




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (9): 112-121.DOI: 10.13304/j.nykjdb.2023.0407
• ANIMAL AND PLANT HEALTH • Previous Articles
Huiting WENG1,2( ), Haiyang LIU3, Huiming GUO4, Hongmei CHENG4, Jun LI1,2, Chao ZHANG1,2(
), Haiyang LIU3, Huiming GUO4, Hongmei CHENG4, Jun LI1,2, Chao ZHANG1,2( ), Xiaofeng SU4(
), Xiaofeng SU4( )
)
Received:2023-05-25
Accepted:2023-07-26
Online:2024-09-15
Published:2024-09-13
Contact:
Chao ZHANG,Xiaofeng SU
翁慧婷1,2( ), 刘海洋3, 郭惠明4, 程红梅4, 李君1,2, 张超1,2(
), 刘海洋3, 郭惠明4, 程红梅4, 李君1,2, 张超1,2( ), 苏晓峰4(
), 苏晓峰4( )
)
通讯作者:
张超,苏晓峰
作者简介:翁慧婷 E-mail:wht-bio@163.com
基金资助:CLC Number:
Huiting WENG, Haiyang LIU, Huiming GUO, Hongmei CHENG, Jun LI, Chao ZHANG, Xiaofeng SU. Preliminary Function Analysis of GhERF020 Gene in Response to Verticillium Wilt in Cotton[J]. Journal of Agricultural Science and Technology, 2024, 26(9): 112-121.
翁慧婷, 刘海洋, 郭惠明, 程红梅, 李君, 张超, 苏晓峰. 棉花抗黄萎病相关基因GhERF020功能的初步分析[J]. 中国农业科技导报, 2024, 26(9): 112-121.
引物名称 Primer name | |
|---|---|
| p1132 | |
| p1132 | |
| GhUBQ7-RT-F | |
| GhUBQ7-RT-R | |
| Vd | |
| Vd |
Table 1 Primers used in this study
引物名称 Primer name | |
|---|---|
| p1132 | |
| p1132 | |
| GhUBQ7-RT-F | |
| GhUBQ7-RT-R | |
| Vd | |
| Vd |
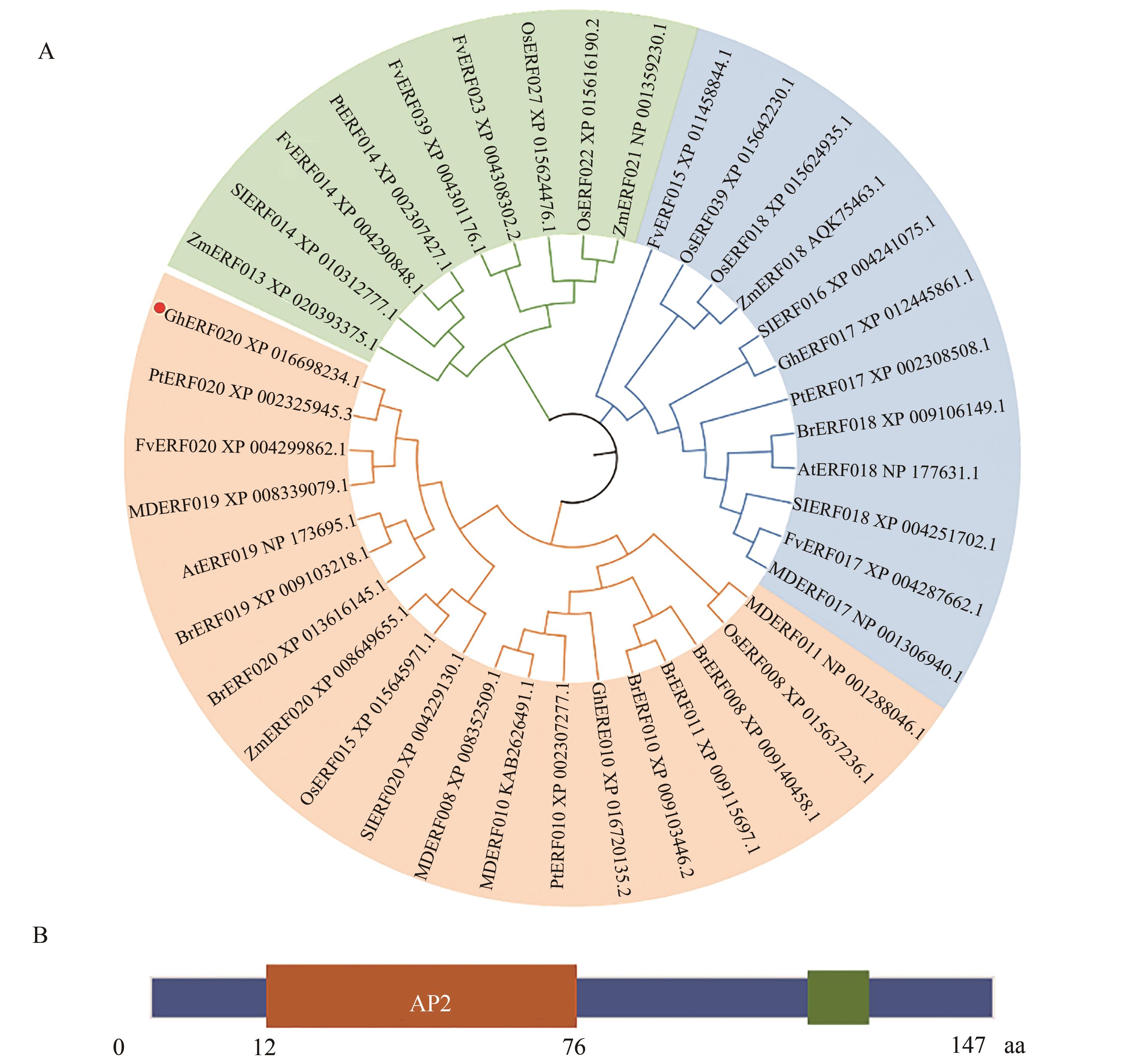
Fig. 1 Homology and structure analysis of GhERF020 in cottonA: Phylogenetic tree analysis of GhERF020 homologous protein in different species; B: GhERF020 domain analysis
| 指标Index | 值Value |
|---|---|
| 氨基酸数量Number of amino acid/aa | 147 |
| 分子量Molecular weight/kD | 16.16 |
| 分子式Formula | C691H1095N207O221S10 |
| 理论等电点Theoretical pI | 7.79 |
| 脂肪族氨基酸指数Aliphatic index | 60.48 |
| 不稳定性指数Instability index | 73.88 |
| N端N-terminal | Met |
总平均亲水性 Grand average of hydropathicity | -0.586 |
Table 2 Physical and chemical properties of GhERF020
| 指标Index | 值Value |
|---|---|
| 氨基酸数量Number of amino acid/aa | 147 |
| 分子量Molecular weight/kD | 16.16 |
| 分子式Formula | C691H1095N207O221S10 |
| 理论等电点Theoretical pI | 7.79 |
| 脂肪族氨基酸指数Aliphatic index | 60.48 |
| 不稳定性指数Instability index | 73.88 |
| N端N-terminal | Met |
总平均亲水性 Grand average of hydropathicity | -0.586 |
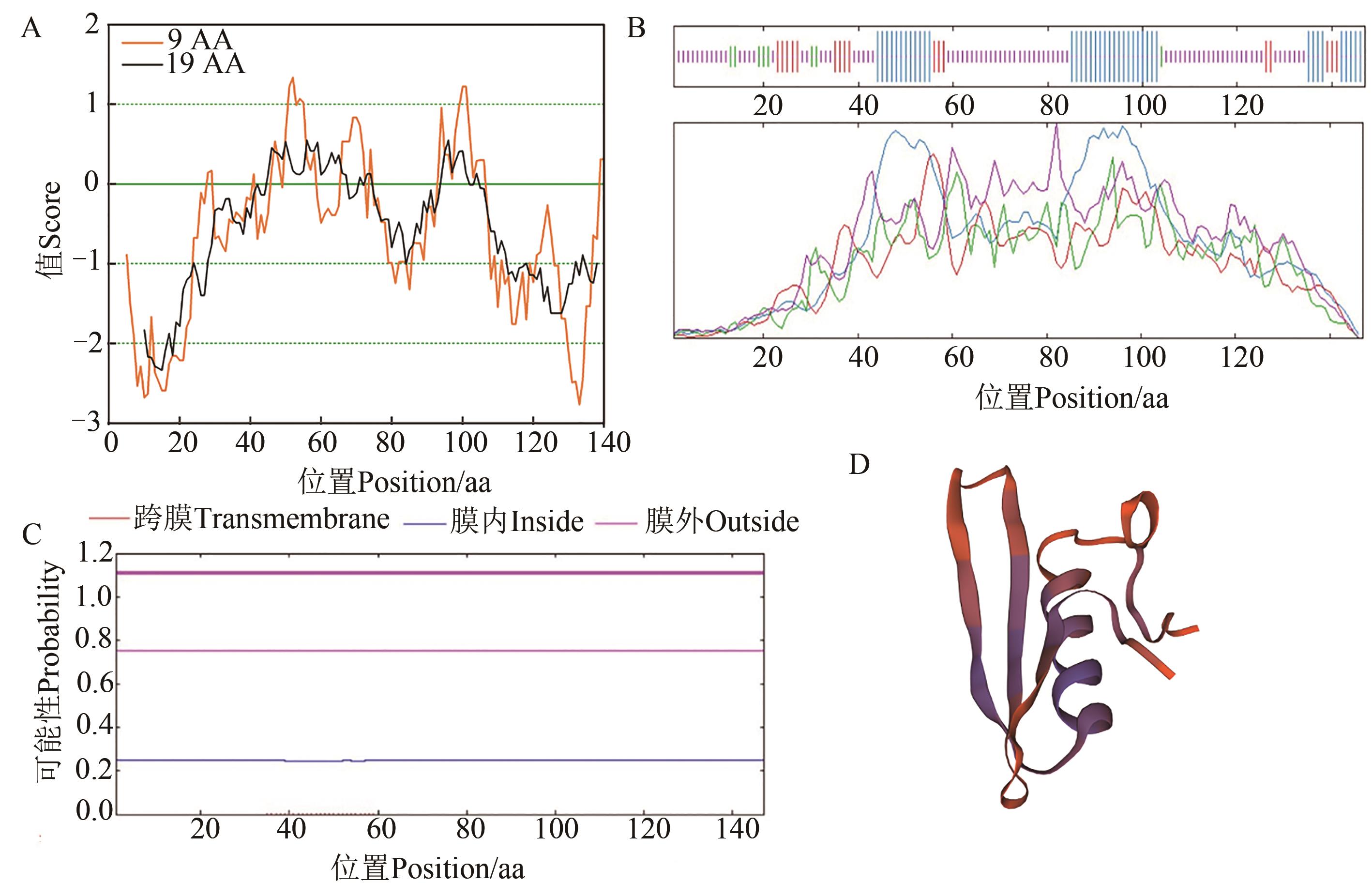
Fig. 2 Physicochemical properties of GhERF020A: Hydrophilic analysis of GhERF020; B: Transmembrane prediction of amino acid sequence of GhERF020; C: Secondary structure of GhERF020; D: Tertiary structure of GhERF020
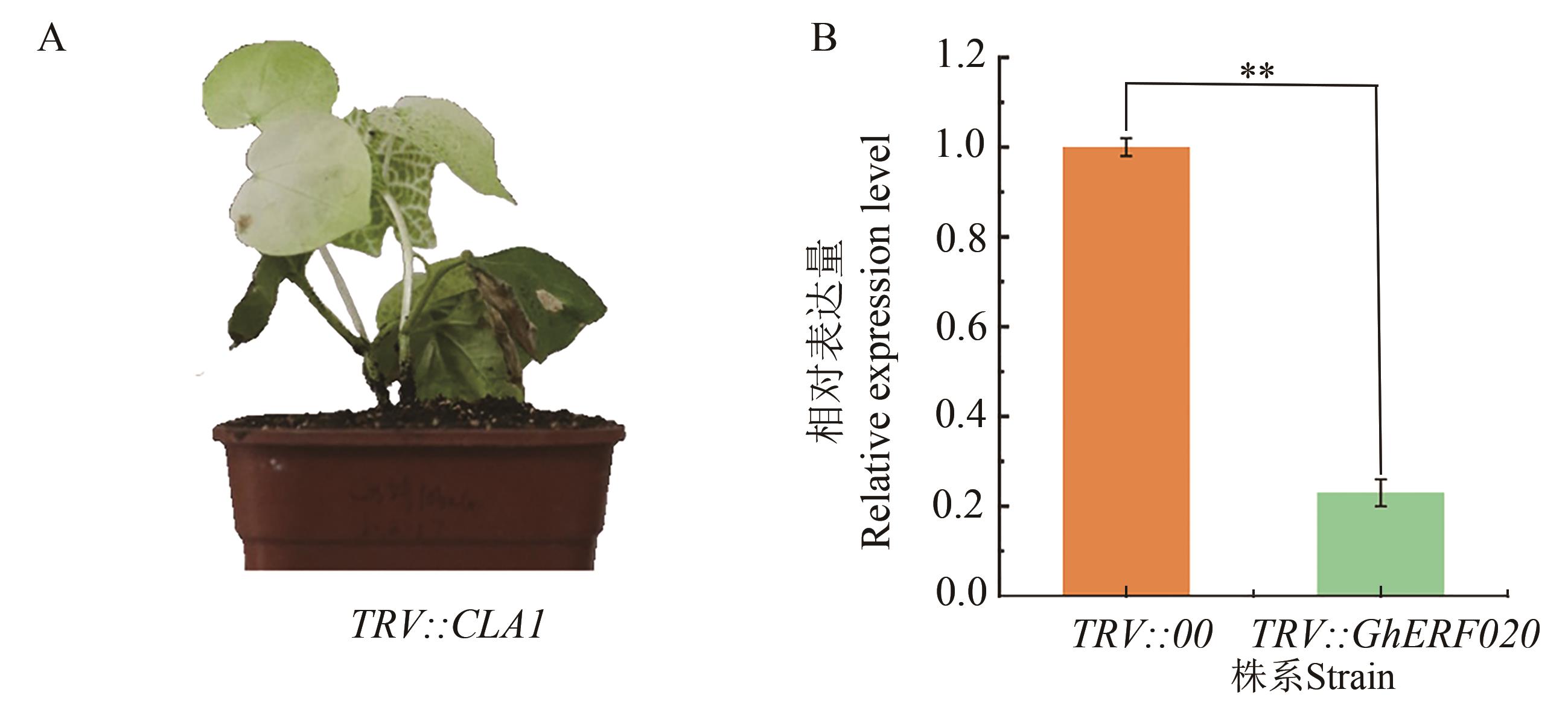
Fig. 6 GhERF020 silencing efficiencyA: TRV:: CLA1 plant; B: Gene expression of GhERF020 in cotton plants; ** indicates significant difference at P<0.01 level
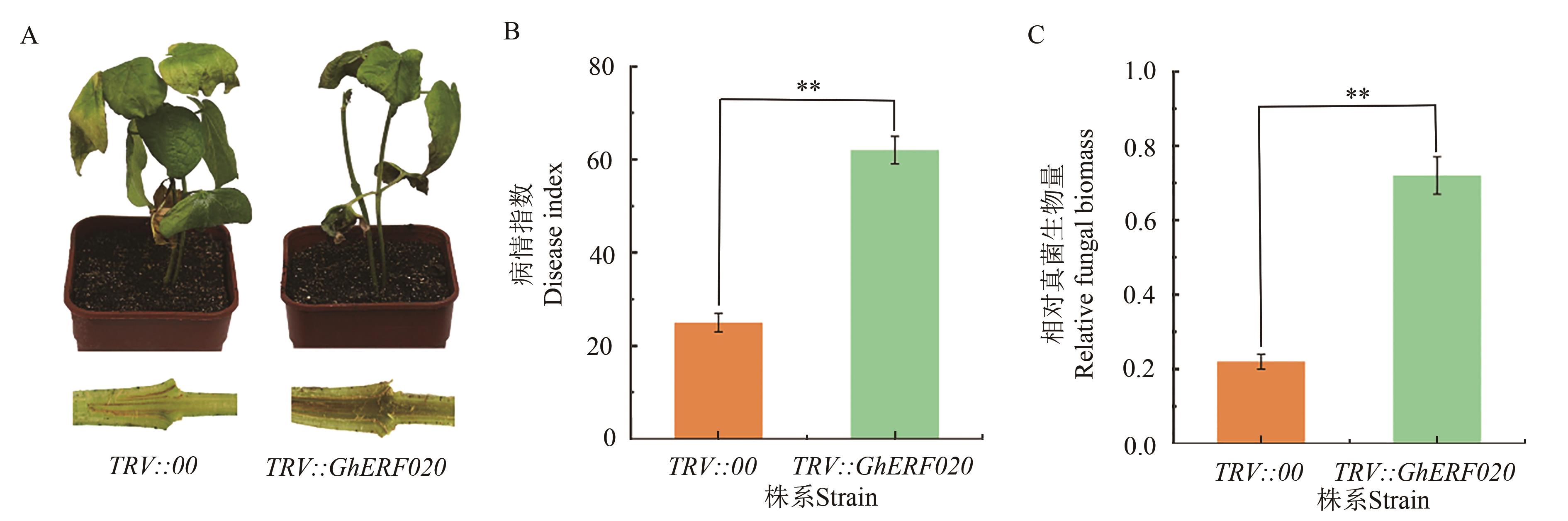
Fig. 7 Identification of GhERF020 gene-silenced cotton in resistance to VWA: Phenotype of TRV:: 00 and TRV:: GhERF020 plants after infection 10 d with V991; B: Disease indices of TRV:: 00 and TRV:: GhERF020 cotton plants after 10 d post inoculation; C: Fungal biomass of TRV:: 00 and TRV:: GhERF020 plants after infection 10 d with V991. ** indicates significant difference at P<0.01 level
| 1 | 范鑫,赵雷霖,翟红红,等. AtNEK6在棉花旱盐胁迫响应中的功能分析[J].中国农业科学,2018,51(22):4230-4240. |
| FAN X, ZHAO L L, ZHAI H H, et al.. Functional characterization of AtNEK6 overexpression in cotton under drought and salt stress [J]. Sci. Agric. Sin., 2018, 51(22):4230-4240. | |
| 2 | WANG P, ZHOU L, JAMIESON P, et al.. The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors [J]. Plant Cell, 2020, 32(12):3978-4001. |
| 3 | 李秀青,李月,刘超,等.棉花黄萎病相关基因GhAAT的克隆与功能鉴定[J].分子植物育种,2020,18(4):1048-1053. |
| LI X Q, LI Y, LIU C, et al.. Cloning and functional identification of cotton Verticillium wilt related gene GhAAT [J]. Mol. Plant. Breed., 2020, 18(4):1048-1053. | |
| 4 | 张计育,王庆菊,郭忠仁.植物AP2/ERF类转录因子研究进展[J].遗传,2012,34(7):835-847. |
| ZHANG J Y, WANG Q J, GUO Z R. Progresses on plant AP2/ERF transcription factors [J]. Hereditas, 2012, 34(7):835-847. | |
| 5 | NAKANO T, SUZUKI K, FUJIMURA T, et al.. Genome-wide analysis of the ERF gene family in Arabidopsis and rice [J]. Plant Physiol., 2006, 140(2):411-432. |
| 6 | 莫纪波,李大勇,张慧娟,等.ERF转录因子在植物对生物和非生物胁迫反应中的作用[J].植物生理学报,2011,47(12):1145-1154. |
| MO J B, LI D Y, ZHANG H J, et al.. Roles of ERF transcription factors in biotic and abiotic stress response in plants [J]. Acta Phytophysiol. Sin., 2011, 47(12):1145-1154. | |
| 7 | ZHANG G Y, CHEN M, CHEN X P, et al.. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) [J]. J. Exp. Bot., 2008, 59(15):4095-4107. |
| 8 | SHARMA M K, KUMAR R, SOLANKE A U, et al.. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato [J]. Mol. Genet. Genom., 2010, 284(6):455-475. |
| 9 | ZHUANG J, CAI B, PENG R H, et al.. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa [J]. Biochem. Biophys. Res. Commun., 2008, 371(3):468-474. |
| 10 | BERROCAL-LOBO M, MOLINA A, SOLANO R. Constitutive expression of ethylene‐response‐factor1 in Arabidopsis confers resistance to several necrotrophic fungi [J]. Plant J., 2002, 29(1):23-32. |
| 11 | CHARFEDDINE M, SAMET M, CHARFEDDINE S, et al.. Ectopic expression of StERF94 transcription factor in potato plants improved resistance to Fusarium solani infection [J]. Plant Mol. Biol. Rep., 2019, 37(5):450-463. |
| 12 | LIU D F, CHEN X J, LIU J Q, et al.. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance [J]. J. Exp. Bot., 2012, 63(10):3899-3911. |
| 13 | WANG F J, WANG C L, LIU P Q, et al.. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922 [J/OL]. PLoS One, 2016, 11(4):e0154027 [2023-04-30]. . |
| 14 | LU W Q, DENG F Y, JIA J B, et al.. The Arabidopsis thaliana gene AtERF019 negatively regulates plant resistance to Phytophthora parasitica by suppressing PAMP-triggered immunity [J]. Mol. Plant Pathol., 2020, 21(9):1179-1193. |
| 15 | FEYS B J, PARKER J E. Interplay of signaling pathways in plant disease resistance [J]. Trends Genet., 2000, 16(10):449-455. |
| 16 | GLAZEBROOK J. Genes controlling expression of defense responses in Arabidopsis [J]. Curr. Opin. Plant Biol., 2001, 4(4):301-308. |
| 17 | 吴朝峰. 棉花腺体形成相关基因GhERF105的功能分析[D]. 武汉:华中农业大学, 2020. |
| WU C F. Functional analysis of GhERF105 gene related to gland formation in cotton [D]. Wuhan: Huazhong Agricultural University, 2020. | |
| 18 | LORENZO O, PIQUERAS R, SANCHEZ-SERRANO J J, et al.. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense [J]. Plant Cell, 2003, 15(1):165-178. |
| 19 | CATINOT J, HUANG J B, HUANG P Y, et al.. Ethylene respense factor 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate-and ethylen-responsive defence genes [J]. Plant Cell Environ., 2015, 38(12):2721-2734. |
| 20 | ZAFAR M M, REHMAN A, RAZZAQ A, et al.. Genome-wide characterization and expression analysis of Erf gene family in cotton [J]. BMC Plant Biol., 2022, 22(1):1-18. |
| 21 | KENNETH J, LIVAK, THOMAS D, et al.. Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCt method [J]. Methods, 2001, 25 (4):402-408. |
| 22 | LIU L, WANG D, ZHANG C, et al.. The heat shock factor GhHSFA4a positively regulates cotton resistance to Verticillium dahliae [J]. Front. Plant Sci., 2022, 13:1050216-1050230. |
| 23 | GAO X, BRITT JR R C, SHAN L, et al.. Agrobacterium-mediated virus-induced gene silencing assay in cotton [J]. J. Vis. Exp., 2011, 20(54):2938-2943. |
| 24 | SPARKES I A, RUNIONS J, KEARNS A, et al.. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants [J]. Nat. Prot., 2006, 1(4):2019-2025. |
| 25 | LI X K, SU X F, LU G Q, et al.. VdOGDH is involved in energy metabolism and required for virulence of Verticillium dahliae [J]. Curr. Genet., 2020, 66(2):345-359. |
| 26 | 许世达,耿兴敏,王露露.植物乙烯响应因子(ERF)的结构、功能及表达调控研究进展[J].浙江农林大学学报,2021,38(3):624-633. |
| XU S D, GENG X M, WANG L L. A review of the structure, function and expression regulation of ethylene response factors (ERF) in plant [J]. J. Zhejiang A&F Univ., 2021, 38(3):624-633. | |
| 27 | 刘茜,王爱云,荣玮.ERF转录因子在作物抗病基因工程中的研究进展[J].种子,2014,33(1):48-53. |
| LIU Q, WANG A Y, RONG W. Progress of ERF transcription factors in genetic engineering for disease resistance in crops [J]. Seed, 2014, 33(1):48-53. | |
| 28 | PIETERSEC M, LEON-REYES A, VAN DER ENT S, et al.. Networking by small-molecule hormones in plant immunity [J]. Nat. Chem. Biol., 2009, 5(5):308-316. |
| 29 | 黄榆普.茉莉酸参与缺硼抑制拟南芥生长的分子机制[D].武汉:华中农业大学,2020. |
| HUANG Y P. Molecular mechnism of jasmonate mediated low-boron-induced growth inhibition in Arabidopsis [D]. Wuhan: Huazhong Agricultural University, 2020. | |
| 30 | OHOME-TAKAGI M, SHINSHI H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element [J]. Plant Cell, 1995, 7(2):173-182. |
| 31 | ZHANG H J, HONG Y B, HUANG L, et al.. Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea [J/OL]. Sci. Rep., 2016, 6(1):30251 [2023-04-30]. . |
| 32 | ONATE-SANCHEZ L, ANDERSON J P, YOUNG J, et al.. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense [J]. Plant Physiol., 2007, 143(1):400-409. |
| [1] | Man ZHANG, Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG. Cloning and Functional Analysis of GhNAC1 in Upland Cotton Involved in Verticillium Wilt Resistance [J]. Journal of Agricultural Science and Technology, 2023, 25(10): 35-44. |
| [2] | Guangke LUO, Rongbo MU, Bing XUE, Hua ZHANG, Yanping REN, Hao MA. Function and Characteristic Analysis of Haloxylon ammodendron NAC Transcription Factor HaNAC38 [J]. Journal of Agricultural Science and Technology, 2023, 25(10): 65-73. |
| [3] | Zhiyong WU, Hong GU, Dawei CHENG, Lan LI, Shasha HE, Ming LI, Jinyong CHEN. Advances in Regulatory Mechanism of Brassinolide on Plant Root Development [J]. Journal of Agricultural Science and Technology, 2022, 24(2): 68-76. |
| [4] | Mengyuan HAO, Qi HANG, Gongyao SHI. Application and Prospect of Virus-induced Gene Silencing in Crop Gene Function Research [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 1-13. |
| [5] | Qinqin WANG, Xiugui CHEN, Xuke LU, Shuai WANG, Yuexin ZHANG, Yapeng FAN, Quanjia CHEN, Wuwei YE. Bioinformatics Analysis and Functional Verification of GhPKE1 inUpland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 38-45. |
| [6] | WAN Dongpu1,2, YU Zhuo1, WU Yanmin3, DING Mengqi2, LI Jinbo2, ZHOU Meiliang2*. Regulation of Anthocyanin Metabolism on Colored Leaves of Plants [J]. Journal of Agricultural Science and Technology, 2020, 22(2): 30-38. |
| [7] | DONG Jie, LI Gezi, HAN Qiaoxia, XIE Yingxin, WANG Yonghua, FENG Wei, MA Dongyun, WANG Chenyang, GUO Tiancai, KANG Guozhang*. Isolation and Function of TabHLH39 Transcription Factor Regulating Expression of the TaAGPL1 Gene in Bread Wheat [J]. Journal of Agricultural Science and Technology, 2020, 22(10): 18-26. |
| [8] | XU Mengjun, GAO Tian, WANG Pengfei, LI Gezi, KANG Guozhang*. Function of Calcium Dependent Protein Kinase 34 in Grain Starch Synthesis of Wheat (Triticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2019, 21(2): 26-33. |
| [9] | YANG Xiaomin, RUI Cun, ZHANG Yuexin, WANG Delong, WANG Junjuan, LU Xuke, CHEN Xiugui, GUO Lixue, WANG Shuai, CHEN Chao, YE Wuwei*. Cloning and Stress Resistance Analysis of Cotton DNA Methyltransferase GhDMT9 Gene [J]. Journal of Agricultural Science and Technology, 2019, 21(10): 12-19. |
| [10] | JIN Qiaochun1, YU Fang2, YU Zongxia1*, FENG Baomin1*. Functional Characterization of PatPTS Gene Using Virus Induced Gene Silencing (VIGS) System in Patchouli (Pogostemon cablin) [J]. Journal of Agricultural Science and Technology, 2018, 20(3): 39-45. |
| [11] | WANG Dezhou, MO Xiaoting, ZHANG Xia, XU Miaoyun, ZHANG Lan, ZHAO Jun*, WANG Lei*. Cloning and Functional Analysis of Transcription Factors Gene ZmbHLH4 From Zea mays [J]. Journal of Agricultural Science and Technology, 2018, 20(12): 16-25. |
| [12] | HAN Song, DUAN Xingpeng, ZHANG Zhidong, ZUO Kaijing*. Functional Verification and Pathogenesis Analysis of VdTri15 Gene from Verticillium dahliae on Cotton [J]. Journal of Agricultural Science and Technology, 2018, 20(12): 26-35. |
| [13] | ZHAO Yanpeng1, LIANG Wei1, WANG Dan1, WANG Yumei2, LIU Zhengjie1, CUI Yupeng1, HUA Jinping1*. Regulation of Oil Biosynthesis and Genetic Improvement in Plant: Advances and Prospects [J]. Journal of Agricultural Science and Technology, 2018, 20(1): 14-24. |
| [14] | GAO Li-hua1, LIU Bo-xin1,2, LI Jin-bo3, WU Yan-min1, TANG Yi-xiong1*. Cloning and Function Analysis of bHLH Transcription Factor Gene GhMYC4 from Gossypium hirsutism L. [J]. Journal of Agricultural Science and Technology, 2016, 18(5): 33-41. |
| [15] | XIE Zheng-wen1, WANG Lian-jun2, CHEN Jin-yang1, WANG Jiao1, SU Yi-jun1, . Studies on WRKY Transcription Factors and Their Biological Functions in Plants [J]. Journal of Agricultural Science and Technology, 2016, 18(3): 46-54. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号