




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (2): 48-59.DOI: 10.13304/j.nykjdb.2021.0699
张曼( ), 王志城, 刘正文, 王国宁, 王省芬, 张艳(
), 王志城, 刘正文, 王国宁, 王省芬, 张艳( )
)
收稿日期:2021-08-14
接受日期:2021-09-22
出版日期:2023-02-15
发布日期:2023-05-17
通讯作者:
张艳
作者简介:张曼 E-mail:zhangman920601@163.com;
基金资助:
Man ZHANG( ), Zhicheng WANG, Zhengwen LIU, Guoning WANG, Xingfen WANG, Yan ZHANG(
), Zhicheng WANG, Zhengwen LIU, Guoning WANG, Xingfen WANG, Yan ZHANG( )
)
Received:2021-08-14
Accepted:2021-09-22
Online:2023-02-15
Published:2023-05-17
Contact:
Yan ZHANG
摘要:
植物BGLU基因在生物和非生物胁迫防御中起着重要作用,利用生物信息学手段对陆地棉(Gossypium hirsutum) BGLU家族成员进行全基因组鉴定,对序列特征、保守域结构、染色体定位、启动子元件等家族特征进行了分析,并进一步结合转录组数据和qRT-PCR分析了BGLU基因家族的病菌胁迫响应模式。结果表明,共鉴定出53个陆地棉BGLU基因,根据系统发育进化关系分为6个亚族,部分基因在病菌胁迫下特异性表达。荧光定量和转录组测序结果表明,GhBGLU5、GhBGLU14、GhBGLU18、GhBGLU26、GhBGLU34在抗病棉花中的表达水平显著高于感病品种,推测这些基因正向调控棉花黄萎病抗性。上述结果为进一步分析棉花BGLU家族基因的功能以及分子机制提供一定的理论基础。
中图分类号:
张曼, 王志城, 刘正文, 王国宁, 王省芬, 张艳. 陆地棉BGLU基因家族成员的全基因组鉴定与表达分析[J]. 中国农业科技导报, 2023, 25(2): 48-59.
Man ZHANG, Zhicheng WANG, Zhengwen LIU, Guoning WANG, Xingfen WANG, Yan ZHANG. Genome-Wide Identification and Analysis of BGLU Genes Family in Gossypium hirsutum[J]. Journal of Agricultural Science and Technology, 2023, 25(2): 48-59.
引物名称 Primer name | 引物序列 Primer sequence(3’-5’) |
|---|---|
| GhUBQ14-F | CAACGCTCCATCTTGTCCTT |
| GhUBQ14-R | TGATCGTCTTTCCCGTAAGC |
| GhBGLU9-RT-F | GGCGGTGATGGACACTGGA |
| GhBGLU9-RT-R | GCCACCCTTATTAGCCATCC |
| GhBGLU19-RT-F | CGTAGAACATGATGATTCCAACACT |
| GhBGLU19-RT-R | CCACCTTCGTTCACACCTCCTT |
| GhBGLU05-RT-F | GTTTTATTTTCGGTGCCGGAT |
| GhBGLU05-RT-R | GATGTTGTCTAACGAGAATGTCCC |
| GhBGLU23-RT-F | CGAGGGAGCAGTGAAGGAGGAT |
| GhBGLU23-RT-R | TATCTTCAACATAGCGGTGGTACT |
| GhBGLU26-RT-F | AAGATGGAAAAGGCCTAAGCAAC |
| GhBGLU26-RT-R | GCATCATCTCCATTCTCATTGTTTT |
| GhBGLU34-RT-F | TGTGTTCTATCTGTTGTTCCTTTGG |
| GhBGLU34-RT-R | CTGTATCTCCATTGCTTCCATCCAT |
| GhBGLU52-RT-F | CTGAAACAAAGCCTGGGTAATGGTA |
| GhBGLU52-RT-R | CTTCATACTGGTAAGCAGAGGAGGC |
| GhBGLU35-RT-F | GTGAATGAAGGAAACAAGGGAGATA |
| GhBGLU35-RT-R | AAGCATCCATTCCCAGATCTTTCAT |
| GhBGLU14-RT-F | GTTCTTCTTCTTACTCAACTTCATTTC |
| GhBGLU14-RT-R | TTCATACTGGTAAGCGGAAGAGGC |
表1 本研究中所用qRT?PCR引物序列
Table 1 Primer sequences for qRT?PCR in this study
引物名称 Primer name | 引物序列 Primer sequence(3’-5’) |
|---|---|
| GhUBQ14-F | CAACGCTCCATCTTGTCCTT |
| GhUBQ14-R | TGATCGTCTTTCCCGTAAGC |
| GhBGLU9-RT-F | GGCGGTGATGGACACTGGA |
| GhBGLU9-RT-R | GCCACCCTTATTAGCCATCC |
| GhBGLU19-RT-F | CGTAGAACATGATGATTCCAACACT |
| GhBGLU19-RT-R | CCACCTTCGTTCACACCTCCTT |
| GhBGLU05-RT-F | GTTTTATTTTCGGTGCCGGAT |
| GhBGLU05-RT-R | GATGTTGTCTAACGAGAATGTCCC |
| GhBGLU23-RT-F | CGAGGGAGCAGTGAAGGAGGAT |
| GhBGLU23-RT-R | TATCTTCAACATAGCGGTGGTACT |
| GhBGLU26-RT-F | AAGATGGAAAAGGCCTAAGCAAC |
| GhBGLU26-RT-R | GCATCATCTCCATTCTCATTGTTTT |
| GhBGLU34-RT-F | TGTGTTCTATCTGTTGTTCCTTTGG |
| GhBGLU34-RT-R | CTGTATCTCCATTGCTTCCATCCAT |
| GhBGLU52-RT-F | CTGAAACAAAGCCTGGGTAATGGTA |
| GhBGLU52-RT-R | CTTCATACTGGTAAGCAGAGGAGGC |
| GhBGLU35-RT-F | GTGAATGAAGGAAACAAGGGAGATA |
| GhBGLU35-RT-R | AAGCATCCATTCCCAGATCTTTCAT |
| GhBGLU14-RT-F | GTTCTTCTTCTTACTCAACTTCATTTC |
| GhBGLU14-RT-R | TTCATACTGGTAAGCGGAAGAGGC |
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU01 | Ghir_A01G020950 | 359 | 42.213 | 8.740 | 35~36 | 细胞质膜Plasma membrane |
| GhBGLU02 | Ghir_A01G021100 | 495 | 57.410 | 6.123 | 23~24 | 液泡Vacuole |
| GhBGLU03 | Ghir_A01G021370 | 502 | 58.129 | 7.219 | 23~24 | 液泡Vacuole |
| GhBGLU04 | Ghir_A02G012630 | 390 | 44.336 | 5.309 | 38~39 | 细胞质Cytoplasmic |
| GhBGLU05 | Ghir_A05G007460 | 476 | 54.519 | 5.022 | 21~22 | 液泡Vacuole |
| GhBGLU06 | Ghir_A06G012310 | 491 | 55.875 | 9.314 | 23~24 | 溶酶体Lysosomal |
| GhBGLU07 | Ghir_A06G015860 | 527 | 59.958 | 6.555 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU08 | Ghir_A06G015870 | 494 | 56.087 | 6.519 | 21~22 | 液泡Vacuole |
| GhBGLU09 | Ghir_A07G008330 | 331 | 37.350 | 9.361 | 19~20 | 胞外Extracellular |
| GhBGLU10 | Ghir_A07G008340 | 497 | 56.916 | 8.338 | 19~20 | 液泡Vacuole |
| GhBGLU11 | Ghir_A08G024630 | 520 | 58.768 | 7.434 | 28~29 | 细胞质Cytoplasmic |
| GhBGLU12 | Ghir_A08G025080 | 371 | 41.575 | 6.883 | 26~27 | 细胞质膜Plasma membrane |
表2 53个陆地棉BGLU蛋白的理化性质
Table 2 Characteristics of 53 BGLU proteins in G. hirsutum
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU01 | Ghir_A01G020950 | 359 | 42.213 | 8.740 | 35~36 | 细胞质膜Plasma membrane |
| GhBGLU02 | Ghir_A01G021100 | 495 | 57.410 | 6.123 | 23~24 | 液泡Vacuole |
| GhBGLU03 | Ghir_A01G021370 | 502 | 58.129 | 7.219 | 23~24 | 液泡Vacuole |
| GhBGLU04 | Ghir_A02G012630 | 390 | 44.336 | 5.309 | 38~39 | 细胞质Cytoplasmic |
| GhBGLU05 | Ghir_A05G007460 | 476 | 54.519 | 5.022 | 21~22 | 液泡Vacuole |
| GhBGLU06 | Ghir_A06G012310 | 491 | 55.875 | 9.314 | 23~24 | 溶酶体Lysosomal |
| GhBGLU07 | Ghir_A06G015860 | 527 | 59.958 | 6.555 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU08 | Ghir_A06G015870 | 494 | 56.087 | 6.519 | 21~22 | 液泡Vacuole |
| GhBGLU09 | Ghir_A07G008330 | 331 | 37.350 | 9.361 | 19~20 | 胞外Extracellular |
| GhBGLU10 | Ghir_A07G008340 | 497 | 56.916 | 8.338 | 19~20 | 液泡Vacuole |
| GhBGLU11 | Ghir_A08G024630 | 520 | 58.768 | 7.434 | 28~29 | 细胞质Cytoplasmic |
| GhBGLU12 | Ghir_A08G025080 | 371 | 41.575 | 6.883 | 26~27 | 细胞质膜Plasma membrane |
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU13 | Ghir_A09G005920 | 506 | 58.313 | 9.323 | 20~21 | 溶酶体Lysosomal |
| GhBGLU14 | Ghir_A10G001000 | 536 | 61.583 | 6.510 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU15 | Ghir_A10G001010 | 500 | 57.229 | 7.590 | 27~28 | 液泡Vacuole |
| GhBGLU16 | Ghir_A10G001020 | 536 | 61.339 | 6.976 | 24~25 | 溶酶体Lysosomal |
| GhBGLU17 | Ghir_A11G032270 | 514 | 58.455 | 6.337 | 26~27 | 细胞质Cytoplasmic |
| GhBGLU18 | Ghir_A11G033190 | 199 | 23.264 | 4.694 | 40~41 | 细胞质Cytoplasmic |
| GhBGLU19 | Ghir_A11G033200 | 195 | 22.294 | 6.799 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU20 | Ghir_A12G017270 | 132 | 15.366 | 7.963 | 19~20 | 胞外Extracellular |
| GhBGLU21 | Ghir_A12G026530 | 482 | 55.028 | 9.470 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU22 | Ghir_A13G008250 | 498 | 56.644 | 5.970 | 23~24 | 液泡Vacuole |
| GhBGLU23 | Ghir_A13G008260 | 136 | 15.630 | 9.499 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU24 | Ghir_A13G008270 | 504 | 56.842 | 7.635 | 24~25 | 液泡Vacuole |
| GhBGLU25 | Ghir_A13G014930 | 383 | 43.308 | 4.834 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU26 | Ghir_A13G015150 | 546 | 62.303 | 6.561 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU27 | Ghir_D01G022500 | 445 | 51.828 | 8.334 | 18~19 | 细胞质膜Plasma membrane |
| GhBGLU28 | Ghir_D01G022510 | 500 | 57.851 | 7.275 | 19~20 | 液泡Vacuole |
| GhBGLU29 | Ghir_D01G022660 | 499 | 58.085 | 6.203 | 23~24 | 液泡Vacuole |
| GhBGLU30 | Ghir_D01G022670 | 511 | 59.569 | 6.400 | 23~24 | 液泡Vacuole |
| GhBGLU31 | Ghir_D03G006350 | 328 | 37.578 | 6.139 | 27~28 | 细胞质Cytoplasmic |
| GhBGLU32 | Ghir_D05G007520 | 482 | 55.193 | 5.549 | 21~22 | 液泡Vacuole |
| GhBGLU33 | Ghir_D06G012190 | 437 | 50.036 | 9.662 | 21~22 | 溶酶体Lysosomal |
| GhBGLU34 | Ghir_D06G016620 | 483 | 54.692 | 6.788 | 16~17 | 细胞质膜Plasma membrane |
| GhBGLU35 | Ghir_D06G016640 | 534 | 60.754 | 6.782 | 21~22 | 细胞质膜Plasma membrane |
| GhBGLU36 | Ghir_D07G008440 | 139 | 15.520 | 8.918 | 19~20 | 胞外Extracellular |
| GhBGLU37 | Ghir_D07G008450 | 479 | 54.695 | 8.085 | 19~20 | 溶酶体Lysosomal |
| GhBGLU38 | Ghir_D08G025520 | 514 | 57.962 | 6.515 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU39 | Ghir_D08G025990 | 463 | 52.616 | 7.511 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU40 | Ghir_D09G005490 | 341 | 39.515 | 9.728 | 24~25 | 胞外Extracellular |
| GhBGLU41 | Ghir_D10G001780 | 466 | 53.328 | 5.882 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU42 | Ghir_D10G001790 | 509 | 58.366 | 7.255 | 27~28 | 细胞质膜Plasma membrane |
| GhBGLU43 | Ghir_D10G001800 | 518 | 58.984 | 6.416 | 24~25 | 溶酶体Lysosomal |
| GhBGLU44 | Ghir_D11G032750 | 514 | 58.322 | 6.206 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU45 | Ghir_D11G033820 | 490 | 56.008 | 5.179 | 38~29 | 细胞质Cytoplasmic |
| GhBGLU46 | Ghir_D12G017490 | 121 | 13.931 | 7.500 | 47~48 | 胞外Extracellular |
| GhBGLU47 | Ghir_D12G026540 | 482 | 54.786 | 9.368 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU48 | Ghir_D12G026620 | 472 | 53.808 | 9.573 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU49 | Ghir_D13G008490 | 504 | 56.847 | 6.680 | 24~25 | 液泡Vacuole |
| GhBGLU50 | Ghir_D13G008500 | 205 | 23.526 | 8.424 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU51 | Ghir_D13G008510 | 498 | 56.553 | 5.218 | 23~24 | 液泡Vacuole |
| GhBGLU52 | Ghir_D13G015630 | 488 | 55.822 | 8.565 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU53 | Ghir_D13G015870 | 546 | 61.929 | 6.658 | 26~27 | 细胞质膜Plasma membrane |
表2 53个陆地棉BGLU蛋白的理化性质 (续表Continued)
Table 2 Characteristics of 53 BGLU proteins in G. hirsutum
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU13 | Ghir_A09G005920 | 506 | 58.313 | 9.323 | 20~21 | 溶酶体Lysosomal |
| GhBGLU14 | Ghir_A10G001000 | 536 | 61.583 | 6.510 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU15 | Ghir_A10G001010 | 500 | 57.229 | 7.590 | 27~28 | 液泡Vacuole |
| GhBGLU16 | Ghir_A10G001020 | 536 | 61.339 | 6.976 | 24~25 | 溶酶体Lysosomal |
| GhBGLU17 | Ghir_A11G032270 | 514 | 58.455 | 6.337 | 26~27 | 细胞质Cytoplasmic |
| GhBGLU18 | Ghir_A11G033190 | 199 | 23.264 | 4.694 | 40~41 | 细胞质Cytoplasmic |
| GhBGLU19 | Ghir_A11G033200 | 195 | 22.294 | 6.799 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU20 | Ghir_A12G017270 | 132 | 15.366 | 7.963 | 19~20 | 胞外Extracellular |
| GhBGLU21 | Ghir_A12G026530 | 482 | 55.028 | 9.470 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU22 | Ghir_A13G008250 | 498 | 56.644 | 5.970 | 23~24 | 液泡Vacuole |
| GhBGLU23 | Ghir_A13G008260 | 136 | 15.630 | 9.499 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU24 | Ghir_A13G008270 | 504 | 56.842 | 7.635 | 24~25 | 液泡Vacuole |
| GhBGLU25 | Ghir_A13G014930 | 383 | 43.308 | 4.834 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU26 | Ghir_A13G015150 | 546 | 62.303 | 6.561 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU27 | Ghir_D01G022500 | 445 | 51.828 | 8.334 | 18~19 | 细胞质膜Plasma membrane |
| GhBGLU28 | Ghir_D01G022510 | 500 | 57.851 | 7.275 | 19~20 | 液泡Vacuole |
| GhBGLU29 | Ghir_D01G022660 | 499 | 58.085 | 6.203 | 23~24 | 液泡Vacuole |
| GhBGLU30 | Ghir_D01G022670 | 511 | 59.569 | 6.400 | 23~24 | 液泡Vacuole |
| GhBGLU31 | Ghir_D03G006350 | 328 | 37.578 | 6.139 | 27~28 | 细胞质Cytoplasmic |
| GhBGLU32 | Ghir_D05G007520 | 482 | 55.193 | 5.549 | 21~22 | 液泡Vacuole |
| GhBGLU33 | Ghir_D06G012190 | 437 | 50.036 | 9.662 | 21~22 | 溶酶体Lysosomal |
| GhBGLU34 | Ghir_D06G016620 | 483 | 54.692 | 6.788 | 16~17 | 细胞质膜Plasma membrane |
| GhBGLU35 | Ghir_D06G016640 | 534 | 60.754 | 6.782 | 21~22 | 细胞质膜Plasma membrane |
| GhBGLU36 | Ghir_D07G008440 | 139 | 15.520 | 8.918 | 19~20 | 胞外Extracellular |
| GhBGLU37 | Ghir_D07G008450 | 479 | 54.695 | 8.085 | 19~20 | 溶酶体Lysosomal |
| GhBGLU38 | Ghir_D08G025520 | 514 | 57.962 | 6.515 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU39 | Ghir_D08G025990 | 463 | 52.616 | 7.511 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU40 | Ghir_D09G005490 | 341 | 39.515 | 9.728 | 24~25 | 胞外Extracellular |
| GhBGLU41 | Ghir_D10G001780 | 466 | 53.328 | 5.882 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU42 | Ghir_D10G001790 | 509 | 58.366 | 7.255 | 27~28 | 细胞质膜Plasma membrane |
| GhBGLU43 | Ghir_D10G001800 | 518 | 58.984 | 6.416 | 24~25 | 溶酶体Lysosomal |
| GhBGLU44 | Ghir_D11G032750 | 514 | 58.322 | 6.206 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU45 | Ghir_D11G033820 | 490 | 56.008 | 5.179 | 38~29 | 细胞质Cytoplasmic |
| GhBGLU46 | Ghir_D12G017490 | 121 | 13.931 | 7.500 | 47~48 | 胞外Extracellular |
| GhBGLU47 | Ghir_D12G026540 | 482 | 54.786 | 9.368 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU48 | Ghir_D12G026620 | 472 | 53.808 | 9.573 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU49 | Ghir_D13G008490 | 504 | 56.847 | 6.680 | 24~25 | 液泡Vacuole |
| GhBGLU50 | Ghir_D13G008500 | 205 | 23.526 | 8.424 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU51 | Ghir_D13G008510 | 498 | 56.553 | 5.218 | 23~24 | 液泡Vacuole |
| GhBGLU52 | Ghir_D13G015630 | 488 | 55.822 | 8.565 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU53 | Ghir_D13G015870 | 546 | 61.929 | 6.658 | 26~27 | 细胞质膜Plasma membrane |
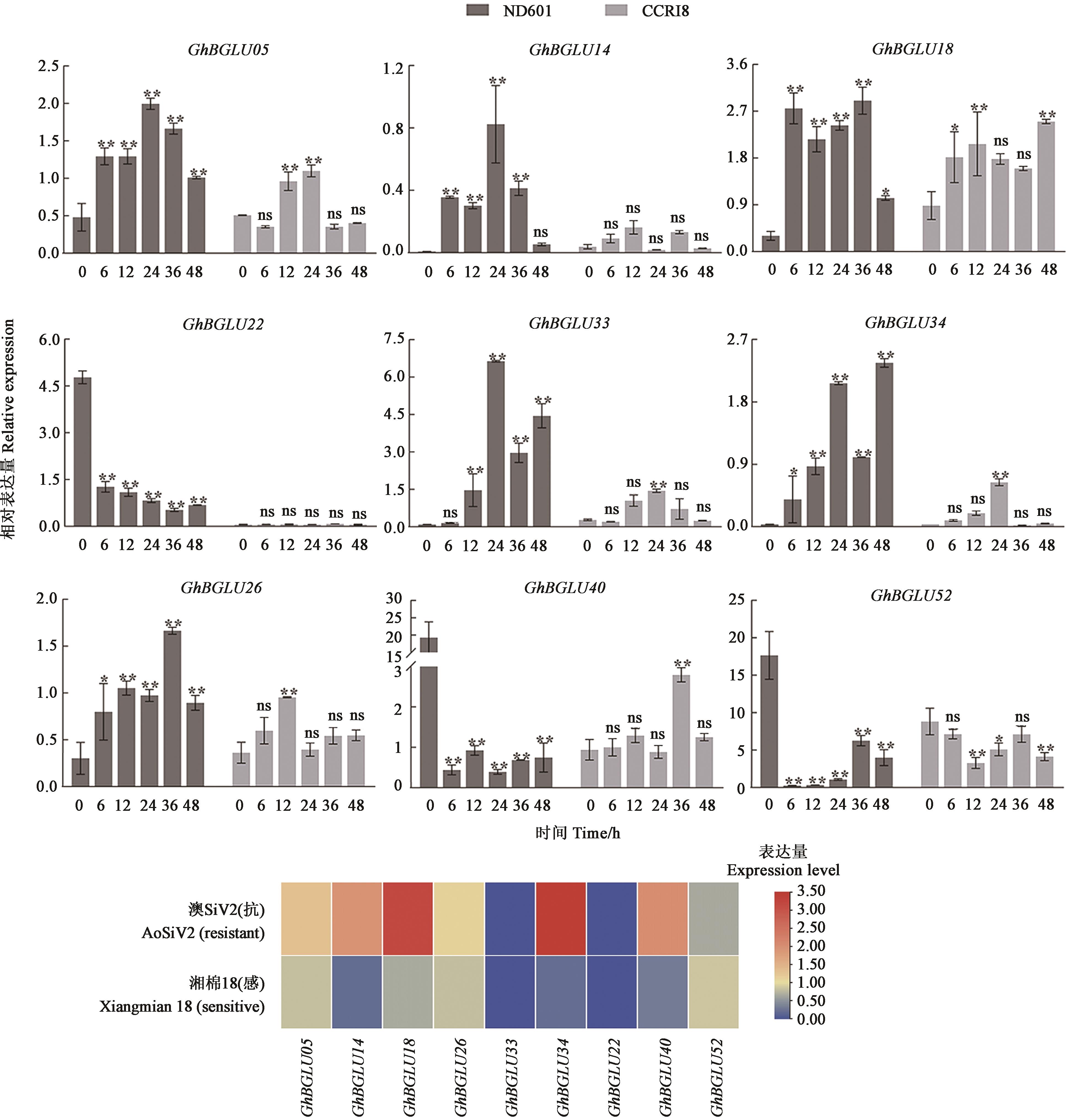
图7 黄萎病菌胁迫下9个GhBGLU基因的表达分析注:**和*分别表示在P<0.01和 P<0.05水平差异显著。
Fig. 7 Analysis of the expression of 9 GhBGLU genes under the stress of V. dahliaeNote: ** and * indicate significant differences at P<0.01 and P<0.05 levels, respectively.
| 1 | WANG K, WANG Z, LI F, et al.. The draft genome of a diploid cotton Gossypium raimondii [J]. Nat. Genet., 2012, 44(10): 1098-1103. |
| 2 | LI F G, FAN G Y, WANG K B, et al.. Genome sequence of the cultivated cotton Gossypium arboreum [J]. Nat. Genet., 2014, 46(6): 567-572. |
| 3 | PATERSON A H, WENDEL J F, GUNDLACH H, et al.. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers [J]. Nature, 2012, 492(7429): 423-427. |
| 4 | HU Y, CHEN J, FANG L, et al.. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton [J]. Nat. Genet., 2019: 51(54): 739-748. |
| 5 | 王晨,李家儒.植物β-葡萄糖苷酶的研究进展[J].生物资源, 2021, 43(2): 101-109. |
| WANG C, LI J. Research progress of plant β⁃glucosidase [J]. Biotic. Resour., 2021, 43(2): 101-109. | |
| 6 | HENRISSAT B A. A classification of glycosyl hydrolases based on amino acid sequence similarities [J]. Biochem. J., 1992, 280(2): 309-316. |
| 7 | DUROUX L, DELMOTTE F M, LANCELIN J M, et al.. Insight into naphthoquinone metabolism: β-glucosidase-catalysed hydrolysis of hydrojuglone β-D-glucopyranoside [J]. Biochem. J., 1998, 333(2): 275-283. |
| 8 | DHARMAWARDHANA D. A beta-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin [J]. Plant Physiol., 1995, 107(2): 331-339. |
| 9 | BRZOBOHATY B, MOORE I, KRISTOFFERSEN P, et al.. Release of active cytokinin by a beta-glucosidase localized to the maize root meristem [J]. Science, 1993, 262(5136): 1051-1054. |
| 10 | SUN H, XUE Y, LIN Y, et al.. Enhanced catalytic efficiency in quercetin-4'-glucoside hydrolysis of Thermotoga maritima β-glucosidase a by site-directed mutagenesis [J]. J. Agric. Food Chem., 2014, 62(28):6763-6770. |
| 11 | SAMPEDRO J, VALDIVIA E R, FRAGA P, et al.. Soluble and membrane-bound b-glucosidases are involved in trimming the xyloglucan backbone1 [J]. Plant Physiol., 2017, 173: 1017-1030. |
| 12 | LUIS L E-T W, CHEN, MARCELLA L, et al.. Arabidopsis thaliana β-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides science direct [J]. Phytochemistry, 2006, 67(15): 1651-1660. |
| 13 | WANG C, CHEN S, DONG Y, et al.. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice [J]. New Phytol., 2020, 226(4): 1042-1054. |
| 14 | WANG H, ZHANG Y, ZHAO Y, et al.. Genome-wide comparative analysis of the β-glucosidase family in five rosaceae species and their potential role on lignification of stone cells in Chinese white pear [J/OL]. Peer J., 2019, 7(3):46971 [2021-12-07] . |
| 15 | KONG W L, ZHONG H, DENG X X, et al.. Evolutionary analysis of GH3 genes in six oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica [J]. Plants, 2019: 8(2):30. |
| 16 | QANMBER G, DAOQIAN Y U, JIE L I, et al.. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses [J]. J. Cotton Res., 2018, 1(1): 3-19. |
| 17 | YANG, XIU, XU, et al.. Genome-wide identification of OSCA gene family and their potential function in the regulation of dehydration and salt stress in Gossypium hirsutum [J]. J. Cotton Res., 2019, 2(02): 14-31. |
| 18 | XU Q F, DUNBRACK RL. Assignment of protein sequences to existing domain and family classification systems: Pfam and the PDB [J]. Bioinformatics, 2012, 28(21): 2763-2772. |
| 19 | LARKIN M. Clustal W and Clustal X v. 2.0 [J]. Bioinformatics, 2007, 23: 2947-2948. |
| 20 | CHENG H Q, HAN L B, YANG C L, et al.. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection [J]. J. Exp. Bot., 2016(6): 1935-1950. |
| 21 | ZHAO Y L, ZHOU T T, GUO H S, et al.. Hyphopodium-specific VdNoxB/VdPls1-dependent ROS-Ca2+ signaling is required for plant infection by Verticillium dahliae [J/OL]. PLoS Pathog., 2016, 12(7): e1005793 [2021-09-10]. . |
| 22 | YANG C L, LIANG S, WANG H Y, et al.. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae [J]. Mol. Plant, 2015, 8(3): 399-411. |
| 23 | WANG G, WANG X, ZHANG Y, et al.. Dynamic characteristics and functional analysis provide new insights into long non-coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum [J/OL]. BMC Plant Biol., 2021, 21(1):8 [2021-09-10]. . |
| 24 | LIVAK K J, SCHIMTTGEN T D. Analysis of relative gene expression data using real-time quantitative and the 2-ΔΔ CT method [J]. Methods, 2001(25):492-408. |
| 25 | DONG O X, METEIGNIER L V, PLOURDE M B, et al.. Arabidopsis TAF15b localizes to RNA processing bodies and contributes to snc1-mediated autoimmunity [J]. Mol. Plant Microbe Interact., 2015, 29(4):247-257. |
| 26 | ROEPKE J, BOZZO G G. Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides [J]. Phytochemistry, 2015, 109(1): 14-24. |
| 27 | NAKAZAKI A, YAMADA K, KUNIEDA T, et al.. Biogenesis of leaf endoplasmic reticulum body is regulated by both jasmonate-dependent and independent pathways [J]. Plant Signal Behav., 2019, 14(8): 1-3. |
| 28 | XU Z Y, LEE K H, DONG T, et al.. A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis [J]. Plant Cell, 2012, 24(5): 2184-2199. |
| 29 | 柯丹霞,刘永辉,张静静,等.大豆BGLU基因家族全基因组鉴定与表达分析[J].信阳师范学院学报, 2019, 32(3): 1003-0972. |
| KE D X, LIU Y H, ZHANG J J, et al.. Genome-wide identification and expression analysis of BGLU family genes in soybean [J]. J. Xinyang Norm.Univ., 2019, 32(3): 1003-0972. | |
| 30 | OPASSIRI R, POMTHONG B, ONKOKSOONG T, et al.. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase [J/OL]. BMC Plant Biol., 2006, 6(1):33 [2021-09-10]. . |
| 31 | CHANG H C, TSAI M C, WU S S, et al.. Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana [J/OL]. Bot. Stud., 2019, 60(1): 16 [2021-09-10]. . |
| 32 | YANG J F, MA L, JIANG W B, et al.. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula [J]. Plant Physiol. Biochem., 2020, 158: 21-33. |
| 33 | GOMEZ A G, CENICEROS E A, CASADOS-V L E, et al.. Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73) [J]. Plant Mol. Biol., 2011, 77(1-2): 159-183. |
| 34 | KWAK G Y, GOO E, JEONG H, et al.. Adverse effects of adaptive mutation to survive static culture conditions on successful fitness of the rice pathogen Burkholderia glumae in a host [J/OL]. PLoS ONE, 2020, 15(8): e0238151 [2021-09-10]. . |
| 35 | CHAPELLE A, MORREEL K, VANHOLME R, et al.. Impact of the absence of stem-specific β-glucosidases on lignin and monolignols [J]. Plant Physiol., 2012, 160(3): 1204-1217. |
| 36 | ZHANG Z Q, LI Z M, STASWICKPAUL E, et al.. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction [J]. Plant Physiol., 2007, 145(2): 450-464. |
| 37 | ISHIHARA H, TOHGE T V, PRISCA F, et al.. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6 [J]. J. Exp. Bot. 2016, 67(5): 1505-1517. |
| 38 | HASS C, LOHRMANN J, ALBRECHT V, et al.. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis [J]. Embo. J., 2014, 23(16): 3290-3302. |
| 39 | BOLLER T. Antimicrobial functions of the plant hydrolases, chitinase and -1,3-glucanase [J]. Plant and Soil., 1984, 2: 391-400. |
| 40 | DUMAS G E, SLEZACK S, DASSI B, et al.. Plant hydrolytic enzymes (chitinases and beta-1, 3-glucanases) in root reactions to pathogenic and symbiotic microorganisms [J]. Plant Soil, 1996, 185(2): 211-221. |
| 41 | 王妮妍,蒋德安.茉莉酸及其甲酯与植物诱导抗病性[J].植物生理学报, 2002, 38(3): 279-284. |
| WANG N Y, JIANG D A. The role of jasmonic acid and methyl jasmonate in plant induced disease resistance [J]. Plant Physiol. Commun., 2002, 38(3): 279-284. |
| [1] | 李美丽, 宿俊吉, 杨永林, 秦江鸿, 李鲜鲜, 杨德龙, 马麒, 王彩香. 陆地棉COI家族基因鉴定及在干旱和盐胁迫下的表达分析[J]. 中国农业科技导报, 2022, 24(4): 63-74. |
| [2] | 黄祥, 楚光明, 郑新开, 程锦涛, 陈健豪, 徐迎春, 金奇江, 杨梅花. 睡莲属叶绿体基因组密码子偏好性及系统发育分析[J]. 中国农业科技导报, 2022, 24(4): 75-84. |
| [3] | 默韶京, 王志城, 王省芬, 刘正文, 吴立强, 张桂寅, 马峙英, 张艳, 段会军. 陆地棉GELP家族基因鉴定及其响应胁迫的表达分析[J]. 中国农业科技导报, 2022, 24(2): 93-103. |
| [4] | 赵晋锋, 余爱丽, 李颜方, 杜艳伟, 王高鸿, 王振华. 谷子SiCBL3对非生物胁迫响应特征分析[J]. 中国农业科技导报, 2022, 24(11): 68-75. |
| [5] | 郑巨云, 桑志伟, 王俊铎, 龚照龙, 梁亚军, 张泽良, 郭江平, 莫明, 李雪源. 棉花品种抗旱性相关指标分析与综合评价[J]. 中国农业科技导报, 2022, 24(10): 23-34. |
| [6] | 王琴琴, 陈修贵, 陆许可, 王帅, 张悦新, 范亚朋, 陈全家, 叶武威. 陆地棉GhPKE1的生物信息学分析及功能验证[J]. 中国农业科技导报, 2022, 24(1): 38-45. |
| [7] | 刘正文, 王省芬, 孟成生, 张艳, 孙正文, 吴立强, 马峙英, 张桂寅. 海岛棉GH9基因家族成员鉴定及分析[J]. 中国农业科技导报, 2021, 23(9): 30-45. |
| [8] | 李生梅, 杨涛, 黄雅婕, 任丹, 耿世伟, 李典鹏, 芮存, 高文伟. 海陆回交群体主要农艺性状与纤维品质关系的探讨[J]. 中国农业科技导报, 2021, 23(8): 16-24. |
| [9] | 蒲全明, 杨鹏, 雍磊, 邓榆川, 何自涵, 林邦民, 施松梅, 向承勇, 方芳. 萝卜紫红叶色突变体的色素含量及光合特性研究[J]. 中国农业科技导报, 2021, 23(8): 45-54. |
| [10] | 苏玥§, 刘娟娟§, 完斌, 张鹏举, 陈正根, 宿俊吉, 王彩香. 乳苣叶绿体基因组特征及其系统发育分析[J]. 中国农业科技导报, 2021, 23(6): 33-42. |
| [11] | 崔江慧§,杨溥原§,常金华*. 高粱GRF基因家族鉴定及在非生物胁迫下的表达分析[J]. 中国农业科技导报, 2021, 23(4): 37-46. |
| [12] | 陈斌, 石荣康, 王志城, 刘松, 李青, 刘正文, 孙正文, 王国宁, 吴金华, 马峙英, 张艳, 王省芬. 陆地棉核心种质抗黄萎病鉴定与优异种质筛选[J]. 中国农业科技导报, 2021, 23(10): 45-51. |
| [13] | 刘鹏,韦杰1,杨毅清1,张娜1,温晓蕾1,2,范学锋1,杨文香1*,刘大群1*. 小麦类钙调素新亚型基因TaCML25/26调控抗叶锈性[J]. 中国农业科技导报, 2020, 22(4): 120-128. |
| [14] | 王俊铎1,曾辉2,龚照龙1,梁亚军1,艾先涛1,郭江平1,莫明1,李雪源1,郑巨云1*. 陆地棉品种资源耐复合盐碱性综合评价分析[J]. 中国农业科技导报, 2019, 21(10): 1-11. |
| [15] | 杨笑敏,芮存,张悦新,王德龙,王俊娟,陆许可,陈修贵,郭丽雪,王帅,陈超,叶武威*. 棉花DNA甲基转移酶GhDMT9抗逆性分析[J]. 中国农业科技导报, 2019, 21(10): 12-19. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号