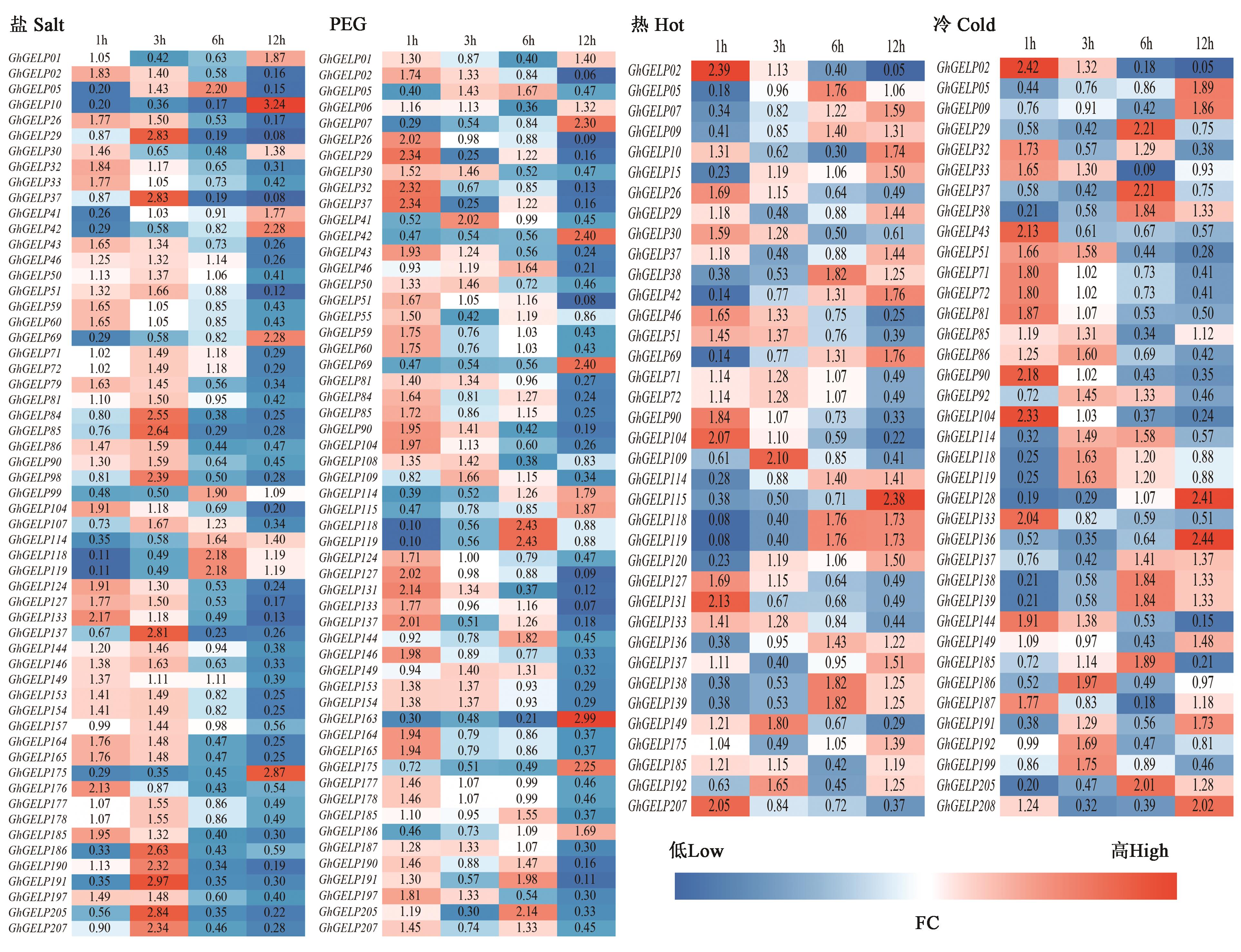
Fig.5 Expression profiles of GhGELPs in response to different abiotic stressNote:FC is the ratio of treatment FPKM to control FPKM. Red color indicates up-regulation expression, blue color indicates down-regulation expression. 1 h, 3 h, 6 h, and 12 h represent the processing time under four abiotic stress conditions.
| 基因名称 Gene name | 盐 Salt | PEG | 热 Hot | 冷 Cold | 基因名称 Gene name | 盐 Salt | PEG | 热 Hot | 冷 Cold |
|---|---|---|---|---|---|---|---|---|---|
| GhGELP1 | √ | √ | GhGELP109 | √ | √ | ||||
| GhGELP2 | √ | √ | √ | √ | GhGELP114 | √ | √ | √ | √ |
| GhGELP5 | √ | √ | √ | √ | GhGELP115 | √ | √ | ||
| GhGELP6 | √ | GhGELP118 | √ | √ | √ | √ | |||
| GhGELP7 | √ | √ | GhGELP119 | √ | √ | √ | √ | ||
| GhGELP9 | √ | √ | GhGELP120 | √ | |||||
| GhGELP10 | √ | √ | GhGELP124 | √ | |||||
| GhGELP15 | √ | GhGELP127 | √ | √ | √ | ||||
| GhGELP26 | √ | √ | √ | GhGELP128 | √ | ||||
| GhGELP29 | √ | √ | √ | √ | GhGELP131 | √ | √ | ||
| GhGELP30 | √ | √ | √ | GhGELP133 | √ | √ | √ | √ | |
| GhGELP32 | √ | √ | √ | GhGELP136 | √ | √ | |||
| GhGELP33 | √ | √ | GhGELP137 | √ | √ | √ | √ | ||
| GhGELP37 | √ | √ | √ | √ | GhGELP138 | √ | √ | ||
| GhGELP38 | √ | √ | GhGELP139 | √ | √ | ||||
| GhGELP41 | √ | √ | GhGELP144 | √ | √ | √ | |||
| GhGELP42 | √ | √ | √ | GhGELP146 | √ | √ | |||
| GhGELP43 | √ | √ | √ | GhGELP149 | √ | √ | √ | √ | |
| GhGELP46 | √ | √ | √ | GhGELP153 | √ | √ | |||
| GhGELP50 | √ | √ | GhGELP154 | √ | √ | ||||
| GhGELP51 | √ | √ | √ | √ | GhGELP157 | √ | |||
| GhGELP55 | √ | GhGELP163 | √ | ||||||
| GhGELP59 | √ | √ | GhGELP164 | √ | √ | ||||
| GhGELP60 | √ | √ | GhGELP165 | √ | √ | ||||
| GhGELP69 | √ | √ | √ | GhGELP175 | √ | √ | √ | ||
| GhGELP71 | √ | √ | √ | GhGELP176 | √ | ||||
| GhGELP72 | √ | √ | √ | GhGELP177 | √ | √ | |||
| GhGELP79 | √ | GhGELP178 | √ | √ | |||||
| GhGELP81 | √ | √ | √ | GhGELP185 | √ | √ | √ | √ | |
| GhGELP84 | √ | √ | GhGELP186 | √ | √ | √ | |||
| GhGELP85 | √ | √ | √ | GhGELP187 | √ | √ | |||
| GhGELP86 | √ | √ | GhGELP190 | √ | √ | ||||
| GhGELP90 | √ | √ | √ | √ | GhGELP191 | √ | √ | √ | |
| GhGELP92 | √ | GhGELP192 | √ | √ | |||||
| GhGELP98 | √ | GhGELP197 | √ | √ | |||||
| GhGELP99 | √ | GhGELP199 | √ | ||||||
| GhGELP104 | √ | √ | √ | √ | GhGELP205 | √ | √ | √ | |
| GhGELP107 | √ | GhGELP207 | √ | √ | √ | ||||
| GhGELP108 | √ | GhGELP208 | √ |
Table 1 GhGELP genes response to different abiotic stress
| 基因名称 Gene name | 盐 Salt | PEG | 热 Hot | 冷 Cold | 基因名称 Gene name | 盐 Salt | PEG | 热 Hot | 冷 Cold |
|---|---|---|---|---|---|---|---|---|---|
| GhGELP1 | √ | √ | GhGELP109 | √ | √ | ||||
| GhGELP2 | √ | √ | √ | √ | GhGELP114 | √ | √ | √ | √ |
| GhGELP5 | √ | √ | √ | √ | GhGELP115 | √ | √ | ||
| GhGELP6 | √ | GhGELP118 | √ | √ | √ | √ | |||
| GhGELP7 | √ | √ | GhGELP119 | √ | √ | √ | √ | ||
| GhGELP9 | √ | √ | GhGELP120 | √ | |||||
| GhGELP10 | √ | √ | GhGELP124 | √ | |||||
| GhGELP15 | √ | GhGELP127 | √ | √ | √ | ||||
| GhGELP26 | √ | √ | √ | GhGELP128 | √ | ||||
| GhGELP29 | √ | √ | √ | √ | GhGELP131 | √ | √ | ||
| GhGELP30 | √ | √ | √ | GhGELP133 | √ | √ | √ | √ | |
| GhGELP32 | √ | √ | √ | GhGELP136 | √ | √ | |||
| GhGELP33 | √ | √ | GhGELP137 | √ | √ | √ | √ | ||
| GhGELP37 | √ | √ | √ | √ | GhGELP138 | √ | √ | ||
| GhGELP38 | √ | √ | GhGELP139 | √ | √ | ||||
| GhGELP41 | √ | √ | GhGELP144 | √ | √ | √ | |||
| GhGELP42 | √ | √ | √ | GhGELP146 | √ | √ | |||
| GhGELP43 | √ | √ | √ | GhGELP149 | √ | √ | √ | √ | |
| GhGELP46 | √ | √ | √ | GhGELP153 | √ | √ | |||
| GhGELP50 | √ | √ | GhGELP154 | √ | √ | ||||
| GhGELP51 | √ | √ | √ | √ | GhGELP157 | √ | |||
| GhGELP55 | √ | GhGELP163 | √ | ||||||
| GhGELP59 | √ | √ | GhGELP164 | √ | √ | ||||
| GhGELP60 | √ | √ | GhGELP165 | √ | √ | ||||
| GhGELP69 | √ | √ | √ | GhGELP175 | √ | √ | √ | ||
| GhGELP71 | √ | √ | √ | GhGELP176 | √ | ||||
| GhGELP72 | √ | √ | √ | GhGELP177 | √ | √ | |||
| GhGELP79 | √ | GhGELP178 | √ | √ | |||||
| GhGELP81 | √ | √ | √ | GhGELP185 | √ | √ | √ | √ | |
| GhGELP84 | √ | √ | GhGELP186 | √ | √ | √ | |||
| GhGELP85 | √ | √ | √ | GhGELP187 | √ | √ | |||
| GhGELP86 | √ | √ | GhGELP190 | √ | √ | ||||
| GhGELP90 | √ | √ | √ | √ | GhGELP191 | √ | √ | √ | |
| GhGELP92 | √ | GhGELP192 | √ | √ | |||||
| GhGELP98 | √ | GhGELP197 | √ | √ | |||||
| GhGELP99 | √ | GhGELP199 | √ | ||||||
| GhGELP104 | √ | √ | √ | √ | GhGELP205 | √ | √ | √ | |
| GhGELP107 | √ | GhGELP207 | √ | √ | √ | ||||
| GhGELP108 | √ | GhGELP208 | √ |
| 1 | ARPIGNY J, JAEGER K. Bacterial lipolytic enzymes:classification and properties [J]. Biochem. J., 1999, 343(1): 177-183. |
| 2 | UPTON C, BUCKLEY J T. A new family of lipolytic enzymes? [J]. Trends Biochem. Sci., 1995, 20(5): 178-179. |
| 3 | AKOH C C, LEE G C, LIAW Y C, et al.. GDSL family of serine esterases/lipases [J]. Prog. Lipid Res., 2004, 43(6): 534-552. |
| 4 | JIANG Y Y, CHEN R J, DONG J L, et al.. Analysis of GDSL lipase (GLIP) family genes in rice (Oryza sativa) [J]. Plant Omics, 2012, 5(4): 351-358. |
| 5 | CHEPYSHKO H L, CHIA P H, LI M, et al.. Multifunctionality and diversity of GDSL esterase/ lipase gene family in rice (Oryza sativa L. japonica) genome: new insights from bioinformatics analysis [J/OL]. BMC Genomics, 2012, 13(1):309 [2021-06-09]. . |
| 6 | KWON S J, JIN H C, LEE S, et al.. GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis [J]. Plant J., 2009, 58(2): 235-245. |
| 7 | KIM H G, KWON S J, JANG Y J, et al.. GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling [J]. Plant Physiol., 2013, 163(4): 1776-1791. |
| 8 | KIM H G, KWON S J, JANG Y J, et al.. GDSL lipase 1 regulates ethylene signaling and ethylene-associated systemic immunity in Arabidopsis [J]. FEBS Lett., 2014, 588(9): 1652-1658. |
| 9 | LEE D S, KIM B K, KWON S J, et al.. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling [J]. Biochem. Biophys. Res. Commun., 2009, 379(4): 1038-1042. |
| 10 | RAJARAMMOAN S, PRADHAN A K, PENTAL D, et al.. Genome-wide association mapping in Arabidopsis identifies novel genes underlying quantitative disease resistance to Alternaria brassicae [J]. Mol. Plant Pathol., 2018, 19(7): 1719-1732. |
| 11 | DING L N, LI M G, XIAO J, et al.. ArabidopsisGDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus [J]. Plant Biotechnol. J., 2020, 18(5): 1255-1270. |
| 12 | HONG J K, CHOI H W, HWANG I S, et al.. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance [J]. Planta, 2008, 227(3): 539-558. |
| 13 | NARANJO M A, FORMENT J, ROLDAN M, et al.. Overexpression of Arabidopsis thaliana LTL1, a salt-induced gene encoding a GDSL-motif lipase, increases salt tolerance in yeast and transgenic plants [J]. Plant Cell Environ., 2006, 29(10): 1890-1900. |
| 14 | KIM K J, LIM J H, KIM M J, et al.. GDSL-lipase1 (CaGL1) contributes to wound stress resistance by modulation of CaPR-4 expression in hot pepper [J].Biochem. Biophys. Res. Commun., 2008, 374(4): 693-698. |
| 15 | LAI C P, HUANG L M, CHRN L F O, et al.. Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis [J]. Plant Mol. Biol., 2017, 95: 181-197. |
| 16 | DONG X, YI H, HAN C T, et al.. GDSL esterase/lipase genes in Brassica rapa L.: genome-wide identification and expression analysis [J]. Mol. Genet. Genomics, 2016, 291(2): 531-542. |
| 17 | NI P Y, JI X R, GUO D L, et al.. Genome-wide identification, characterization, and expression analysis of GDSL-type esterases/lipases gene family in relation to grape berry ripening [J]. Sci. Hortic., 2020, 264: 109162. |
| 18 | SU H G, ZHANG X H, WANG T T, et al.. Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybean [J/OL]. Front. Plant Sci., 2020, 11: 726[2021-06-09]. . |
| 19 | BOLEK Y, EL-ZIK K M, PEPPER A E, et al.. Mapping of verticillium wilt resistance genes in cotton [J]. Plant Sci., 2005. 168(6): 1581-1590. |
| 20 | CAI Y F, HE X H, MO J C, et al.. Molecular research and genetic engineering of resistance to Verticillium wilt in cotton [J]. Afr. J. Biotechnol., 2009, 8(25): 7363-7372. |
| 21 | SHABAN M, MIAO Y, ULLAH A, et al.. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae [J]. Plant Physiol. Biochem., 2018, 125: 193-204. |
| 22 | JIMENEZ-DIAZ R M, GARCIA C O, TRAPERO-CASAS J L, et al.. Variation of pathotypes and races and their correlations with clonal lineages in Verticillium dahliae [J]. Plant Pathol., 2017, 66(4): 651-666. |
| 23 | OH I S, PARK A R, BAE M S, et al.. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola [J]. Plant Cell, 2005, 17(10): 2832-2847. |
| 24 | WANG G N, WANG X F, ZHANG Y, et al.. Dynamic characteristics and functional analysis provide new insights into long non-coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum [J]. BMC Plant Biol., 2021, 21(1): 68 [2021-06-09]. . |
| 25 | YADAV V K, YADAV V K, PANT P, et al.. GhMYB1 regulates SCW stage-specific expression of the GhGDSL promoter in the fibres of Gossypium hirsutum L [J]. Plant Biotechnol. J., 2017, 15(9): 1163-1174. |
| 26 | ZHANG J S, MA R, YUAN H, et al.. A Gossypium hirsutum GDSL lipase/hydrolase gene (GhGLIP) appears to be involved in promoting seed growth in Arabidopsis [J/OL]. PLoS One, 2018, 13(4): e0195556 [2021-06-09]. . |
| 27 | TAKAHASHI K, SHIMADA T, KONDO M, et al.. Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana [J]. Plant Cell Physiol., 2010, 51(1): 123-131. |
| 28 | GAO M, YIN X, YANG W, et al.. GDSL lipases modulate immunity through lipid homeostasis in rice [J/OL]. PLoS Pathog., 2017, 13(11): e1006724[2021-06-09]. . |
| 29 | ZHANG B C, ZHANG L J, LI F, et al.. Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase [J]. Nat. Plants, 2017, 3(3): 17017 [2021-06-09]. . |
| 30 | UPDEGRAFF E P, Zhao F, PREUSS D, et al.. The extracellular lipase EXL4 is required for efficient hydration of Arabidopsis pollen [J]. Sex. Plant Reprod., 2009, 22(3): 197-204. |
| 31 | ZHAO J, LONG T, WANG Y F, et al.. RMS2 Encoding a GDSL lipase mediates lipid homeostasis in anthers to determine rice male fertility [J]. Plant Physiol., 2020, 182(4): 2047-2064. |
| 32 | YU Y, WOO M O, RIHUA P, et al.. The DROOPING LEAF (DR) gene encoding GDSL esterase is involved in silica deposition in rice (Oryza sativa L.) [J/OL]. PLoS One, 2020, 15(9): e0238887 [2021-06-09]. . |
| 33 | LIM G H, SINGHAL R, KACHROO A, et al.. Fatty acid-and lipid-mediated signaling in plant defense [J]. Annu. Rev. Phytopathol., 2017, 55(1): 505-536. |
| 34 | WALLEY J W, KLIEBENSTEIN D J, BOSTOCK R M, et al.. Fatty acids and early detection of pathogens [J]. Curr. Opin. Plant Biol., 2013, 16(4): 520-526. |
| [1] | SU Yue§, LIU Juanjuan§, WAN Bin, ZHANG Pengju, CHEN Zhenggen, SU Junji, WANG Caixiang. Chloroplast Genome Structure Characteristic and Phylogenetic Analysis of Mulgedium tataricum [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 33-42. |
| [2] | PU Weijun, TAN Binglan, ZHU Li*. Progress on the Biological Functions of Argonaute Proteins in Response to Stress in Plants [J]. Journal of Agricultural Science and Technology, 2021, 23(2): 17-26. |
| [3] | SHI Mengmeng1, WEN Siyu1, ZHAO Jiajia2, QIAO Ling2, WU Bangbang2, ZHENG Xingwei1,2*, ZHENG Jun1,2*. Identification, Evolution and Stress Response of RCAR Family Genes in Wheat (Trticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2020, 22(8): 14-24. |
| [4] | GE Chuan1, YANG Rong2, LI Liujun2, ZHANG Jiancheng2, ZHENG Xingwei2*. Genome-Wide Identification and Characterization of the YABBY Family Genes of Wheat (Trticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2019, 21(8): 11-18. |
| [5] | HUANG Fei1,2, LI Xuemei1*, WANG Wensheng2*, FU Binying2. Research Progress of DNA Methylation in Stress Response and Breeding in Plant [J]. , 2013, 15(6): 83-91. |
| [6] | SONG Peiyong, ZHENG Yaqiang, LI Bin, XIAO Zhongjiu. Studies on Antimicrobial Activity of Actinomycetes Isolated from Soils of Chishui River Basin [J]. , 2013, 15(1): 136-143. |
| [7] | FENG Lei, ZHANG Hai-wen*, HUANG Rong-feng. Research Progress on LRR Receptorlike Protein Kinase in Plant [J]. , 2012, 14(6): 43-48. |
| [8] | SHI Gong-yao1, WANG Yu-mei2, HUA Jin-ping1. Aquaporins and Salt Tolerance of Higher Plant [J]. , 2012, 14(4): 31-38. |
| [9] | XU Tao, WANG Lei. Research Progress on Plant RNA-dependant RNA Polymerase [J]. , 2011, 13(6): 46-53. |
| [10] | ZHAO Jin-feng1, YU Ai-li2, WANG Gao-hong1, TIAN Gang1, WANG Han-yu2, Du Yan-wei1. Progress of CBL/CIPK Signal System in Response to Stresses in Plant [J]. , 2011, 13(4): 32-38. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号