




















中国农业科技导报 ›› 2022, Vol. 24 ›› Issue (6): 36-46.DOI: 10.13304/j.nykjdb.2021.0806
吴楠( ), 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬(
), 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬( )
)
收稿日期:2021-09-13
接受日期:2021-11-22
出版日期:2022-06-15
发布日期:2022-06-21
通讯作者:
王省芬
作者简介:吴楠 E-mail:wn2013@126.com;
基金资助:
Nan WU( ), Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG(
), Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG( )
)
Received:2021-09-13
Accepted:2021-11-22
Online:2022-06-15
Published:2022-06-21
Contact:
Xingfen WANG
摘要:
纤维品质改良一直是棉花育种的重要目标。基于纤维不同发育时期的RNA-seq数据,发现1个在海岛棉和陆地棉间差异表达的葡萄糖醛酸激酶基因(glucuronic acid kinase/glucuronokinase,GlcAK)。从海岛棉(Gossypium barbadense)Pima90-53纤维中克隆了GbGlcAK,其ORF为1 101 bp,编码366个氨基酸。GbGlcAK具有GHMP 激酶 N和C结构域,为GHMP超家族成员,属于可溶性非分泌蛋白,与拟南芥GlcAK亲缘关系最近。亚细胞定位结果显示GbGlcAK分布在细胞质中。超表达GbGlcAK的拟南芥叶表皮毛、下胚轴、下胚轴细胞和根的长度较野生型均极显著增加,UDP-D-GlcA代谢相关基因表达量增加,果胶含量显著增加,说明过表达GbGlcAK能够上调UDP-D-GlcA代谢通路相关基因的表达从而提高果胶含量,进而促进细胞伸长,这为进一步研究GbGlcAK在棉纤维发育中的功能提供了参考。
中图分类号:
吴楠, 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬. 过表达棉花葡萄糖醛酸激酶基因GbGlcAK促进拟南芥细胞伸长[J]. 中国农业科技导报, 2022, 24(6): 36-46.
Nan WU, Jun YANG, Yan ZHANG, Zhengwen SUN, Dongmei ZHANG, Lihua LI, Jinhua WU, Zhiying MA, Xingfen WANG. Overexpression of a Cotton Glucuronokinase Gene GbGlcAK Promotes Cell Elongation in Arabidopsis thaliana[J]. Journal of Agricultural Science and Technology, 2022, 24(6): 36-46.
引物名称 Primer names | 登录号 Accession No. | 引物序列 Primer sequences (5’-3’) | 用途 Purpose |
|---|---|---|---|
| GlcAK-F | — | TGGTATTGGCTTGGTGCAGT | 基因克隆 Gene cloning |
| GlcAK-R | CAAAAACAATCCCTAAAATGCTCTT | ||
| sGbGlcAK-R | — | GGTGGTACCCTGCTTAGACAATGT | 亚细胞定位Subcellular location |
| GbGlcAK-F | — | GGCGTCTAGAATGGATCAAAATATG | 亚细胞定位和载体构建 Subcellular location and vector construction |
| GbGlcAK-R | GGTACCCGCCTACTTAGACAATG | 载体构建 Vector construction | |
| qGbGlcAK-F | — | CATCATCAACCCTCACCCCATTC | 拟南芥实时定量PCR Real-time PCR for A. thaliana |
| qGbGlcAK-R | GCCGCATACAATAGCACTGGACC | ||
| qAtGlcAK-F | AT3G01640 | GGATAGAGCATCGGTCCTTCGC | |
| qAtGlcAK-R | CGCCATTAGCAACCTTACCCCA | ||
| qAtMIPS-F | AT4G39800 | AAGGAGAAGAATAAGGTGGATAAGGTT | |
| qAtMIPS-R | GCAATCGCATAAAGTGTTGAAGG | ||
| qAtIMP-F | AT3G02870 | GACAGAGACTGATAAAGGATGTGAAGA | |
| qAtIMP-R | AGGGAACCCGTGAACGAAAT | ||
| qAtMIOX-F | AT4G26260 | CGAAGCCATCCGCAAAGATTACC | |
| qAtMIOX-R | CCAACAACAGCCCATTGAGGAAGTC | ||
| qAtPGM-F | AT1G23190 | GCTACCTACGGTCGTCACTATTACACTCG | |
| qAtPGM-R | GTAACGGATTCCCTGGTGCTTCG | ||
| qAtUGP-F | AT5G17310 | GCCCAGCAAGGGAAAGACCG | |
| qAtUGP-R | CGATGGCACCCAAGTTGTCTGAAT | ||
| qAtUGD-F | AT5G15490 | GATTGCGGTTCTCGGCTTCG | |
| qAtUGD-R | TGCTTCACAGTGGTGGGGCTC | ||
| qAtUXS-F | AT5G59290 | TGAGAAGAATGAGGTGGTTGTTGC | |
| qAtUXS-R | GGTTGTATTTGTAGAAGATAGGAGAGGC | ||
| qAtGAE-F | AT3G23820 | GCGACGGCGGATACAAGCA | |
| qAtGAE-R | GATGGCGAAGATGAGGACGAGG | ||
| qAtUB5-F | AT3G62250 | CCTCGCCGACTACAACATCCAG | |
| qAtUB5-R | CTTCTTCCTCTTCTTAGCACCACCA |
表1 试验中用到的PCR和qPCR引物
Table 1 PCR and qPCR primers used in this study
引物名称 Primer names | 登录号 Accession No. | 引物序列 Primer sequences (5’-3’) | 用途 Purpose |
|---|---|---|---|
| GlcAK-F | — | TGGTATTGGCTTGGTGCAGT | 基因克隆 Gene cloning |
| GlcAK-R | CAAAAACAATCCCTAAAATGCTCTT | ||
| sGbGlcAK-R | — | GGTGGTACCCTGCTTAGACAATGT | 亚细胞定位Subcellular location |
| GbGlcAK-F | — | GGCGTCTAGAATGGATCAAAATATG | 亚细胞定位和载体构建 Subcellular location and vector construction |
| GbGlcAK-R | GGTACCCGCCTACTTAGACAATG | 载体构建 Vector construction | |
| qGbGlcAK-F | — | CATCATCAACCCTCACCCCATTC | 拟南芥实时定量PCR Real-time PCR for A. thaliana |
| qGbGlcAK-R | GCCGCATACAATAGCACTGGACC | ||
| qAtGlcAK-F | AT3G01640 | GGATAGAGCATCGGTCCTTCGC | |
| qAtGlcAK-R | CGCCATTAGCAACCTTACCCCA | ||
| qAtMIPS-F | AT4G39800 | AAGGAGAAGAATAAGGTGGATAAGGTT | |
| qAtMIPS-R | GCAATCGCATAAAGTGTTGAAGG | ||
| qAtIMP-F | AT3G02870 | GACAGAGACTGATAAAGGATGTGAAGA | |
| qAtIMP-R | AGGGAACCCGTGAACGAAAT | ||
| qAtMIOX-F | AT4G26260 | CGAAGCCATCCGCAAAGATTACC | |
| qAtMIOX-R | CCAACAACAGCCCATTGAGGAAGTC | ||
| qAtPGM-F | AT1G23190 | GCTACCTACGGTCGTCACTATTACACTCG | |
| qAtPGM-R | GTAACGGATTCCCTGGTGCTTCG | ||
| qAtUGP-F | AT5G17310 | GCCCAGCAAGGGAAAGACCG | |
| qAtUGP-R | CGATGGCACCCAAGTTGTCTGAAT | ||
| qAtUGD-F | AT5G15490 | GATTGCGGTTCTCGGCTTCG | |
| qAtUGD-R | TGCTTCACAGTGGTGGGGCTC | ||
| qAtUXS-F | AT5G59290 | TGAGAAGAATGAGGTGGTTGTTGC | |
| qAtUXS-R | GGTTGTATTTGTAGAAGATAGGAGAGGC | ||
| qAtGAE-F | AT3G23820 | GCGACGGCGGATACAAGCA | |
| qAtGAE-R | GATGGCGAAGATGAGGACGAGG | ||
| qAtUB5-F | AT3G62250 | CCTCGCCGACTACAACATCCAG | |
| qAtUB5-R | CTTCTTCCTCTTCTTAGCACCACCA |
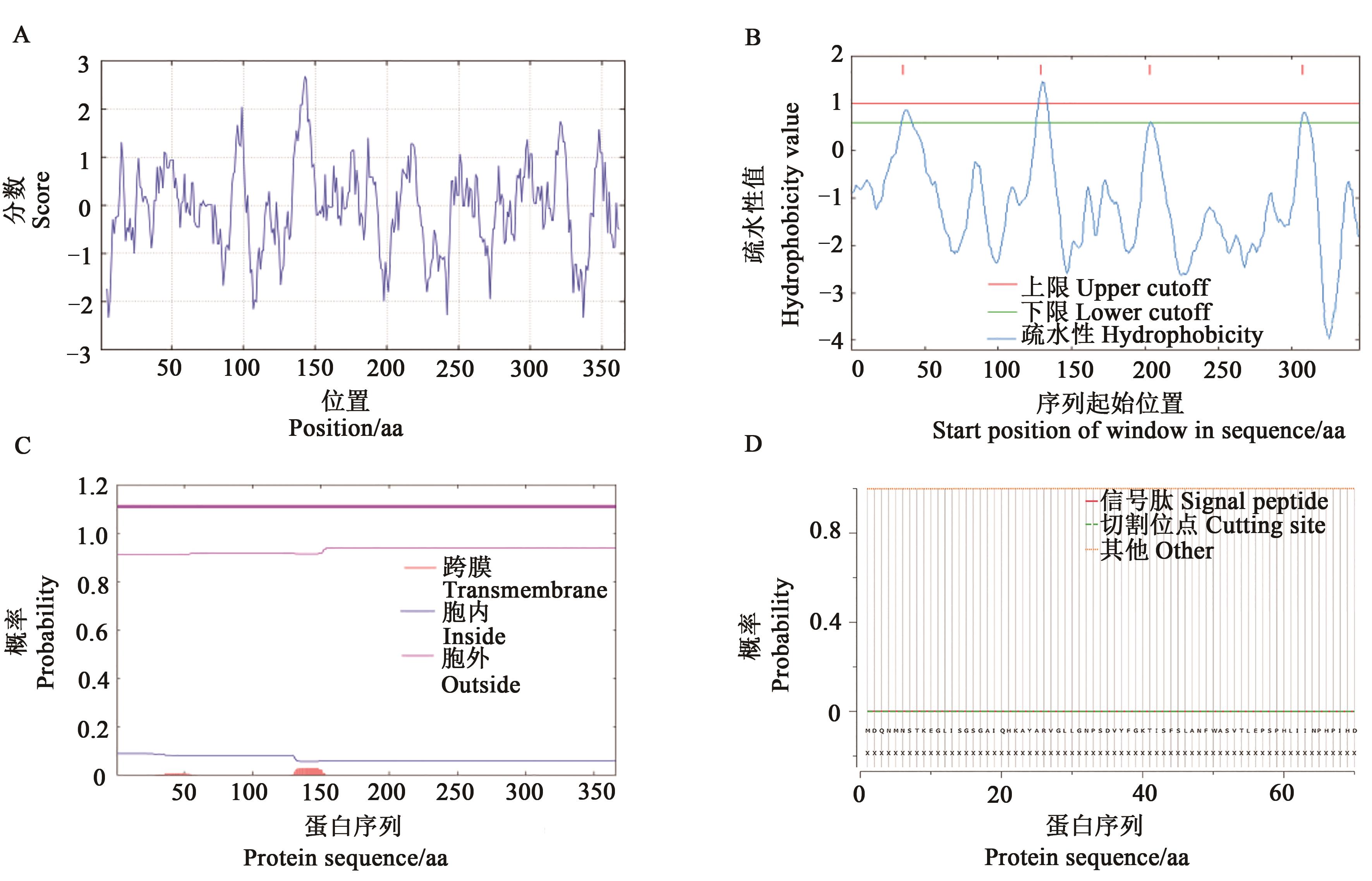
图1 GbGlcAK蛋白特征A:ProtScale分析GbGlcAK肽链亲/疏水性分布曲线;B、C:TOPpred2和TMHMM 2.0工具对GbGlcAK氨基酸序列的跨膜区预测;D:GbGlcAK蛋白信号肽分析
Fig. 1 Characterization of GbGlcAK proteinA: Hydrophilic/hydrophobic distribution curve of the GbGlcAK peptide analyzed by ProtScale; B, C: Transmembrane region prediction of the GbGlcAK amino acid sequence by TOPpred2 and TMHMM 2.0; D: Signal peptide analysis of the GbGlcAK protein

图2 GbGlcAK系统进化分析注:Rc—蓖麻;Pt—毛果杨;Vv—葡萄;At—拟南芥;Gm—大豆;Os—水稻;Zm—玉米;Sb—高粱。
Fig.2 Evolutionary analysis of GbGlcAKNote: Rc—Ricinus communis; Pt—Populus trichocarpa; Vv—Vitis vinifera; At—Arabidopsis thaliana; Gm—Glycine max; Os—Oryza sativa; Zm—Zea mays; Sb—Sorghum bicolor.
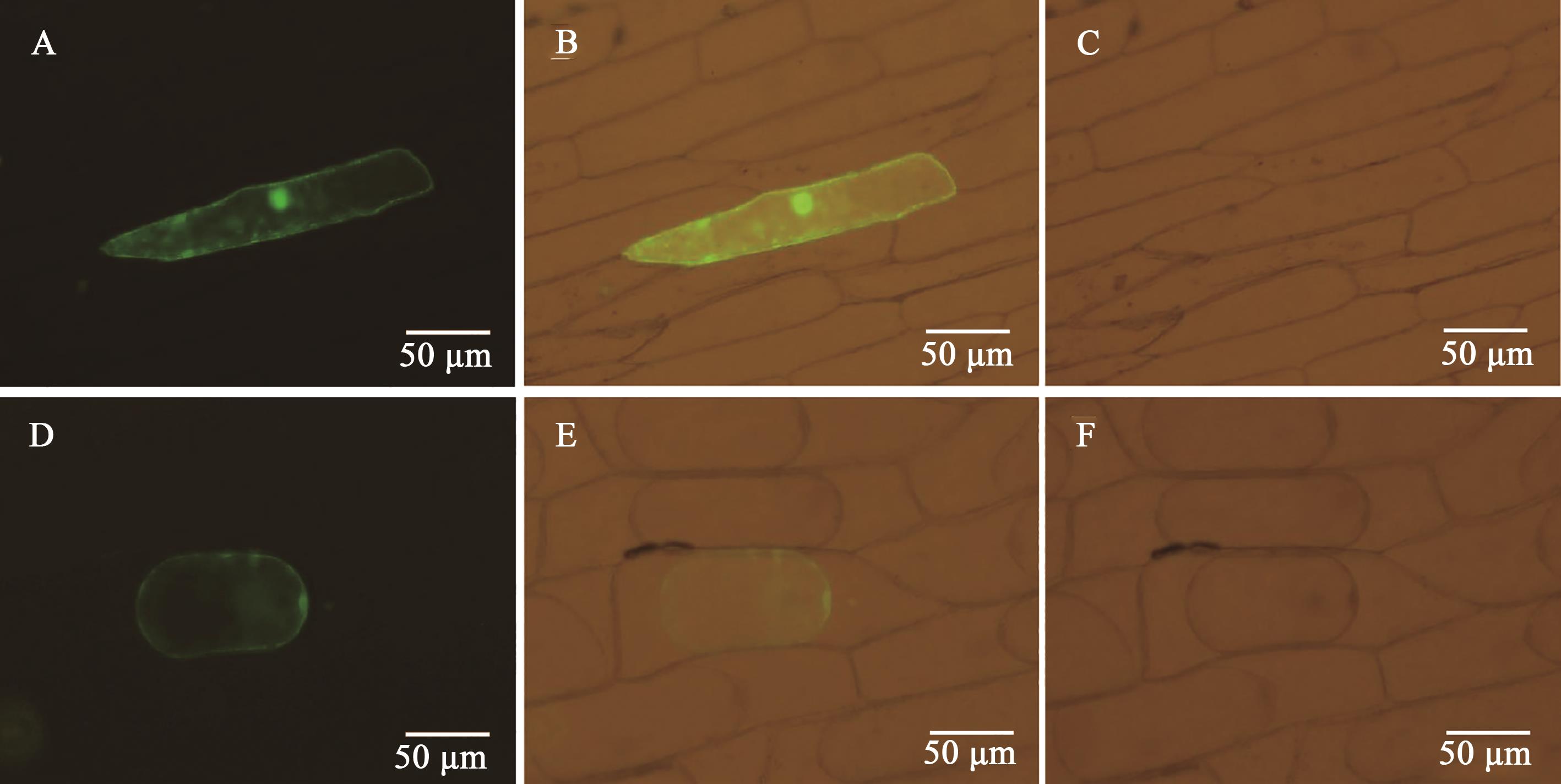
图3 GbGlcAK的亚细胞定位A、B、C:蓝光、可见光叠加蓝光、可见光下的转pCam::GFP细胞;D、E、F:质壁分离后蓝光、可见光叠加蓝光、可见光下的转pCam::GbGlcAK-GFP细胞
Fig. 3 Subcellular localization of GbGlcAKA, B, C: Cell transfected with pCam::GFP under Blu-ray, visible light superimposed Blu-ray, and visible light; D, E, F: Cell transfected with pCam::GbGlcAK-GFP under Blu-ray, visible light superimposed Blu-ray, and visible light after plasma-wall separation
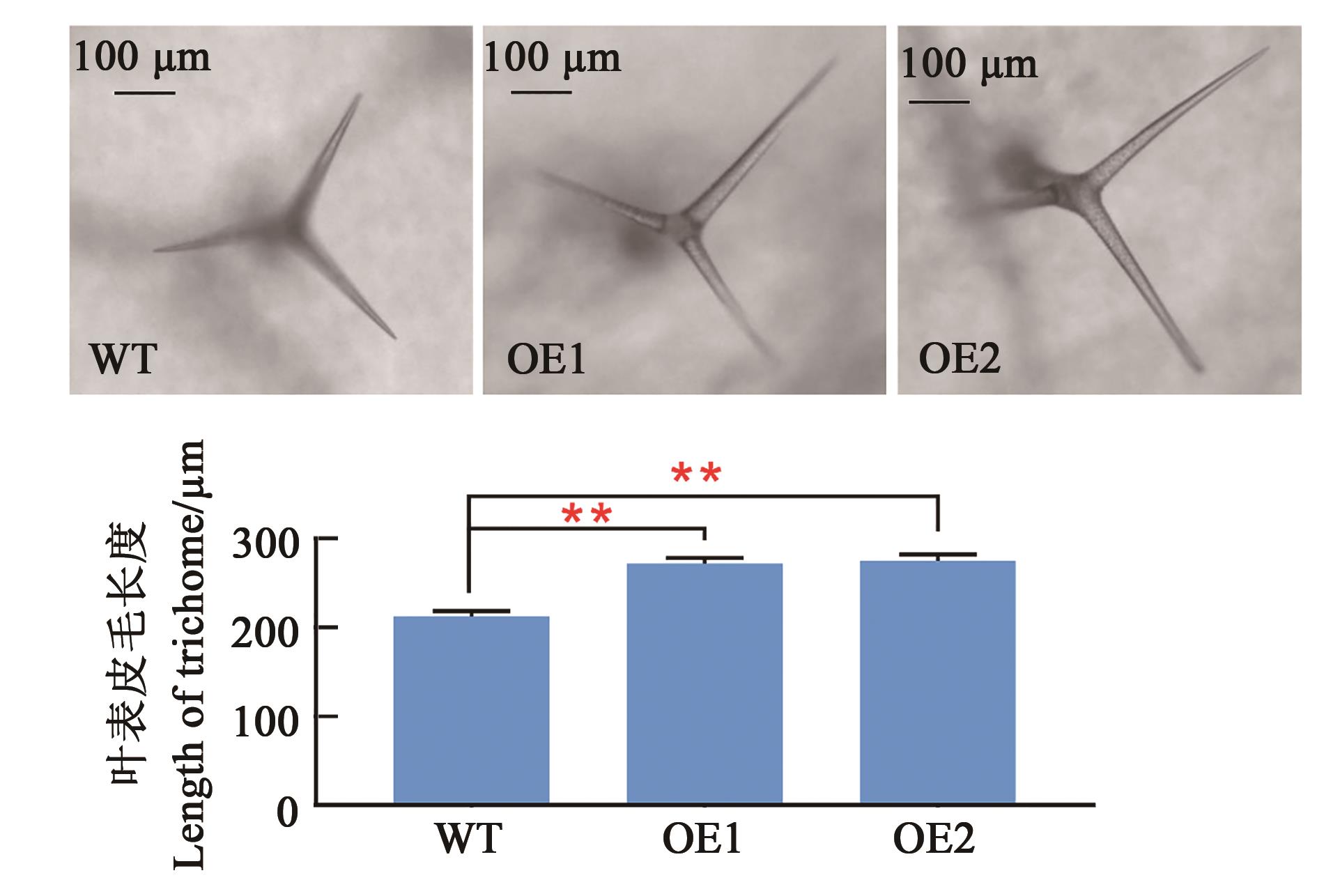
图4 转基因拟南芥叶表皮毛长度分析注:**表示在P<0.01水平差异显著(双尾t检验)。
Fig. 4 Analysis of leaf trichome length in transgenic A. thalianaNote:** indicates significant difference at the P<0.01 level (2-tailed t-test).
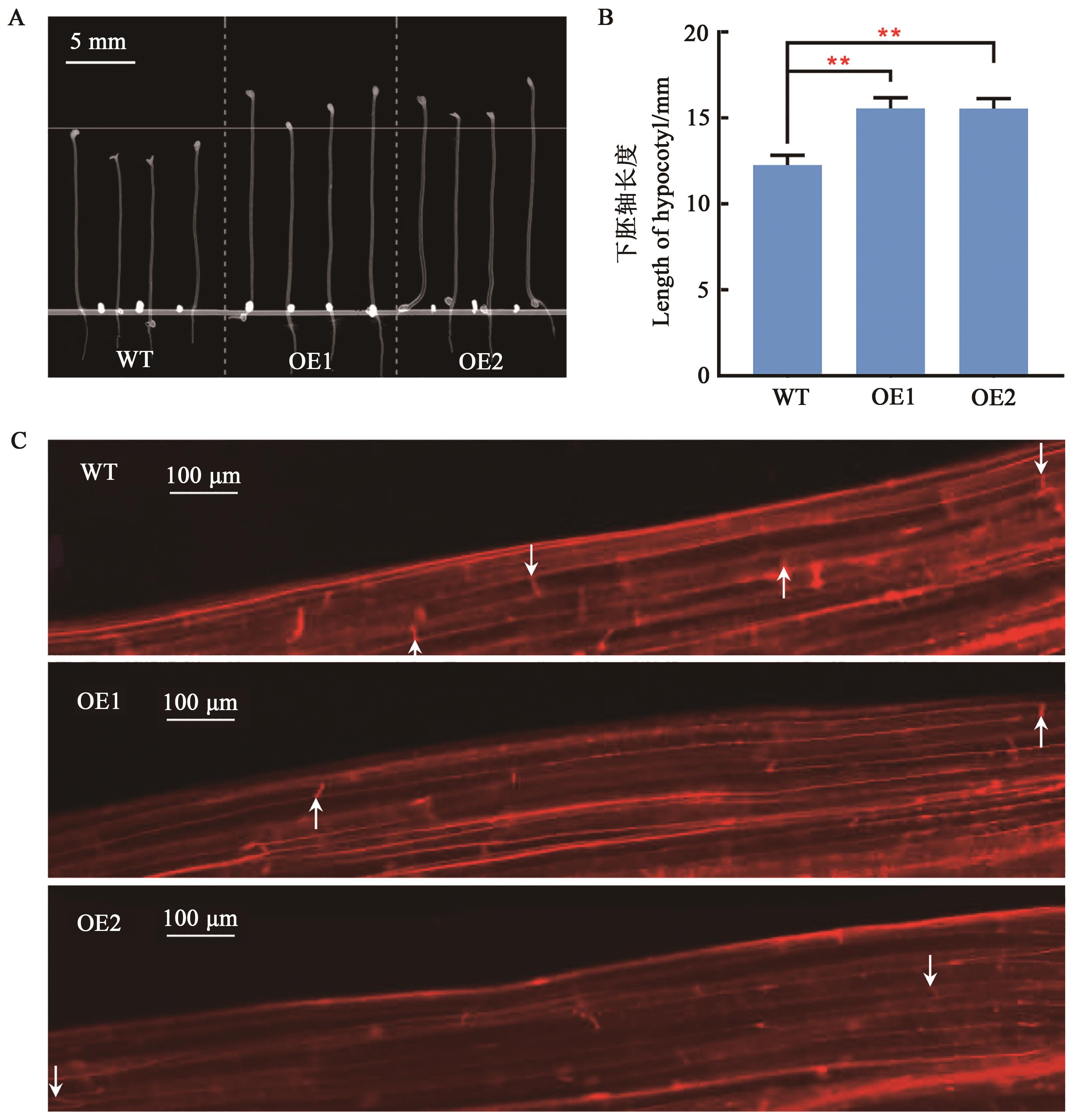
图5 转基因拟南芥下胚轴长度分析A、B:超表达GbGlcAK拟南芥与野生型拟南芥下胚轴长度比较;C:显微镜下观察超表达GbGlcAK拟南芥与野生型拟南芥叶下胚轴细胞。**表示P<0.01水平的显著性(双尾t检验);白色箭头所示细胞边界
Fig. 5 Analysis of hypocotyl length in transgenic A. thalianaA, B: Comparison of hypocotyl length between transgenic and wild-type Arabidopsis; C: Microscopic observation of hypocotyl cells; ** indicates significant difference at the P<0.01 level (2-tailed t-test);white arrows show the ends of cells
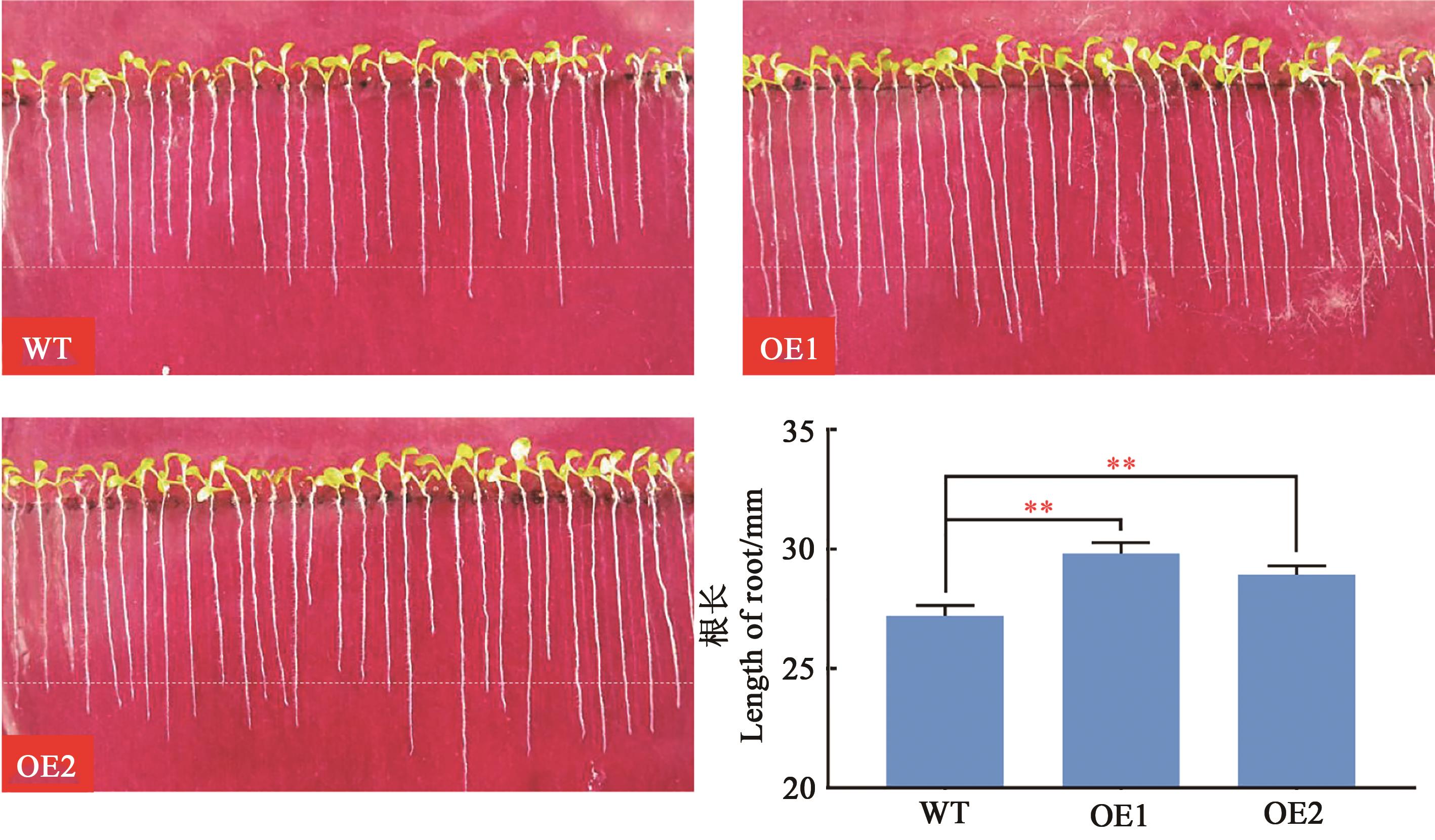
图6 转基因拟南芥根长分析注:**表示P<0.01水平的显著性(双尾t检验)。
Fig. 6 Analysis of root length in transgenic A. thalianaNote:** indicates significant difference at the P<0.01 level (2-tailed t-test).
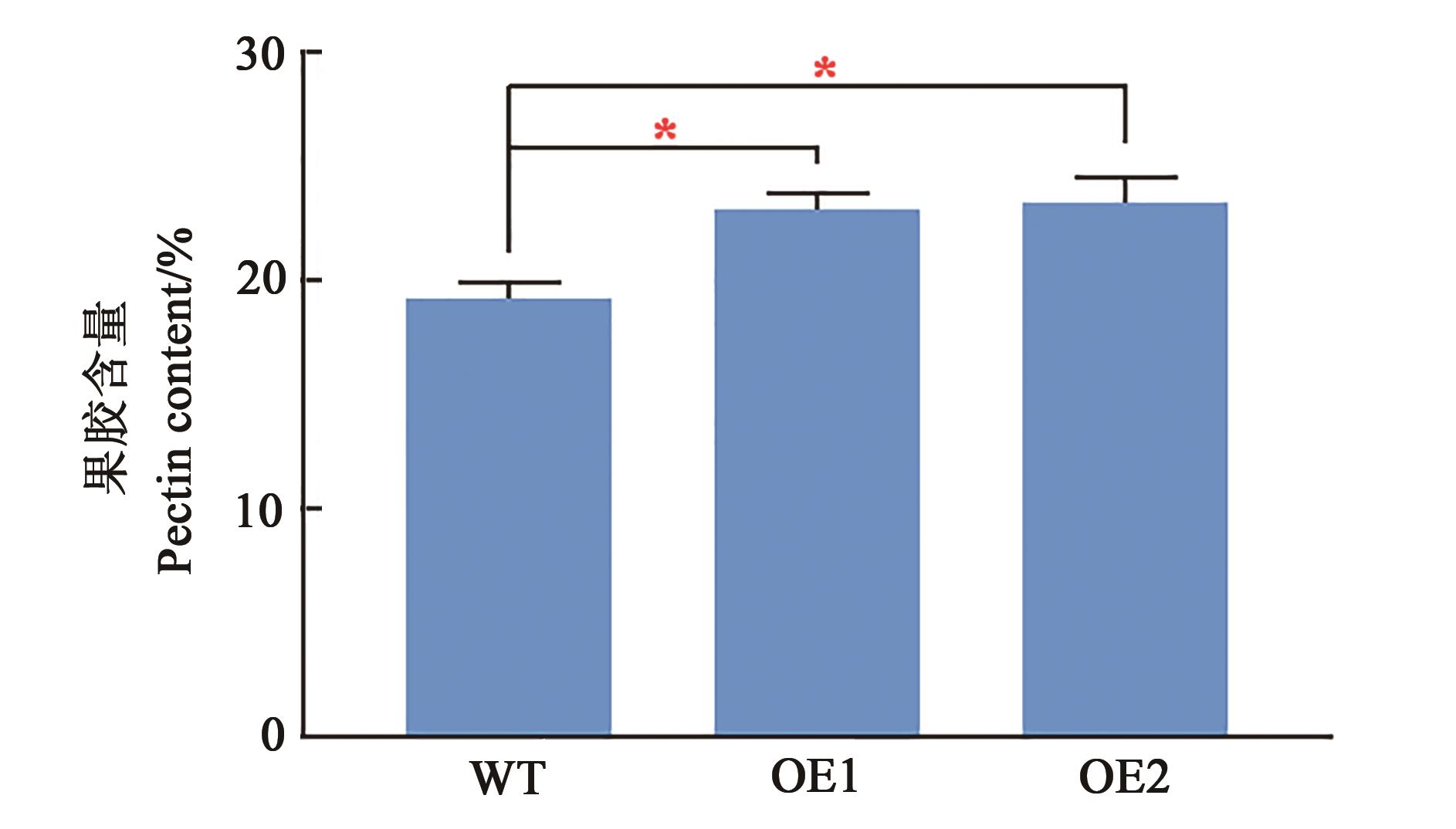
图8 转基因拟南芥果胶含量注:*表示P<0.05水平的显著性(双尾t检验)。
Fig. 8 Pectin content in transgenic ArabidopsisNote:* indicates significant difference at P<0.05 level (2-tailed t-test).
| 1 | 喻树迅,魏晓文,赵新华.中国棉花生产与科技发展 [J].棉花学报, 2000, 12(6): 327-329. |
| YU S X, WEI X W, ZHAO X H. Cotton production and technical development in China [J]. Acta Gossypii Sin., 2000, 12(6): 327-329. | |
| 2 | MA Z, ZHANG Y, WU L, et al.. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement [J]. Nat. Genet., 2021, 53(9): 1385-1391. |
| 3 | BASRA A S, MALIK C P. Development of the cotton fiber [J]. Int. Rev. Cytol., 1984, 89(6): 65-113. |
| 4 | HAIGLER C H, BETANCUR L, STIFF M R, et al.. Cotton fiber: a powerful single-cell model for cell wall and cellulose research [J/OL]. Front. Plant Sci., 2012, 3: 104 [2021-11-09]. . |
| 5 | QIN Y M, ZHU Y X. How cotton fibers elongate: a tale of linear cell-growth mode [J]. Curr. Opin. Plant Biol., 2011, 14(1): 106-111. |
| 6 | SINGH B, AVCI U, EICHLER INWOOD S E, et al.. A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles [J]. Plant Physiol., 2009, 150(2): 684-699. |
| 7 | ZABLACKIS E, JING H, MüLLER B, et al.. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves [J]. Plant Physiol., 1995, 107(4): 1129-1138. |
| 8 | KANTER U, USADEL B, GUERINEAU F, et al.. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides [J]. Planta, 2005, 221(2): 243-254. |
| 9 | LOEWUS F A, KELLY S, NEUFELD E F. Metabolism of myo-inositol in plants: conversion to pectin, hemicellulose, D-xylose, and sugar acids [J]. Proc. Natl. Acad. Sci. USA, 1962, 48(3): 421-425. |
| 10 | NEUFELD E F, FEINGOLD D S, HASSID W Z. Enzymic phosphorylation of D-glucuronic acid by extracts from seedlings of Phaseolus aureus [J]. Arch. Biochem. Biophys., 1959, 83(1): 96-100. |
| 11 | LEIBOWITZ M D, DICKINSON D B, LOEWUS F A, et al.. Partial purification and study of pollen glucuronokinase [J]. Arch. Biochem. Biophys., 1977, 179(2): 559-564. |
| 12 | PIESLINGER A M, HOEPFLINGER M C, TENHAKEN R. Nonradioactive enzyme measurement by high-performance liquid chromatography of partially purified sugar-1-kinase (glucuronokinase) from pollen of Lilium longiflorum [J]. Anal. Biochem., 2009, 388(2): 254-259. |
| 13 | PIESLINGER A M, HOEPFLINGER M C, TENHAKEN R. Cloning of Glucuronokinase from Arabidopsis thaliana, the last missing enzyme of the myo-inositol oxygenase pathway to nucleotide sugars [J]. J. Biol. Chem., 2010, 285(5): 2902-2910. |
| 14 | HOLDEN H M, THODEN J B, TIMSON D J, et al.. Galactokinase: structure, function and role in type Ⅱ galactosemia [J]. Cell. Mol. Life Sci., 2004, 61(19-20): 2471-2484. |
| 15 | XIAO W, HU S, ZHOU X, et al.. A glucuronokinase gene in Arabidopsis, AtGlcAK, is involved in drought tolerance by modulating sugar metabolism [J]. Plant Mol. Biol. Rep., 2017, 35(2): 1-14. |
| 16 | IVANOV KAVKOVA E, BLöCHL C, TENHAKEN R. The Myo-inositol pathway does not contribute to ascorbic acid synthesis [J]. Plant Biol., 2019, 21 (): 95-102. |
| 17 | LIU Z, WANG X, SUN Z, et al.. Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes [J/OL]. BMC Plant Biol., 2021, 21(1): 89[2022-03-07]. . |
| 18 | WANG Z, YANG Z, LI F. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton [J]. Plant Biotechnol. J., 2019, 17(9): 1706-1722. |
| 19 | 吴立柱,王省芬,李喜焕,等.通用型植物表达载体pCamE的构建及功能验证[J].农业生物技术学报, 2014, 22(6): 661-671. |
| WU L Z, WANG X F, LI X H, et al.. Construction and function identification of universal plant expression vector pCamE [J]. J. Agric. Biotechnol., 2014, 22(6): 661-671. | |
| 20 | WANG M, TU L, YUAN D, et al.. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense [J]. Nat. Genet., 2019, 51(2): 224-229. |
| 21 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al.. Pfam: The protein families database in 2021 [J]. Nucleic Acids Res., 2021, 49(D1): D412-D419. |
| 22 | ARTIMO P, JONNALAGEDDA M, ARNOLD K, et al.. ExPASy: SIB bioinformatics resource portal [J]. Nucleic Acids Res., 2012, 40(W1): W597-W603. |
| 23 | CUTHBERTSON J M, DOYLE D A, SANSOM M S. Transmembrane helix prediction: a comparative evaluation and analysis [J]. Protein Eng. Des. Sel., 2005, 18(6): 295-308. |
| 24 | KROGH A, LARSSON B, GVON HEIJNE, et al.. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes [J]. J. Mol. Biol., 2001, 305(3): 567-580. |
| 25 | MÖLLER S, CRONING M D, APWEILER R. Evaluation of methods for the prediction of membrane spanning regions [J]. Bioinformatics, 2001, 17(7): 646-653. |
| 26 | ALMAGRO ARMENTEROS J J, TSIRIGOS K D, SøNDERBY C K, et al.. SignalP 5.0 improves signal peptide predictions using deep neural networks [J]. Nat. Biotechnol., 2019, 37(4): 420-423. |
| 27 | KUMAR S, STECHER G, TAMURA K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets [J]. Mol. Biol. Evol., 2016, 33(7): 1870-1874. |
| 28 | YANG J, JI L, WANG X, et al.. Overexpression of 3-deoxy-7-phosphoheptulonate synthase gene from Gossypium hirsutum enhances Arabidopsis resistance to Verticillium wilt [J]. Plant Cell Rep., 2015, 34(8): 1429-1441. |
| 29 | CHEN H, NELSON R S, SHERWOOD J L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection [J]. Biotechniques, 1994, 16(4): 664-668, 670. |
| 30 | CLOUGH S J, BENT A F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana [J]. Plant J., 1998, 16(6): 735-743. |
| 31 | An C, Wang C, Mou Z. The Arabidopsis Elongator complex is required for nonhost resistance against the bacterial pathogens Xanthomonas citri subsp. citri and Pseudomonas syringae pv. phaseolicola NPS3121 [J]. New Phytol., 2017, 214(3): 1245-1259. |
| 32 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method [J]. Methods, 2001, 25(4): 402-408. |
| 33 | JEAN-MARC L, TRUNG-BIEU N, BRIGITTE C, et al.. QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbadense backcross generations [J]. BMC Plant Biol., 2005, 45(1): 123-140. |
| 34 | SERNA L, MARTIN C. Trichomes: different regulatory networks lead to convergent structures [J]. Trends Plant Sci., 2006, 11(6): 274-280. |
| 35 | GUAN X, YU N, SHANGGUAN X, et al.. Arabidopsis trichome research sheds light on cotton fiber development mechanisms [J]. Chin Sci. Bull., 2007, 52(13): 1734-1741. |
| 36 | GUAN X, PANG M, NAH G, et al.. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development [J/OL]. Nat. Commun., 2014, 5: 3050[2022-03-07]. . |
| 37 | MA Z, HE S, WANG X, et al.. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield [J]. Nat. Genet., 2018, 50(6): 803-813. |
| 38 | GENDREAU E, TRAAS J, DESNOS T, et al.. Cellular basis of hypocotyl growth in Arabidopsis thaliana [J]. Plant Physiol., 1997, 114(1): 295-305. |
| 39 | BORON A K, VISSENBERG K. The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion [J]. Plant Cell Rep., 2014, 33(5): 697-706. |
| 40 | 武耀廷,刘进元.棉纤维细胞发育过程中非纤维素多糖的生物合成 [J].棉花学报, 2004, 16(3): 189-192. |
| WU Y, LIU J. Noncellulosic polysaccharides biosynthesis in cotton fiber developing [J]. Cotton Sci., 2004, 16(3): 189-193. | |
| 41 | 喻树迅,朱玉贤,陈晓亚.棉花纤维发育生物学 [M].北京:科学出版社, 2016: 169. |
| 42 | KROH M, MIKI-HIROSIGE H, ROSEN W, et al.. Incorporation of label into pollen tube walls from myoinositol-labeled Lilium longiflorum pistils [J]. Plant Physiol., 1970, 45(1): 92-94. |
| 43 | SEITZ B, KLOS C, WURM M, et al.. Matrix polysaccharide precursors in Arabidopsis cell walls are synthesized by alternate pathways with organ-specific expression patterns [J]. Plant J., 2000, 21(6): 537-546. |
| [1] | 黄雅婕, 任丹, 李生梅, 崔进鑫, 杨涛, 任姣姣, 高文伟. 陆地棉苗期的耐盐碱性评价及鉴定指标筛选[J]. 中国农业科技导报, 2022, 24(5): 46-55. |
| [2] | 周雨青, 杨永飞, 葛常伟, 沈倩, 张思平, 刘绍东, 马慧娟, 陈静, 刘瑞华, 李士丛, 赵新华, 李存东, 庞朝友. 基于WGCNA的棉花子叶抗冷相关共表达模块鉴定[J]. 中国农业科技导报, 2022, 24(4): 52-62. |
| [3] | 刘正文, 王省芬, 孟成生, 张艳, 孙正文, 吴立强, 马峙英, 张桂寅. 海岛棉GH9基因家族成员鉴定及分析[J]. 中国农业科技导报, 2021, 23(9): 30-45. |
| [4] | 李生梅, 张大伟, 迪丽拜尔·迪力买买提, 魏鑫, 芮存, 杨涛, 耿世伟, 高文伟. 减量灌溉对转ScALDH21基因棉花农艺性状、产量和品质的影响[J]. 中国农业科技导报, 2021, 23(9): 152-159. |
| [5] | 张旭1,何俊峰1,陈佛文1,李继福1*,吴启侠1,谭京红1,邹家龙2. 麦秆还田对直播和移栽棉花产量及氮素吸收的影响[J]. 中国农业科技导报, 2021, 23(3): 122-131. |
| [6] | 张特,康正华,赵强*,聂志勇,王蜜蜂,崔延楠. 施氮量及打顶方式对棉花营养积累与分配及产量的影响[J]. 中国农业科技导报, 2021, 23(3): 139-147. |
| [7] | 马盼盼1,2,赵曾强1,2,祝建波2,孙国清3*. 棉花耐旱耐盐碱生理和分子机制研究进展[J]. 中国农业科技导报, 2021, 23(2): 27-36. |
| [8] | 王国宁, 张艳, 宋俊丽, 杨君, 王省芬, 吴立强, 张桂寅. 黄萎病胁迫下抗病陆地棉茎组织lncRNA的鉴定与分析[J]. 中国农业科技导报, 2021, 23(12): 29-41. |
| [9] | 吴雪琴, 崔延楠, 赵强. 化学打顶后使用外源物质对棉花脱叶催熟及产量品质的影响[J]. 中国农业科技导报, 2021, 23(12): 151-160. |
| [10] | 李憬霖, 刘绍东, 张思平, 陈静, 刘瑞华, 沈倩, 李阳, 马慧娟, 赵新华, 庞朝友. 棉花品种资源全生育期抗旱性评价及抗旱指标筛选[J]. 中国农业科技导报, 2021, 23(10): 52-65. |
| [11] | 刘梦丽1,李进2,张军高2,周小云2,杜鹏程1,郭庆元1*,雷斌2*. 棉花红腐病菌不同致病力菌株间毒素活性差异[J]. 中国农业科技导报, 2020, 22(7): 99-105. |
| [12] | 刘松涛1,田春丽1,曹雯梅1,郑贝贝1,李鹏程2,董合林2. 基于不同土壤质地棉花根际微生物和酶活性特征分析[J]. 中国农业科技导报, 2020, 22(2): 73-79. |
| [13] | 段怡红,阎媛媛,陈丽婷,李青,张冬梅,孙正文,张艳,马峙英,王省芬*. 陆地棉开花时间相关基因GhMYB44的克隆与功能验证[J]. 中国农业科技导报, 2020, 22(12): 29-38. |
| [14] | 王潭刚1§,马丽1§,李克富1,王冀川2*,李慧琴1,吉光鹏1,郝全有1,崔建强1,胡宝1. 不同密度下封顶方式对南疆棉花生长及产量性状的影响[J]. 中国农业科技导报, 2019, 21(6): 110-116. |
| [15] | 赵婧文,张庆伟,李政,张文太*. 膜下滴灌施用生物有机肥对土壤盐分及棉花产量的影响[J]. 中国农业科技导报, 2019, 21(3): 102-108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号