




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (10): 35-44.DOI: 10.13304/j.nykjdb.2022.0280
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Man ZHANG( ), Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG(
), Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG( )
)
Received:2022-04-09
Accepted:2022-05-28
Online:2023-10-15
Published:2023-10-27
Contact:
Yan ZHANG
通讯作者:
张艳
作者简介:张曼 E-mail:zhangman920601@163.com;
基金资助:CLC Number:
Man ZHANG, Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG. Cloning and Functional Analysis of GhNAC1 in Upland Cotton Involved in Verticillium Wilt Resistance[J]. Journal of Agricultural Science and Technology, 2023, 25(10): 35-44.
张曼, 张进, 张新雨, 王国宁, 王省芬, 张艳. 陆地棉GhNAC1基因的克隆及抗黄萎病功能分析[J]. 中国农业科技导报, 2023, 25(10): 35-44.
引物名称 Primer name | 引物序列 Primer sequence(5’-3’) | 用途 Utilization |
|---|---|---|
| NAC1-F | ATGAGCTACCAATCAAACC | 基因克隆Gene cloning |
| NAC1-R | TTAAAAGTTGAGGATATTAGC | 基因克隆Gene cloning |
| NAC1-RT-F | GAACACATCTCTTCCTTCATCATCTT | 实时定量PCR Real-time PCR |
| NAC1-RT-R | AGTTGTCCCATATTTTCATTGCCTA | 实时定量PCR Real-time PCR |
| NAC1-V-F | GAATTCGGTTGAACTTCCTGGCTTTA | 载体构建Vector construction |
| NAC1-V-R | GGTACCGCAAAGTAGCATCAGGGAG | 载体构建Vector construction |
| NAC1-G-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGA GCTACCAATCAAACC | 载体构建Vector construction |
| NAC1-G-R | GGGGACCACTTTGTACAAGAAAGCTGGGTGAAAGTT GAGGATATTAGC | 载体构建Vector construction |
| GhUBQ14-F | CAACGCTCCATCTTGTCCTT | 实时定量PCR Real-time PCR |
| GhUBQ14-R | TAGTCGTCTTTCCCGTAAGC | 实时定量PCR Real-time PCR |
| GhNDR1-F | CCCGTAACCAAGGAGGCTGT | 实时定量PCR Real-time PCR |
| GhNDR1-R | CTGCTAAGGGAAGGCAAGGATAG | 实时定量PCR Real-time PCR |
| GhNPR1-F | GTCTGGCTGATGTCAATCTGCG | 实时定量PCR Real-time PCR |
| GhNPR1-R | TCCTTCCCTTGCTCTGTCTTGG | 实时定量PCR Real-time PCR |
| GhPR1-F | GGCACAGAACTACGCTAATCAACG | 实时定量PCR Real-time PCR |
| GhPR1-R | GCTTTACCCTCTCACTAACCCACAT | 实时定量PCR Real-time PCR |
| GhPAD4-F | GGATGGAAGAATGGAAAGAAATGAA | 实时定量PCR Real-time PCR |
| GhPAD4-R | GAACTAGGAAAGCAGACTAAGGAACCA | 实时定量PCR Real-time PCR |
Table 1 Primers used in this experiment
引物名称 Primer name | 引物序列 Primer sequence(5’-3’) | 用途 Utilization |
|---|---|---|
| NAC1-F | ATGAGCTACCAATCAAACC | 基因克隆Gene cloning |
| NAC1-R | TTAAAAGTTGAGGATATTAGC | 基因克隆Gene cloning |
| NAC1-RT-F | GAACACATCTCTTCCTTCATCATCTT | 实时定量PCR Real-time PCR |
| NAC1-RT-R | AGTTGTCCCATATTTTCATTGCCTA | 实时定量PCR Real-time PCR |
| NAC1-V-F | GAATTCGGTTGAACTTCCTGGCTTTA | 载体构建Vector construction |
| NAC1-V-R | GGTACCGCAAAGTAGCATCAGGGAG | 载体构建Vector construction |
| NAC1-G-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGA GCTACCAATCAAACC | 载体构建Vector construction |
| NAC1-G-R | GGGGACCACTTTGTACAAGAAAGCTGGGTGAAAGTT GAGGATATTAGC | 载体构建Vector construction |
| GhUBQ14-F | CAACGCTCCATCTTGTCCTT | 实时定量PCR Real-time PCR |
| GhUBQ14-R | TAGTCGTCTTTCCCGTAAGC | 实时定量PCR Real-time PCR |
| GhNDR1-F | CCCGTAACCAAGGAGGCTGT | 实时定量PCR Real-time PCR |
| GhNDR1-R | CTGCTAAGGGAAGGCAAGGATAG | 实时定量PCR Real-time PCR |
| GhNPR1-F | GTCTGGCTGATGTCAATCTGCG | 实时定量PCR Real-time PCR |
| GhNPR1-R | TCCTTCCCTTGCTCTGTCTTGG | 实时定量PCR Real-time PCR |
| GhPR1-F | GGCACAGAACTACGCTAATCAACG | 实时定量PCR Real-time PCR |
| GhPR1-R | GCTTTACCCTCTCACTAACCCACAT | 实时定量PCR Real-time PCR |
| GhPAD4-F | GGATGGAAGAATGGAAAGAAATGAA | 实时定量PCR Real-time PCR |
| GhPAD4-R | GAACTAGGAAAGCAGACTAAGGAACCA | 实时定量PCR Real-time PCR |

Fig. 2 Structure analysis of GhNAC1 proteinA: Prediction result of GhNAC1 signal peptide; B: Prediction result of GhNAC1 transmembrane domain; C: Prediction of secondary structure of GhNAC1 protein
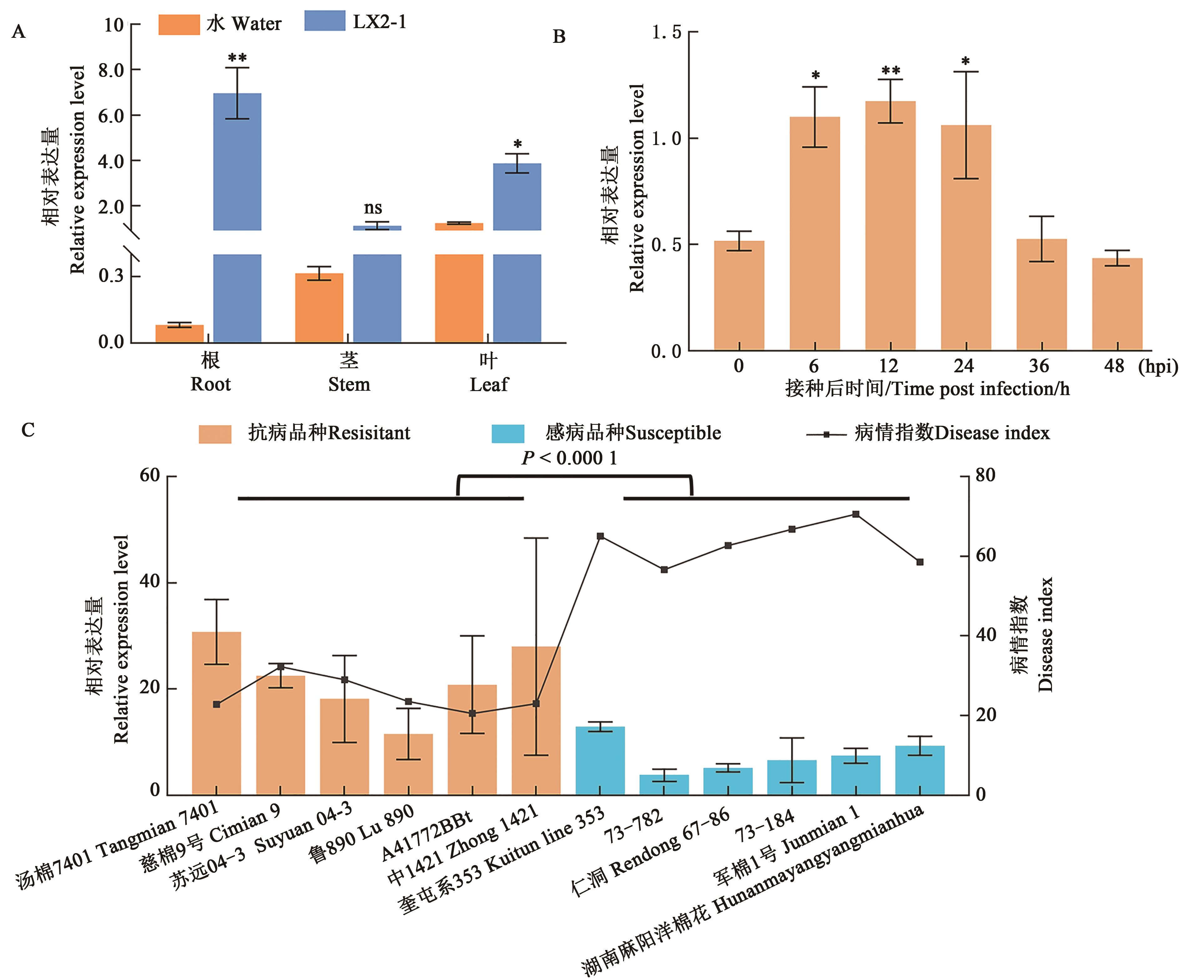
Fig. 3 Expression pattern of GhNAC1A: Expression analysis of GhNAC1 in different tissues and organs of ND601; B: Expression analysis of GhNAC1 in ND601 roots under Verticillium wilt stress; C: Expression analysis of GhNAC1 in roots of 12 different resistant cultivars under Verticillium wilt stress; * and ** indicate significant differences compared with 0 h at P < 0.05 and P < 0.01 levels, respectively

Fig. 4 Silencing GhNAC1 reduced the resistance to V. dahliae in cottonA: CLA1 gene was used as a positive control with an albino phenotype on leave after VIGS in cotton; B: Relative expression levels of GhNAC1 in WT and GhNAC1-silenced plants 7 d after hand-infiltration; C: Disease manifestations of control plants and silent plants at 20 dpi; D: Disease index; E: Phenotype of stem infected by V. dahliae. CK stands for control group, VIGS stands for silent group; ** indicates significant difference compared with CK at P < 0.01 level
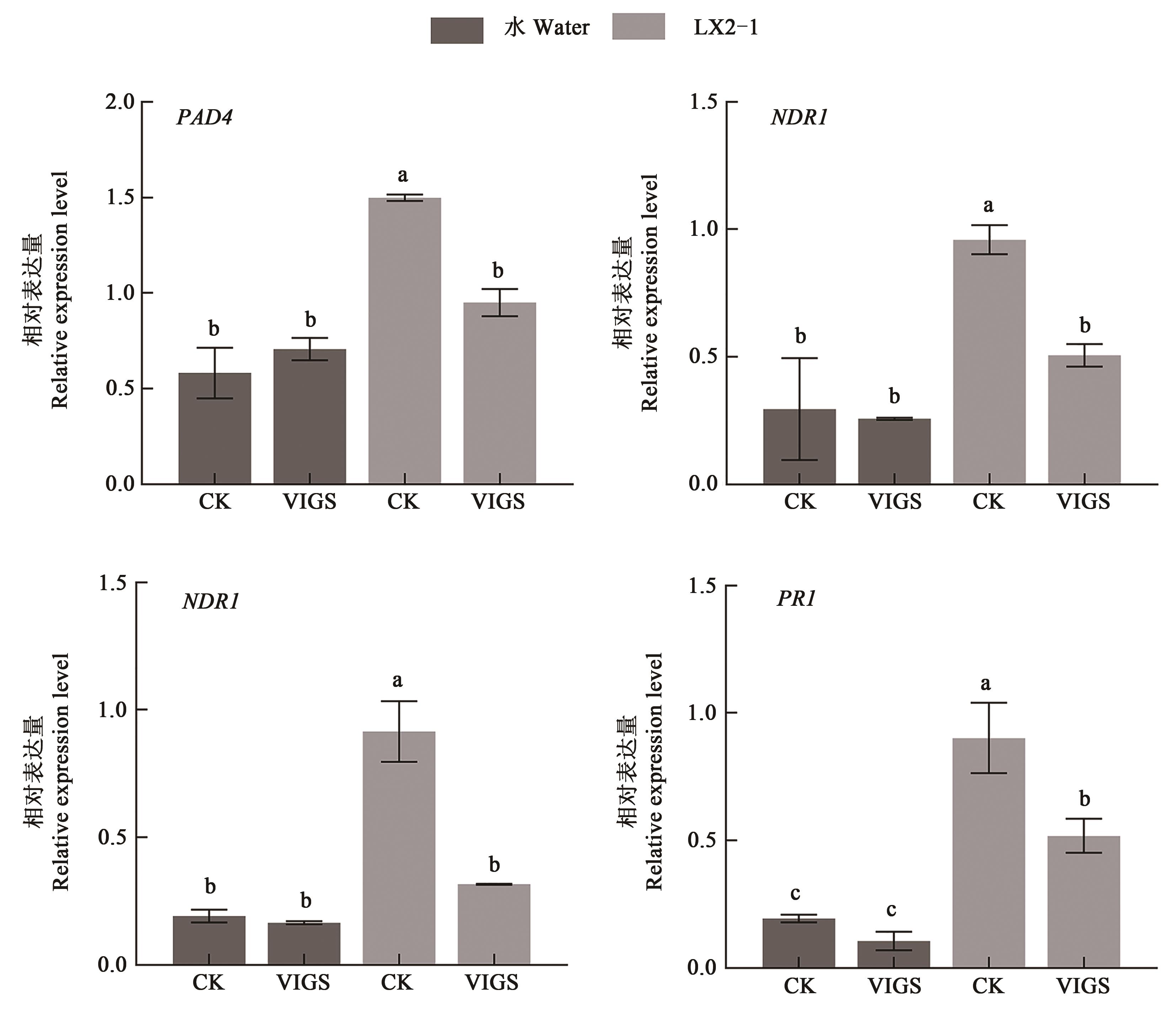
Fig. 5 Analysis of gene expression of salicylic acid pathway in GhNAC1 silenced cotton after inoculationNote:Different lowercase letters represent significant differences at P < 0.05 level.
| 1 | KLOSTERMAN S J, ATALLAH Z K, VALLAD G E, et al.. Diversity, pathogenicity and management of Verticillium species [J]. Annu. Rev. Phytopathol., 2009, 47(1): 39-62. |
| 2 | VALLAD G E, SU B B, ARAO K V. Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae [J/OL]. Phytopathology, 2008, 98(8): 871 [2022-03-19]. . |
| 3 | CHEN J Y, KLOSTERMAN S J, HU X P, et al.. Key insights and research prospects at the dawn of the population genomics era for Verticillium dahliae [J]. Annu. Rev. Phytopathol., 2021, 59(1): 31-51. |
| 4 | SONG R, LI Y S, XIE H Y, et al.. An overview of the molecular genetics of plant resistance to the Verticillium wilt pathogen Verticillium dahliae [J/OL]. Int. J. Mol. Sci., 2020, 21(3): 1120 [2022-03-21]. . |
| 5 | 朱荷琴,李志芳,冯自力,等.我国棉花黄萎病研究十年回顾及展望[J].棉花学报, 2017, 29(z1): 37-50. |
| ZHU H Q, LI Z F, FENG Z L, et al.. Review and prospect of cotton Verticillium wilt research in ten years in China [J]. Cotton Sci., 2017, 029(z1): 37-50. | |
| 6 | GAO W, LONG L, ZHU L F, et al.. Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae [J]. Mol. Cell Proteomics, 2013, 12(12): 3690-3703. |
| 7 | YANG J, MA Q, ZHANG Y, et al.. Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt [J]. Gene, 2016, 575(2): 687-694. |
| 8 | ZHANG Y, WU L, WANG X, et al.. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants [J]. Mol. Plant Pathol., 2019, 20(3): 309-322. |
| 9 | CHEN B, ZHANG Y, YANG J, et al.. The G-protein α subunit GhGPA positively regulates Gossypium hirsutum resistance to Verticillium dahliae via induction of SA and JA signaling pathways and ROS accumulation [J]. Crop J., 2021, 9(4): 823-833. |
| 10 | CHEN B, ZHANG Y, SUN Z W, et al.. Tissue-specific expression of GhnsLTPs identified via GWAS sophisticatedly coordinates disease- and insect-resistance by regulating metabolic flux redirection in cotton [J]. Plant J., 2021, 107(3): 831-846. |
| 11 | 安汶铠,常丹,杨艺,等.利用VIGS技术沉默GhBES1基因对棉花幼苗生理指标的影响[J].分子植物育种, 2016, 14(9): 1055-1061. |
| AN W K, CHANG D, YANG Y, et al.. Effects of GhBES1 gene silencing by VIGS technique on physiological indexes of cotton seedlings [J]. Mol. Plant Breeding, 2016, 14(9): 1055-1061. | |
| 12 | AIDA M, ISHIDA T, FUKAKI H, et al.. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant [J]. Plant Cell, 1997, 9(6): 841-857. |
| 13 | 孙利军,李大勇,张慧娟,等.NAC转录因子在植物抗病和抗非生物胁迫反应中的作用[J].遗传, 2012, 34(8): 993-1002. |
| SUN L J, LI D Y, ZHANG H J, et al.. Functions of NAC transcription factors in biotic and abiotic stress responses in plants [J]. Hereditas, 2012, 34(8): 993-1002. | |
| 14 | ZHONG R, LEE C, HAGHIGHAT M, et al.. Xylem vessel-specific SND5 and its homologs regulate secondary wall biosynthesis through activating secondary wall NAC binding elements [J]. New Phytol., 2021, 231(4): 1496-1509. |
| 15 | FU B L, WANG W Q, LIU X F, et al.. An ethylene-hypersensitive methionine sulfoxide reductase regulated by NAC transcription factors increases methionine pool size and ethylene production during kiwifruit ripening [J]. New Phytol., 2021,231(1): 237-251. |
| 16 | 曲潇玲,焦裕冰,罗健达,等.本氏烟NbNAC062的克隆及对马铃薯Y病毒侵染的抑制作用[J].中国农业科学, 2021, 54(19):84-94. |
| QU X L, JIAO Y B, LUO J D, et al.. Cloning of tobacco NbNAC062 and its inhibition on potato virus Y infection [J]. Sci. Agric. Sin., 2021, 54(19): 84-94. | |
| 17 | 叶鸿鹰,梅双双,戎伟. ATAF2正调控拟南芥对橡胶树白粉菌的抗病性[J].热带生物学报, 2020, 11(1): 61-65. |
| YE H Y, MEI S S, RONG W. ATAF2 positively contributes to the disease resistance against Oidium heveae in Arabidopsis [J]. J. Trop. Biol., 2020, 11(1): 61-65. | |
| 18 | CAI W, YANG S, WU R, et al.. Pepper NAC-type transcription factor NAC2c balances the trade-off between growth and defense responses [J]. Plant Physiol., 2021, 186(4): 4. |
| 19 | 王国宁,赵贵元,岳晓伟,等.河北省棉花黄萎病菌致病性与ISSR遗传分化[J].棉花学报, 2012, 24(4): 348-357. |
| WANG G N, ZHAO G Y, YUE X W, et al.. Pathogenicity and ISSR genetic differentiation of Verticillium dahliae isolates from cotton growing areas of Hebei province [J]. Cotton Sci., 2012, 24(4): 348-357. | |
| 20 | SPARKES I A, RUNIONS J, KEARNS A, et al.. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants [J]. Nature Protoc., 2006, 1(4):2019-2025. |
| 21 | ZHANG M, WANG X F, YANG J, et al.. GhENODL6 isoforms from the phytocyanin gene family regulate Verticillium wilt resistance in cotton [J]. Int. J. Mol. Sci., 2022, 23(6):2913-2921. |
| 22 | ZHANG Y, WANG X F, DING Z G, et al.. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement [J/OL]. BMC Genomics, 2013, 14(1): 637 [2022-03-21]. . |
| 23 | YANG J, MA Q, ZHANG Y, et al.. Molecular cloning and functional analysis of GbRvd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt [J]. Gene, 2016, 575(2016): 687-694. |
| 24 | 李宝笃,沈崇尧. Hoaglands营养液栽苗法快速鉴定甜椒疫霉病菌致病性及对杀菌剂敏性研究初报[J].植物病理学报, 1993, 23(3): 243-244. |
| LI B D, SHEN C Y. Preliminary report on the rapid identification of pathogenicity and fungicide sensitivity of phytophthora capsicum by Hoaglands nutrient solution seedling method [J]. Acta Phytopathol. Sin., 1993, 23(3): 243-244. | |
| 25 | 唐永凯, 贾永义.荧光定量PCR数据处理方法的探讨[J].生物技术, 2008, 18(3): 91-93. |
| TANG Y K, JIA Y Y. Discussion on data processing method of fluorescence quantitative PCR [J]. Biotechnology, 2008, 18(3):91-93. | |
| 26 | ZHANG B, YANG Y, CHEN T, et al.. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae [J/OL]. PLoS One, 2012, 7(12): e51091 [2022-03-21]. . |
| 27 | JYOTHISHWARAN G, KOTRESHA D, SELVARAJ T, et al.. A modified freeze-thaw method for efficient transformation of Agrobacterium tumefaciens [J]. Curr. Sci., 2007, 93(6): 770-772. |
| 28 | GAO X, BRITT R C, SHAN L, et al.. Agrobacterium-mediated virus-induced gene silencing assay in cotton [J/OL]. J. Vis. Exp., 2011, 54(54): e2938 [2022-03-21]. . |
| 29 | LOAKE G, GRANT M. Salicylic acid in plant defence-the players and protagonists [J]. Curr. Opin. Plant Biol., 2007, 10(5): 466-472. |
| 30 | MARQUES D N, REIS S D, SOUZA C D. Plant NAC transcription factors responsive to abiotic stresses [J]. Plant Gene, 2017, 11: 170-179. |
| 31 | PURANIK S, SAHU P P, SRIVASTAVA P S, et al.. NAC proteins: regulation and role in stress tolerance [J/OL]. Trends Plant Sci., 2012, 17(6): 369-381 [2022-03-21]. . |
| 32 | SHEN H, YIN Y, CHEN F, et al.. A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production [J/OL]. Bioenergy Res., 2009, 2(4): 217 [2022-03-21]. |
| 33 | WANG X, BASNAYAKE B, ZHANG H, et al.. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens [J]. Mol. Plant Microbe., 2009, 22(10): 1227-1238. |
| 34 | NAKASHIMA K, TRAN L S P, NGUYEN D V, et al.. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice [J]. Plant J., 2010, 51(4): 617-630. |
| 35 | SANG R P, KIM H S, LEE K S, et al.. Overexpression of rice NAC transcription factor OsNAC58 on increased resistance to bacterial leaf blight [J]. J. Plant Biochem. Biot., 2017, 44(2): 149-155. |
| 36 | ZHANG X M, ZHANG Q, PEI C L, et al.. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage [J]. Physiol. Mol. Plant Pathol., 2018, 102: 144-153. |
| 37 | LIU B, OUYANG Z, ZHANG Y, et al.. Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response [J]. PloS One, 2014, 9(7): e102067 [2022-03-21]. . |
| 38 | CHANG Y, YU R, FENG J, et al.. NAC transcription factor involves in regulating bacterial wilt resistance in potato [J]. Funct. Plant Biol., 2020, 47: 925-936. |
| 39 | SHIRASU K, NAKAJIMA H, LAMB D C, et al.. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms [J]. Plant Cell, 1997, 9: 261-270. |
| 40 | LONG L, XU F C, ZHAO J R, et al.. GbMPK3 overexpression increases cotton sensitivity to Verticillium dahliae by regulating salicylic acid signaling [J/OL]. Plant Sci., 2020, 292: 110374 [2022-03-21]. . |
| 41 | MÉTRAUX J. Recent breakthroughs in the study of salicylic acid biosynthesis [J]. Trends Plant Sci., 2002, 7(8): 332-334. |
| 42 | CHEN Z, ZHENG Z, HUANG J, et al.. Biosynthesis of salicylic acid in plants [J]. Plant Signal. Behav., 2009, 4(6): 493-496. |
| 43 | ZHENG X Y, SPIVEY N, ZENG W, et al.. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation [J]. Cell Host Microbe, 2012, 11(6): 587-596. |
| [1] | Wei WANG, Qiang ZHAO, Abuduaini Munire·, Alimu·Amuli, Xinxin LI, Yangqing TIAN. Effects of Different Exogenous Substances on Chemical Capping and Yield and Quality of Cotton [J]. Journal of Agricultural Science and Technology, 2023, 25(9): 57-68. |
| [2] | Pengfei LIU, Xiaoshuang LU, Dilimurat Reheman, Tangnur Slay, Yanying QU, Quanjia CHEN, Xiaojuan DENG. Genetic Variation Analysis of Main Quality Traits and Agronomic Traits in Upland Cotton Seed [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 22-32. |
| [3] | Liting CHEN, Yuanyuan YAN. Investigation of Regulatory Mechanism of Floral Integrators in Upland Cotton [J]. Journal of Agricultural Science and Technology, 2023, 25(6): 11-21. |
| [4] | Zhengran SUN, Cuiping ZHANG, Jinli ZHANG, Hao WU, Xiuyan LIU, Zhenkai WANG, Yuzhen YANG, Daohua HE. Effects of Chemical Detopping on Cotton Plant Growth in Guanzhong Cotton Region [J]. Journal of Agricultural Science and Technology, 2023, 25(4): 167-177. |
| [5] | Guoqing LU, Caixia MA, Guoqing SUN, Huiming GUO, Hongmei CHENG. Molecular Characterization and Inheritance Stability Analysis of Herbicide-resistant Cotton GV-2 [J]. Journal of Agricultural Science and Technology, 2023, 25(1): 42-49. |
| [6] | Yan LIU, Hongshuai BAO, Hongyan SHANG, Guoning WANG, Yan ZHANG, Xingfen WANG, Zhiying MA, Jinhua WU. Selection of Cotton Fusarium Wilt and Culture Conditions [J]. Journal of Agricultural Science and Technology, 2022, 24(8): 124-132. |
| [7] | Ling LI, Helin DONG, Pengcheng LI, Liwen TIAN, Chunmei LI, Yunzhen MA, Na ZHANG, Fang WANG, Wenxiu XU. Effects of Machine Harvesting Planting Methods on Photosynthetic Characteristics and Dry Matter Accumulation of Different Plant Types of Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(8): 172-181. |
| [8] | Zhengwen SUN, Qishen GU, Yan ZHANG, Xingfen WANG, Zhiying MA. Research Progress on Cotton Gene Discovery and Molecular Breeding [J]. Journal of Agricultural Science and Technology, 2022, 24(7): 32-38. |
| [9] | Chengchuan YAN, Qingtao ZENG, Qin CHEN, Jincheng FU, Tingwei WANG, Quanjia CHEN, Yanying QU. Screening and Evaluation of Drought Resistance Indicators at Flowering and Boll Stage of Upland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(7): 46-57. |
| [10] | Lijuan ZHANG, Yukun QIN, Huihuang CHENG, Yongqi LI, Haihua LUO. Research on Characteristics of Nitrogen and Phosphorus Loss from Surface Runoff of Cotton Field in Northern Jiangxi Province of Poyang Lake Region [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 166-175. |
| [11] | Nan WU, Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG. Overexpression of a Cotton Glucuronokinase Gene GbGlcAK Promotes Cell Elongation in Arabidopsis thaliana [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 36-46. |
| [12] | Yajie HUANG, Dan REN, Shengmei LI, Jinxin CUI, Tao YANG, Jiaojiao REN, Wenwei GAO. Evaluation and Screening of Salt and Alkali Tolerance Indices of Upland Cotton Seedlings [J]. Journal of Agricultural Science and Technology, 2022, 24(5): 46-55. |
| [13] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [14] | Jing ZHANG, Simeng GUO, Yingchun HAN, Yaping LEI, Fangfang XING, Wenli DU, Yabing LI, Lu FENG. Estimation of Cotton Yield Based on Unmanned Aerial Vehicle RGB Images [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 112-120. |
| [15] | Juan LI, Xuyu YAN, Xiang WU, Ling LI. Research Progress on the Remediation of Cadmium Contaminated Farmland Soil by Fiber Crops [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 171-178. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号