




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (9): 79-91.DOI: 10.13304/j.nykjdb.2024.0208
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Huaqiang TAN1( ), Liping LI1, Manman TIE2, Jiaqin YANG1, Xiaoyun ZHENG1, Shaokun PAN1, Youwan TANG1(
), Liping LI1, Manman TIE2, Jiaqin YANG1, Xiaoyun ZHENG1, Shaokun PAN1, Youwan TANG1( )
)
Received:2024-03-18
Accepted:2024-04-22
Online:2025-09-15
Published:2025-09-24
Contact:
Youwan TANG
谭华强1( ), 李丽平1, 铁曼曼2, 杨家勤1, 郑晓云1, 潘绍坤1, 唐有万1(
), 李丽平1, 铁曼曼2, 杨家勤1, 郑晓云1, 潘绍坤1, 唐有万1( )
)
通讯作者:
唐有万
作者简介:谭华强 E-mail:307927595@qq.com;
基金资助:CLC Number:
Huaqiang TAN, Liping LI, Manman TIE, Jiaqin YANG, Xiaoyun ZHENG, Shaokun PAN, Youwan TANG. Mining and Analysis of Genes Related to Anthocyanin Degradation in Purple Pepper[J]. Journal of Agricultural Science and Technology, 2025, 27(9): 79-91.
谭华强, 李丽平, 铁曼曼, 杨家勤, 郑晓云, 潘绍坤, 唐有万. 紫色辣椒花青素降解相关基因的挖掘与分析[J]. 中国农业科技导报, 2025, 27(9): 79-91.
基因编号 Gene ID | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer(5’-3’) | 大小 Size/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| T459_29804 | TGGAAGAGTTGAACCACCGA | GTGTCCTCGTCACCACTACA | 248 | 59 |
| T459_02736 | GGACCAACAACACACCAACA | CTTGGGTCACTGAAGCATCG | 249 | 59 |
| T459_27911 | AACGGGTTCTTGGGGCTAAT | GTGCATCGGTAGTAGCTCCT | 236 | 59 |
| T459_07162 | CTGCCAGTTGACTTGTTCGT | TGCAAAACGGTCACAGTGAG | 249 | 59 |
| T459_09393 | CTGCCAACACTCAAATCCCC | ATTTGCATCGCCACCAGAAG | 230 | 59 |
| T459_33197 | TTACCCAACAGTTGACCCGA | CAGCAGCCATTTTCTCCACA | 239 | 59 |
| T459_08317 | GTTGCAGCACAGAGAACCAA | ACCAAAGTCCCTGCTATGCT | 234 | 59 |
| T459_08320 | TGCTGGTTCTAGATTGCCCA | CTCCTTGCAGACCTGGTTCT | 233 | 59 |
| T459_27108 | TGCTTGGAGAGGATTTGGGA | TGTACACTTTGCTCCCCAGG | 244 | 59 |
| T459_12184 | ACCCGACTGATGAAGAGCTT | TCGCCTTCCAGTACCCATTT | 231 | 59 |
| T459_30033 | CCCTGTCCTGCTCACTGAAG | GTCACGTCCAGCAAGATCCA | 250 | 60 |
Table 1 Primers used in qRT-PCR for candidate genes
基因编号 Gene ID | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer(5’-3’) | 大小 Size/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| T459_29804 | TGGAAGAGTTGAACCACCGA | GTGTCCTCGTCACCACTACA | 248 | 59 |
| T459_02736 | GGACCAACAACACACCAACA | CTTGGGTCACTGAAGCATCG | 249 | 59 |
| T459_27911 | AACGGGTTCTTGGGGCTAAT | GTGCATCGGTAGTAGCTCCT | 236 | 59 |
| T459_07162 | CTGCCAGTTGACTTGTTCGT | TGCAAAACGGTCACAGTGAG | 249 | 59 |
| T459_09393 | CTGCCAACACTCAAATCCCC | ATTTGCATCGCCACCAGAAG | 230 | 59 |
| T459_33197 | TTACCCAACAGTTGACCCGA | CAGCAGCCATTTTCTCCACA | 239 | 59 |
| T459_08317 | GTTGCAGCACAGAGAACCAA | ACCAAAGTCCCTGCTATGCT | 234 | 59 |
| T459_08320 | TGCTGGTTCTAGATTGCCCA | CTCCTTGCAGACCTGGTTCT | 233 | 59 |
| T459_27108 | TGCTTGGAGAGGATTTGGGA | TGTACACTTTGCTCCCCAGG | 244 | 59 |
| T459_12184 | ACCCGACTGATGAAGAGCTT | TCGCCTTCCAGTACCCATTT | 231 | 59 |
| T459_30033 | CCCTGTCCTGCTCACTGAAG | GTCACGTCCAGCAAGATCCA | 250 | 60 |
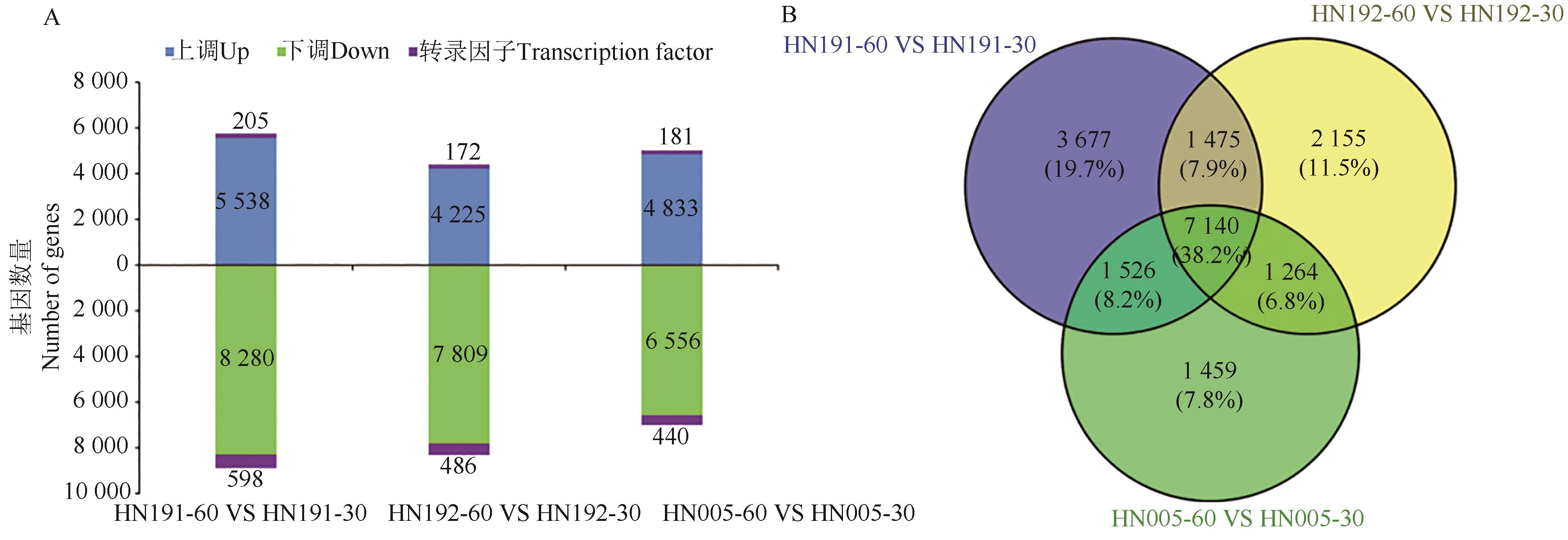
Fig. 1 DEGs between fruits of 30 and 60 d from different varietiesA:Histogram of differentially expressed genes; B. Venn plot of differentially expressed genes

Fig. 6 Construction of a weighted co-expression networkA:Cluster dendrogram of 18 samples;B:Soft threshold selection;C:Gene clustering and module cutting
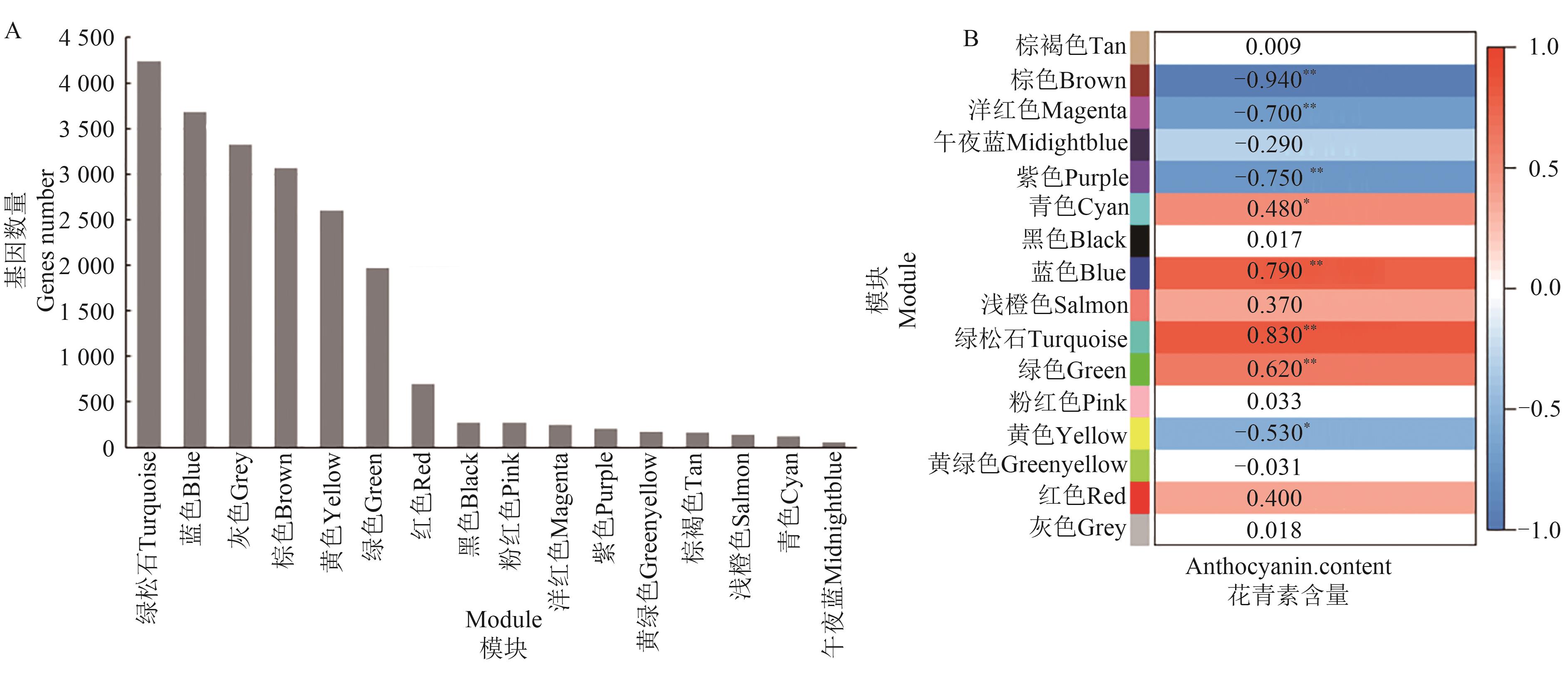
Fig. 7 Analysis of WGCNA modulesA: Statistics of gene numbers in 16 modules; B: Relationship heatmap of 16 modules with anthocyanin content. * and ** indicate significant correlations at P<0.05 and P<0.01 levels, respectively
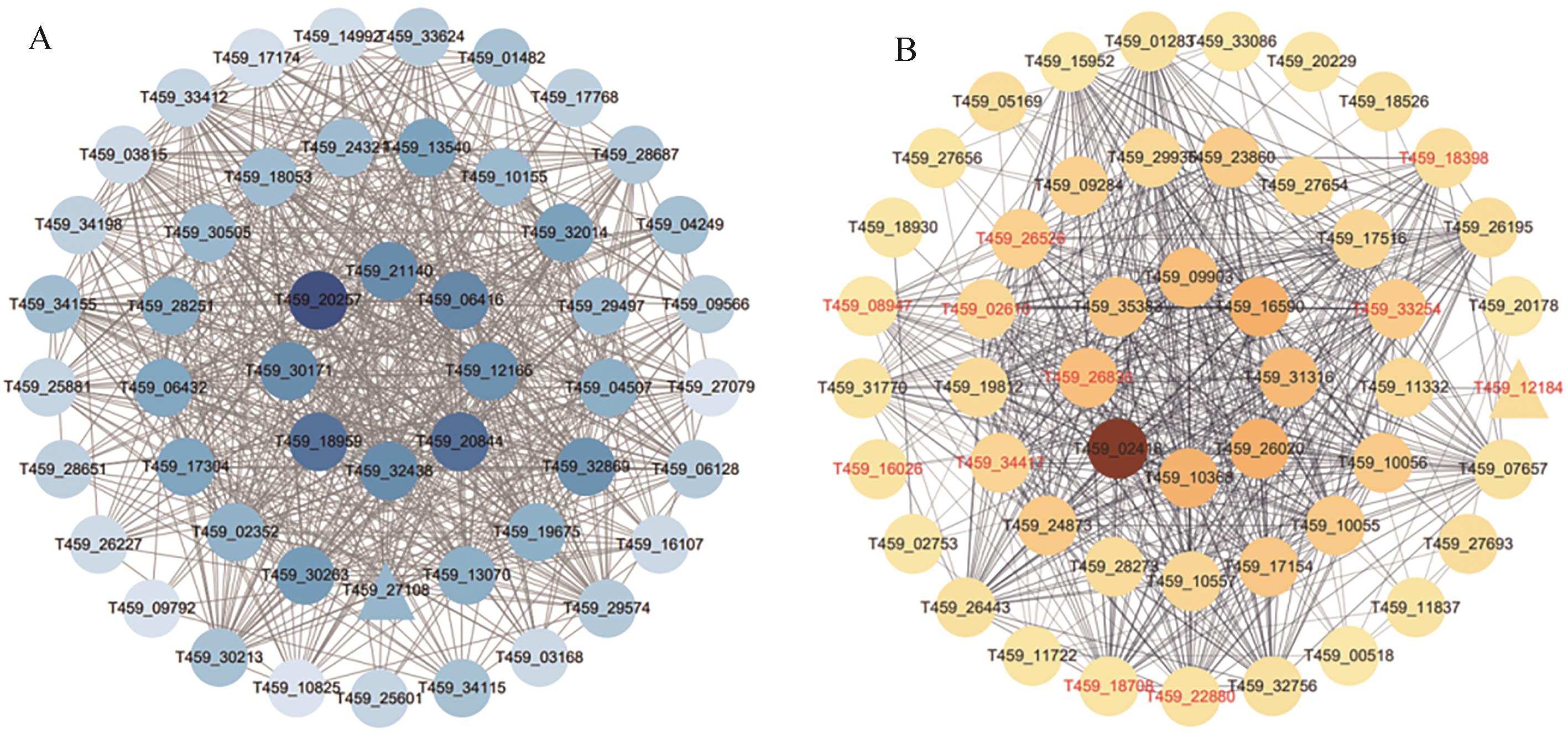
Fig. 8 Analysis of hub genes in WGCNA networkA: Network diagram of degree top 50 genes in the blue module obtained by WGCNA. The color shade on the diagram represents the Degree value, with darker colors indicating higher values and triangular node in the diagram represents transcription factor;B: Network diagram of Degree top 50 genes in PPI network of common DEGs. The red gene names indicate genes share with the blue module, the color shade represents the Degree value, with darker colors indicating higher values. Triangular node in this diagram represents transcription factor
| [1] | PERRY L, DICKAU R, ZARRILLO S,et al..Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp.L.) in the Americas [J]. Science, 2007, 315(5814):986-988. |
| [2] | 邹学校,马艳青,戴雄泽,等.辣椒在中国的传播与产业发展[J].园艺学报,2020,47(9):1715-1726. |
| ZOU X X, MA Y Q, DAI X Z, et al.. Spread and industry development of pepper in China [J]. Acta Hortic. Sin., 2020, 47(9):1715-1726. | |
| [3] | STOMMEL J R, LIGHTBOURN G J, WINKEL B S, et al.. Transcription factor families regulate the anthocyanin biosynthetic pathway in Capsicum annuum [J]. J. Am. Soc. Hortic. Sci., 2009, 134(2): 244-251. |
| [4] | CHALKER-SCOTT L. Environmental significance of anthocyanins in plant stress responses [J]. Photochem. Photobiol., 1999, 70(1):1-9. |
| [5] | DE PASCUAL-TERESA S, MORENO D A, GARCÍA-VIGUERA C.Flavanols and anthocyanins in cardiovascular health:a review of current evidence [J]. Int. J. Mol. Sci., 2010,11(4):1679-1703. |
| [6] | LIU Y, TIKUNOV Y, SCHOUTEN R E,et al.. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables:a review [J/OL].Front.Chem.,2018,6:52 [2024-02-20]. . |
| [7] | JAAKOLA L.New insights into the regulation of anthocyanin biosynthesis in fruits [J]. Trends Plant Sci., 2013, 18(9):477-483. |
| [8] | BOROVSKY Y, OREN-SHAMIR M, OVADIA R, et al.. The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia [J]. Theor. Appl. Genet., 2004, 109(1):23-29. |
| [9] | JUNG S, VENKATESH J, KANG M Y,et al..A non-LTR retrotransposon activates anthocyanin biosynthesis by regulating a MYB transcription factor in Capsicum annuum [J/OL].Plant Sci.,2019,287:110181 [2024-02-20]. . |
| [10] | LIU J Q, AI X Y, WANG Y H, et al..Fine mapping of the Ca3GT gene controlling anthocyanin biosynthesis in mature unripe fruit of Capsicum annuum L. [J]. Theor. Appl. Genet., 2020,133(9):2729-2742. |
| [11] | WANG G, CHEN B, DU H,et al..Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA [J/OL].PLoS One,2018,13(9):e0204690 [2024-02-20]. . |
| [12] | TANG B Y, LI L, HU Z L, et al.. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple pepper [J]. J. Agric. Food Chem., 2020,68(43):12152-12163. |
| [13] | LIU Y H, LYU J H, LIU Z B,et al..Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.) [J/OL]. Food Chem., 2020, 306:125629 [2024-02-20]. . |
| [14] | MENG Y, ZHANG H, FAN Y, et al.. Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum) [J/OL]. BMC Plant Biol., 2022,22(1):358 [2024-02-20]. . |
| [15] | WANG Y, LIU S, WANG H, et al.. Identification of the regulatory genes of UV-B-induced anthocyanin biosynthesis in pepper fruit [J/OL]. Int. J. Mol. Sci., 2022,23(4):1960 [2024-02-20]. . |
| [16] | ZHOU Y, MUMTAZ MALI, ZHANG Y H, et al.. Response of anthocyanin biosynthesis to light by strand-specific transcriptome and miRNA analysis in Capsicum annuum [J/OL]. BMC Plant Biol., 2022,22(1):79 [2024-02-20]. . |
| [17] | TAN H, LI L, TIE M, et al.. Transcriptome analysis of green and purple fruited pepper provides insight into novel regulatory genes in anthocyanin biosynthesis [J/OL]. PeerJ, 2024,12:e16792 [2024-02-20]. . |
| [18] | PASSERI V, KOES R, QUATTROCCHIO F M. New challenges for the design of high value plant products:stabilization of anthocyanins in plant vacuoles [J/OL]. Front. Plant Sci., 2016,7:153 [2024-02-20]. . |
| [19] | KOES R, VERWEIJ W, QUATTROCCHIO F.Flavonoids:a colorful model for the regulation and evolution of biochemical pathways [J]. Trends Plant Sci., 2005,10(5):236-242. |
| [20] | VERWEIJ W, SPELT C, DI SANSEBASTIANO G P, et al.. An H+P-ATPase on the tonoplast determines vacuolar pH and flower colour [J]. Nat. Cell Biol., 2008,10:1456-1462. |
| [21] | FARACO M, SPELT C, BLIEK M, et al..Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color [J]. Cell Rep., 2014,6(1):32-43. |
| [22] | DE VLAMING P, SCHRAM A W, WIERING H. Genes affecting flower colour and pH of flower limb homogenates in Petunia hybrid [J]. Theor. Appl. Genet., 1983, 66(3):271-278. |
| [23] | DE VLAMING P, VAN EEKERES J E M, WIERING H. A gene for flower colour fading in Petunia hybrid [J]. Theor. Appl.Genet., 1982, 61(1):41-46. |
| [24] | CHASSY A W, BUESCHL C, LEE H, et al.. Tracing flavonoid degradation in grapes by MS filtering with stable isotopes [J]. Food Chem., 2015,166:448-455. |
| [25] | OREN-SHAMIR M.Does anthocyanin degradation play a significant role in determining pigment concentration in plants? [J]. Plant Sci., 2009, 177(4):310-316. |
| [26] | FANG F, ZHANG X L, LUO H H, et al.. An intracellular laccase is responsible for epicatechin-mediated anthocyanin degradation in Litchi fruit pericarp [J]. Plant Physiol., 2015, 169(4):2391-2408. |
| [27] | MOVAHED N, PASTORE C, CELLINI A, et al.. The grapevine VviPrx31 peroxidase as a candidate gene involved in anthocyanin degradation in ripening berries under high temperature [J]. J. Plant Res., 2016, 129(3):513-526. |
| [28] | ZIPOR G, DUARTE P, CARQUEIJEIRO I,et al..In planta anthocyanin degradation by a vacuolar class Ⅲ peroxidase in Brunfelsia calycina flowers [J]. N. Phytol., 2015, 205(2):653-665. |
| [29] | WANG L S, BURHENNE K, KRISTENSEN B K, et al.. Purification and cloning of a Chinese red radish peroxidase that metabolise pelargonidin and forms a gene family in Brassicaceae [J]. Gene, 2004, 343(2):323-335. |
| [30] | YAMADA Y, NAKAYAMA M, SHIBATA H, et al.. Anthocyanin production and enzymatic degradation during the development of dark purple and lilac paprika fruit [J]. J. Am. Soc. Hortic. Sci., 2019, 144(5): 329-338. |
| [31] | SAKAMURA S, OBATA Y.Anthocyanase and anthocyanins occurring in eggplant,Solanum melongena L.(I) [J]. Agric. Biol. Chem., 1961, 25(10):750-756. |
| [32] | BARBAGALLO R N, PALMERI R, FABIANO S, et al.. Characteristic of β-glucosidase from Sicilian blood oranges in relation to anthocyanin degradation [J]. Enzyme Microb. Technol., 2007, 41(5):570-575. |
| [33] | DONG B, LUO H, LIU B, et al.. BcXyl,a β-xylosidase isolated from Brunfelsia calycina flowers with anthocyanin-β-glycosidase activity [J/OL]. Investig. N. Drugs, 2019,20(6):E1423 [2024-02-20]. . |
| [34] | ZHAO G, XIANG F, ZHANG S, et al.. PbLAC4-like,activated by PbMYB26,related to the degradation of anthocyanin during color fading in pear [J/OL]. BMC Plant Biol., 2021,21(1):469 [2024-02-20]. . |
| [35] | AZA-GONZÁLEZ C, HERRERA-ISIDRÓN L, NÚÑEZ-PALENIUS H G,et al..Anthocyanin accumulation and expression analysis of biosynthesis-related genes during chili pepper fruit development [J]. Biol. Plant., 2013, 57(1):49-55. |
| [36] | CHEN S F, ZHOU Y Q, CHEN Y R, et al.. Fastp:an ultra-fast all-in-one FASTQ preprocessor [J]. Bioinformatics, 2018,34(17):884-890. |
| [37] | KIM D, LANGMEAD B, SALZBERG S L. HISAT:a fast spliced aligner with low memory requirements [J]. Nat. Methods, 2015,12:357-360. |
| [38] | KIM S, PARK M, YEOM S I, et al.. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species [J]. Nat. Genet., 2014, 46:270-278. |
| [39] | PERTEA M, KIM D, PERTEA G M,et al..Transcript-level expression analysis of RNA-seq experiments with HISAT,StringTie and Ballgown [J]. Nat. Protoc., 2016,11:1650-1667. |
| [40] | VARET H, BRILLET-GUÉGUEN L, COPPÉE J Y,et al..SARTools:a DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data [J/OL].PLoS One, 2016,11(6):e0157022 [2024-02-20]. . |
| [41] | CHEN C J, WU Y, LI J W, et al.. TBtools-II:a “one for all, all for one” bioinformatics platform for biological big-data mining [J].Mol. Plant, 2023,16(11):1733-1742. |
| [42] | ZHENG Y, JIAO C, SUN H, et al.. ITAK:a program for Genome-wide prediction and classification of plant transcription factors,transcriptional regulators,and protein kinases [J]. Mol. Plant, 2016,9(12):1667-1670. |
| [43] | LIVAK K J, SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001,25(4):402-408. |
| [44] | TOGNOLLI M, PENEL C, GREPPIN H,et al..Analysis and expression of the class Ⅲ peroxidase large gene family in Arabidopsis thaliana [J]. Gene, 2002,288(1/2):129-138. |
| [45] | LANGFELDER P, HORVATH S. WGCNA:an R package for weighted correlation network analysis [J/OL].BMC Bioinf.,2008,9:559 [2024-02-20]. . |
| [46] | SEYMOUR G B, ØSTERGAARD L, CHAPMAN N H,et al..Fruit development and ripening [J]. Annu. Rev. Plant Biol., 2013,64:219-241. |
| [47] | GROSS K C, WATADA A E, KANG M S, et al.. Biochemical changes associated with the ripening of hot pepper fruit [J]. Physiol. Plant., 1986, 66(1):31-36. |
| [48] | PRIYA SETHU K M, PRABHA T N, THARANATHAN R N. Post-harvest biochemical changes associated with the softening phenomenon in Capsicum annuum fruits [J].Phytochemistry,1996,42(4):961-966. |
| [49] | JANG Y K, JUNG E S, LEE H A,et al..Metabolomic characterization of hot pepper (Capsicum annuum “CM334”) during fruit development [J]. J. Agric. Food Chem., 2015,63(43):9452-9460. |
| [50] | LIU Z, LYU J, ZHANG Z, et al.. Integrative transcriptome and proteome analysis identifies major metabolic pathways involved in pepper fruit development [J].J. Proteome Res., 2019,18(3):982-994. |
| [51] | SONG J, SUN B, CHEN C, et al.. An R-R-type MYB transcription factor promotes non-climacteric pepper fruit carotenoid pigment biosynthesis [J]. Plant J., 2023,115(3):724-741. |
| [52] | TAO H, GAO F, LINYING LI,et al..WRKY33 negatively regulates anthocyanin biosynthesis and cooperates with PHR1 to mediate acclimation to phosphate starvation [J/OL]. Plant Commun., 2024,5(5):100821 [2024-02-20]. . |
| [53] | DEVAIAH B N, KARTHIKEYAN A S, RAGHOTHAMA K G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis [J].Plant Physiol., 2007,143(4):1789-1801. |
| [54] | DUAN S W, WANG J J, GAO C H, et al..Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana [J]. Plant Sci., 2018,268:47-53. |
| [55] | MAO Z L, JIANG H Y, WANG S,et al..The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple [J/OL]. Plant Sci., 2021,306:110848 [2024-02-20]. . |
| [56] | ZHANG Z Q, PANG X Q, JI Z L, et al.. Role of anthocyanin degradation in litchi pericarp browning [J]. Food Chem., 2001,75(2):217-221. |
| [57] | NORMAN C, RUNSWICK M, POLLOCK R, et al.. Isolation and properties of cDNA clones encoding SRF,a transcription factor that binds to the c-fos serum response element [J]. Cell, 1988,55(6):989-1003. |
| [58] | SAEDLER H, BECKER A, WINTER K U,et al.. MADS-box genes are involved in floral development and evolution [J]. Acta Biochim. Pol., 2001, 48(2):351-358. |
| [59] | LI C, WANG Y, XU L, et al.. Genome-wide characterization of the MADS-box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis [J/OL]. Front. Plant Sci., 2016,7:1390 [2024-02-20]. . |
| [60] | LU W J, CHEN J X, REN X C, et al.. One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening [J]. Sci. Hortic., 2018, 227:124-131. |
| [61] | MA G Y, ZOU Q C, SHI X H, et al.. Ectopic expression of the AaFUL1 gene identified in Anthurium andraeanum affected floral organ development and seed fertility in tobacco [J]. Gene, 2019, 696:197-205. |
| [62] | WU R, WANG T, MCGIE T,et al..Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals,but has no effect on vegetative growth,dormancy,or flowering time [J]. J. Exp.Bot., 2014, 65(17):4985-4995. |
| [63] | QI F T, LIU Y T, LUO Y L, et al.. Functional analysis of the ScAG and ScAGL11 MADS-box transcription factors for anthocyanin biosynthesis and bicolour pattern formation in Senecio cruentus ray florets [J/OL]. Hortic. Res., 2022,9:uhac071 [2024-02-20]. . |
| [64] | OOKA H, SATOH K, DOI K, et al.. Comprehensive analysis of nac family genes in Oryza sativa and Arabidopsis thaliana [J].DNA Res., 2003, 10(6):239-247. |
| [65] | OLSEN A N, ERNST H A, LEGGIO L L, et al.. NAC transcription factors:structurally distinct,functionally diverse [J]. Trends Plant Sci., 2005, 10(2):79-87. |
| [66] | NURUZZAMAN M, SHARONI A M, KIKUCHI S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants [J/OL]. Front. Microbiol., 2013,4:248 [2024-02-20]. . |
| [67] | MAHMOOD K, XU Z, EL-KEREAMY A, et al.. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses [J/OL]. Front. Plant Sci., 2016,7:1548 [2024-02-20]. . |
| [68] | WU A H, ALLU A D, GARAPATI P, et al.. JUNGBRUNNEN1,a reactive oxygen species-responsive NAC transcription factor,regulates longevity in Arabidopsis [J]. Plant Cell, 2012,24(2):482-506. |
| [69] | WANG J F, LIAN W R, CAO Y Y, et al.. Overexpression of BoNAC019,a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis [J/OL]. Sci. Rep., 2018, 8:13349 [2024-02-20]. . |
| [1] | Linghui YANG, Zhiwei DING, Li GONG, Xuejun LI, Yunping DONG, Zhenjiang LYU. Progress in Extraction, Synthetic Metabolic Pathways, and Bioactivity Research of Coffea Alkaloid Main Compounds [J]. Journal of Agricultural Science and Technology, 2025, 27(8): 132-143. |
| [2] | Dongyu LIU-XU, Xiaoxiao GUO, Chenqing FU, Rui HAN, Guohui LI, Xiuping WANG. Prediction of Anthocyanin Content in Perilla frutescens Leaves Based on RGB and CIELab [J]. Journal of Agricultural Science and Technology, 2024, 26(7): 103-110. |
| [3] | Lixia CHEN, Jingze LIU, Lizhu WU, Yinzhu SHEN. Food Security:Current Situation, Problem and Countermeasure [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 7-14. |
| [4] | Mingjie SHAO, Yanqi CHEN, Wenke LIU. Effects of Red-blue LED Bight Supply Modes on Growth and Quality of Purple Leaf Lettuce Under Continuous Light [J]. Journal of Agricultural Science and Technology, 2023, 25(4): 77-85. |
| [5] | Chunyu LIU, Lina MEN, Yaqin LIAN, Yuhong ZHANG, Zhiwei ZHANG, Wei ZHANG. Expression and Characterization of Enzymes for Polyethylene Degradation in the Gut of Galleria mellonella L. larvae [J]. Journal of Agricultural Science and Technology, 2023, 25(3): 132-139. |
| [6] | Wenzhu YANG, Rumei CHEN. Breeding Progress of Anthocyanin Corn [J]. Journal of Agricultural Science and Technology, 2022, 24(8): 18-24. |
| [7] | Yuhong WU, Rongjun GUO, Guizhen MA, Shidong LI. Characteristics of the Growth of Rhodococcuspyridinivorans Rp3 and Ability to Degrade Skatole [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 82-89. |
| [8] | Lu LIU, Xiuping TAO, Jianchao SONG, Bin SHANG, Wenqian XU, Hongmin DONG, Yangyang CAI. Bench-scale Study on Operation Effect and Power Generation Performance Treatment of Dairy Farms Wastewater by Microbial Fuel Cell [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 134-143. |
| [9] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [10] | LI Guowei1, HE Jing1, GUO Kunjie1, JIRIMUTU1,2*. Screening of Cellulose-degrading Bacteria from Bactrian Camel and Their Degradation Effect on Distillers Grains [J]. Journal of Agricultural Science and Technology, 2021, 23(4): 64-75. |
| [11] | HE Yefeng1, ZHANG Chunying2, LIU Qunlu1,2*. Characterization of Flower Coloration in Petals of Rhododendron simsii#br# [J]. Journal of Agricultural Science and Technology, 2021, 23(2): 50-56. |
| [12] | YANG Mengya1, YAN Feifan1, YAN Meichao1, WANG He1, PIAO Renzhe1, CUI Zongjun2, ZHAO Hongyan1*. Decomposition Characteristics of Corn Stover by Microbial Consortium PLC-8 with Lignocellulose-degradation at Low Temperature [J]. Journal of Agricultural Science and Technology, 2021, 23(1): 73-81. |
| [13] | WAN Dongpu1,2, YU Zhuo1, WU Yanmin3, DING Mengqi2, LI Jinbo2, ZHOU Meiliang2*. Regulation of Anthocyanin Metabolism on Colored Leaves of Plants [J]. Journal of Agricultural Science and Technology, 2020, 22(2): 30-38. |
| [14] | GUO Lianghu1, WANG Zhen1, LI Nan1, LU Lahu1, GAO Wei1, GUO Yaxian1, WU Zongxin2*. Effects of Temperature and Humidity on Storage Quality of Purple Sweet Potato [J]. Journal of Agricultural Science and Technology, 2020, 22(2): 107-114. |
| [15] | LIU Yang1, WANG Wei1, ZHU Xuran1, ZHANG Jiaxiao1, LIU Yuan1,2, WANG Jian1,2*. Study on Residue and Degradation Dynamics of Dimethomorph in Potato and Its Soil [J]. Journal of Agricultural Science and Technology, 2019, 21(9): 104-110. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号