




















中国农业科技导报 ›› 2024, Vol. 26 ›› Issue (12): 77-87.DOI: 10.13304/j.nykjdb.2023.0151
陈美欣1,2( ), 顾淑莹2, 刘佳2, 刘浩1, 李金根2, 田朝光2(
), 顾淑莹2, 刘佳2, 刘浩1, 李金根2, 田朝光2( )
)
收稿日期:2023-03-04
接受日期:2023-04-06
出版日期:2024-12-15
发布日期:2024-12-17
通讯作者:
田朝光
作者简介:陈美欣 E-mail:2813841338@qq.com;
基金资助:
Meixin CHEN1,2( ), Shuying GU2, Jia LIU2, Hao LIU1, Jingen LI2, Chaoguang TIAN2(
), Shuying GU2, Jia LIU2, Hao LIU1, Jingen LI2, Chaoguang TIAN2( )
)
Received:2023-03-04
Accepted:2023-04-06
Online:2024-12-15
Published:2024-12-17
Contact:
Chaoguang TIAN
摘要:
为提高发酵菌株的木糖利用速率,以嗜热毁丝霉为体系,利用遗传学手段及转录组学技术,探究丝状真菌嗜热毁丝霉木糖代谢的调控机制。结果表明,在嗜热毁丝霉中,转录因子XlnR能够正向调控木糖代谢和转运以及半纤维酶的表达,是木糖条件下孢子萌发的必需因子。在木糖条件下,突变体ΔxlnR孢子丧失了萌发的能力,并且菌丝木糖利用速率显著降低,而过表达xlnR 使得木糖利用速率提升14.2%。转录组数据表明,敲除xlnR 导致木聚糖水解酶基因、木糖转运蛋白及代谢相关基因的转录水平显著降低。全局调控因子Cre-1是木糖及葡萄糖代谢的抑制因子,敲除cre-1后显著提升嗜热毁丝霉木糖和葡萄糖利用速率;同时,Cre-1可直接抑制xlnR及自身的表达。通过过表达木糖代谢激活因子XlnR编码基因xlnR,同时敲除抑制因子Cre-1编码基因cre-1,嗜热毁丝霉木糖利用速率提高100%。以上研究结果为生物质高效利用底盘细胞的创建提供了良好的出发菌株。
中图分类号:
陈美欣, 顾淑莹, 刘佳, 刘浩, 李金根, 田朝光. 嗜热毁丝霉木糖代谢调控机制研究[J]. 中国农业科技导报, 2024, 26(12): 77-87.
Meixin CHEN, Shuying GU, Jia LIU, Hao LIU, Jingen LI, Chaoguang TIAN. Research on Regulation Mechanism of Xylose Metabolism in Myceliophthora thermophila[J]. Journal of Agricultural Science and Technology, 2024, 26(12): 77-87.
| 引物名称 Prime name | 引物序列 Prime sequence(5’-3’) | 备注 Note |
|---|---|---|
| cre-1-up-F/R | GAGGACTTGACGCTGC/GTCGTTAGACTCTTTCA | cre-1 5’ |
| cre-1-down-F/R | GTGCCCCTGATAATAGC/CGACGGAATTGAGGATG | cre-1 3’ |
| xlnR-up-F/R | CACAGTACACGAGGACTTG/CAATATCAGTTAACGTCG | xlnR 5’ |
| xlnR-down-F/R | GAGTTCTTCTGATAATGAC/GAGGATCAGGACTGG | xlnR 3’ |
| neo-F/R | CTACGACGTTAACTG/CAGAAGAACTCGTC | neo |
| bar-F/R | CAAAGATATTGAAG/GATCACTAGTACAGG | bar |
| OE xlnR-F/R | GTTCTTCAGAATGTTGT/GATCCCTACAGCGCCAGAC | xlnR |
| U6p-cre-1-R | GAGTTGCTCCAACACGAGGAAAGAAAGAAAAGAAG | U6p-cre-1-sgRNA |
| cre-1-gRNA-F | TGTTGGAGCAACTCCGCTTGTTTTAGAGCTAGAAATAG | |
| U6p-xlnR-R | TAGCTGGTTGCACTGATCGCGAGGAAAGAAAG | U6p-xlnR-sgRNA |
| xlnR-gRNA-F | AGTGCAACCAGCTAGTTTTAGAGCTAGAAATAGC | |
| U6-F/gRNA-R | GTGGAGTGAAGTTCGGAA/AAAAAGCACCGACTC | sgRNA |
| cas-F/R | TCCGAGGTTCGACATCAG/GCTCCCTCTAAACAAGTG | cas9 |
| cre-1-emsa-F/R | CCAAAATCGGATCTGGTTCCGCGTGGATCCCAGCAGAAGCAGCAAGAACAACA/GATGCGGCCCTAGTGGTGGTGGTGGTGGTGCTCGAGCATGGCGGCGTGCTGATGATG | cre-1-EMSA |
| xlnR-emsa-F/R | CCAAAATCGGATCTGGTTCCGCGTGGATCCGCTCGGACTGGCCCTCAACCGA/CTAGTGGTGGTGGTGGTGGTGCTCGTGAATTCTCGGTCTTCTGGC | xlnR-EMSA |
表1 扩增目的片段所用引物
Table 1 Primers used to amplify the target fragments
| 引物名称 Prime name | 引物序列 Prime sequence(5’-3’) | 备注 Note |
|---|---|---|
| cre-1-up-F/R | GAGGACTTGACGCTGC/GTCGTTAGACTCTTTCA | cre-1 5’ |
| cre-1-down-F/R | GTGCCCCTGATAATAGC/CGACGGAATTGAGGATG | cre-1 3’ |
| xlnR-up-F/R | CACAGTACACGAGGACTTG/CAATATCAGTTAACGTCG | xlnR 5’ |
| xlnR-down-F/R | GAGTTCTTCTGATAATGAC/GAGGATCAGGACTGG | xlnR 3’ |
| neo-F/R | CTACGACGTTAACTG/CAGAAGAACTCGTC | neo |
| bar-F/R | CAAAGATATTGAAG/GATCACTAGTACAGG | bar |
| OE xlnR-F/R | GTTCTTCAGAATGTTGT/GATCCCTACAGCGCCAGAC | xlnR |
| U6p-cre-1-R | GAGTTGCTCCAACACGAGGAAAGAAAGAAAAGAAG | U6p-cre-1-sgRNA |
| cre-1-gRNA-F | TGTTGGAGCAACTCCGCTTGTTTTAGAGCTAGAAATAG | |
| U6p-xlnR-R | TAGCTGGTTGCACTGATCGCGAGGAAAGAAAG | U6p-xlnR-sgRNA |
| xlnR-gRNA-F | AGTGCAACCAGCTAGTTTTAGAGCTAGAAATAGC | |
| U6-F/gRNA-R | GTGGAGTGAAGTTCGGAA/AAAAAGCACCGACTC | sgRNA |
| cas-F/R | TCCGAGGTTCGACATCAG/GCTCCCTCTAAACAAGTG | cas9 |
| cre-1-emsa-F/R | CCAAAATCGGATCTGGTTCCGCGTGGATCCCAGCAGAAGCAGCAAGAACAACA/GATGCGGCCCTAGTGGTGGTGGTGGTGGTGCTCGAGCATGGCGGCGTGCTGATGATG | cre-1-EMSA |
| xlnR-emsa-F/R | CCAAAATCGGATCTGGTTCCGCGTGGATCCGCTCGGACTGGCCCTCAACCGA/CTAGTGGTGGTGGTGGTGGTGCTCGTGAATTCTCGGTCTTCTGGC | xlnR-EMSA |
引物名称 Prime name | 引物序列 Primer sequence (5’-3’) | 备注 Note |
|---|---|---|
| Orf-cre-1-F/R | CGATACTCGACCAAGAA/GAACCACTCTGGCTGCT | ∆cre-1 |
| Orf-xlnR-F/R | GCTCCCCATCGCCG/GGCCAGAAGACCGAG | ∆xlnR |
| Orf-OExlnR-F/R | GCGTAACCCCACCAAC/GCTGATCTGACCAGT | OexlnR |
表 2 转化体验证引物
Table 2 Verification primer of mutation
引物名称 Prime name | 引物序列 Primer sequence (5’-3’) | 备注 Note |
|---|---|---|
| Orf-cre-1-F/R | CGATACTCGACCAAGAA/GAACCACTCTGGCTGCT | ∆cre-1 |
| Orf-xlnR-F/R | GCTCCCCATCGCCG/GGCCAGAAGACCGAG | ∆xlnR |
| Orf-OExlnR-F/R | GCGTAACCCCACCAAC/GCTGATCTGACCAGT | OexlnR |
引物名称 Prime name | 引物序列 Primer sequence (5’-3’) | 备注 Note |
|---|---|---|
| cre-1-P-F/R | CCGTAGGTACCTGG/ GTTTTAAATGACGC | cre-1探针cre-1 probe |
| xlnR-P-F/R | GGCATCCCTTGGTC/ GCTCCGCCGGTGCTAA | xlnR探针xlnR probe |
表3 探针扩增引物
Table 3 Probe amplification primer
引物名称 Prime name | 引物序列 Primer sequence (5’-3’) | 备注 Note |
|---|---|---|
| cre-1-P-F/R | CCGTAGGTACCTGG/ GTTTTAAATGACGC | cre-1探针cre-1 probe |
| xlnR-P-F/R | GGCATCCCTTGGTC/ GCTCCGCCGGTGCTAA | xlnR探针xlnR probe |
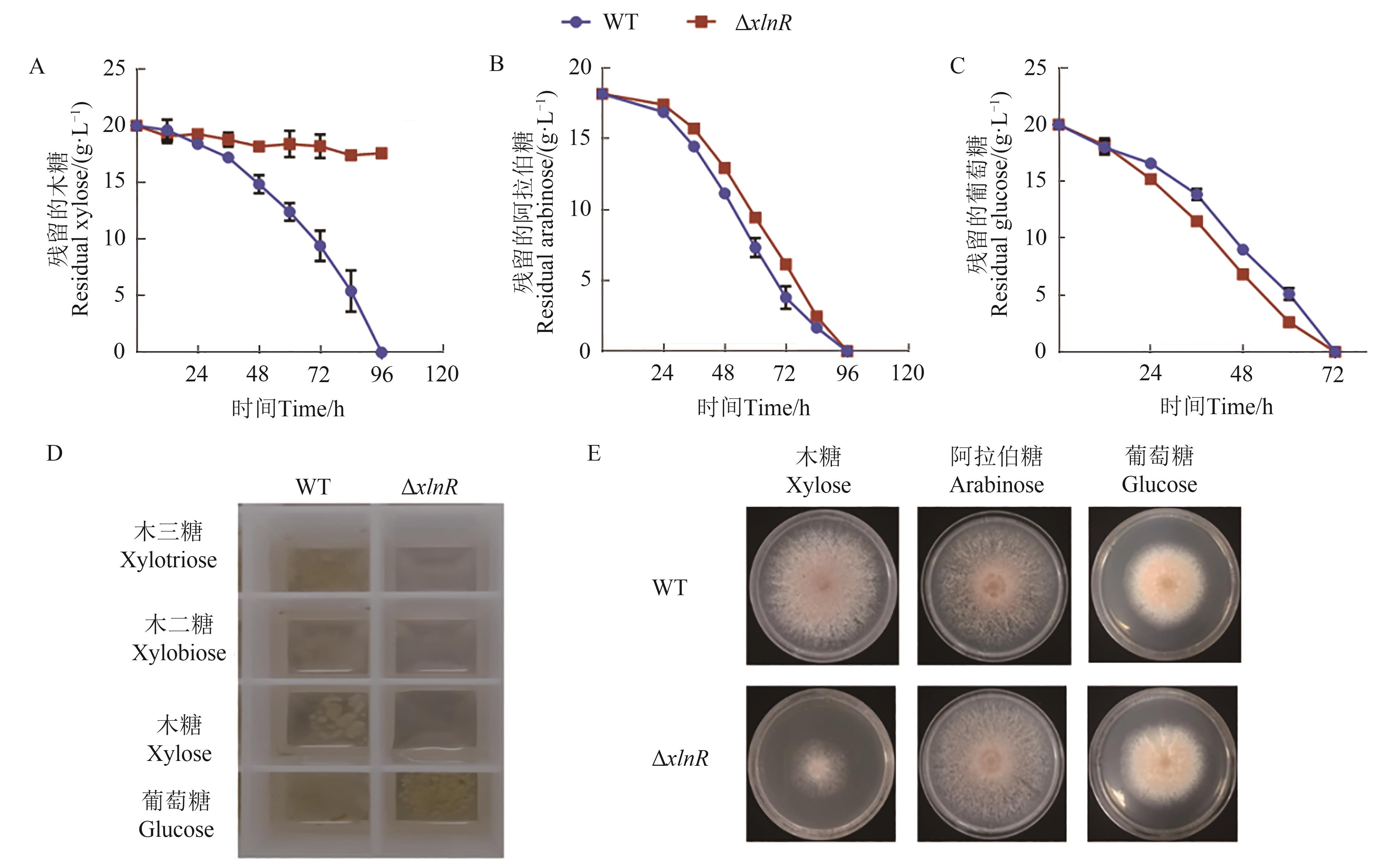
图1 野生型嗜热毁丝霉和突变体ΔxlnR 的表型A:接种突变体ΔxlnR孢子后木糖利用;B:接种突变体ΔxlnR孢子后阿拉伯糖利用;C:接种突变体ΔxlnR孢子后葡萄糖利用;D:突变体ΔxlnR 在木三糖、木二糖、木糖和葡萄糖条件下的生长表型;E:突变体ΔxlnR 在木糖、阿拉伯糖、葡萄糖固体平板上的表型
Fig. 1 Phenotypic analysis of M. thermophila strains WT and ΔxlnRA: Xylose utilization after the inoculation of the conidia from strains WT and ΔxlnR; B: Arabinose utilization after the inoculation of the conidia from strains WT and ΔxlnR; C: Glucose utilization after the inoculation of the conidia from strains WT and ΔxlnR; D: Growth phenotypes of ΔxlnR mutant on xylotriose, xylobiose, xylose and glucose; E: Growth phenotypes of ΔxlnR mutant on the agar plate with xylose, arabinose or glucose as the carbon source
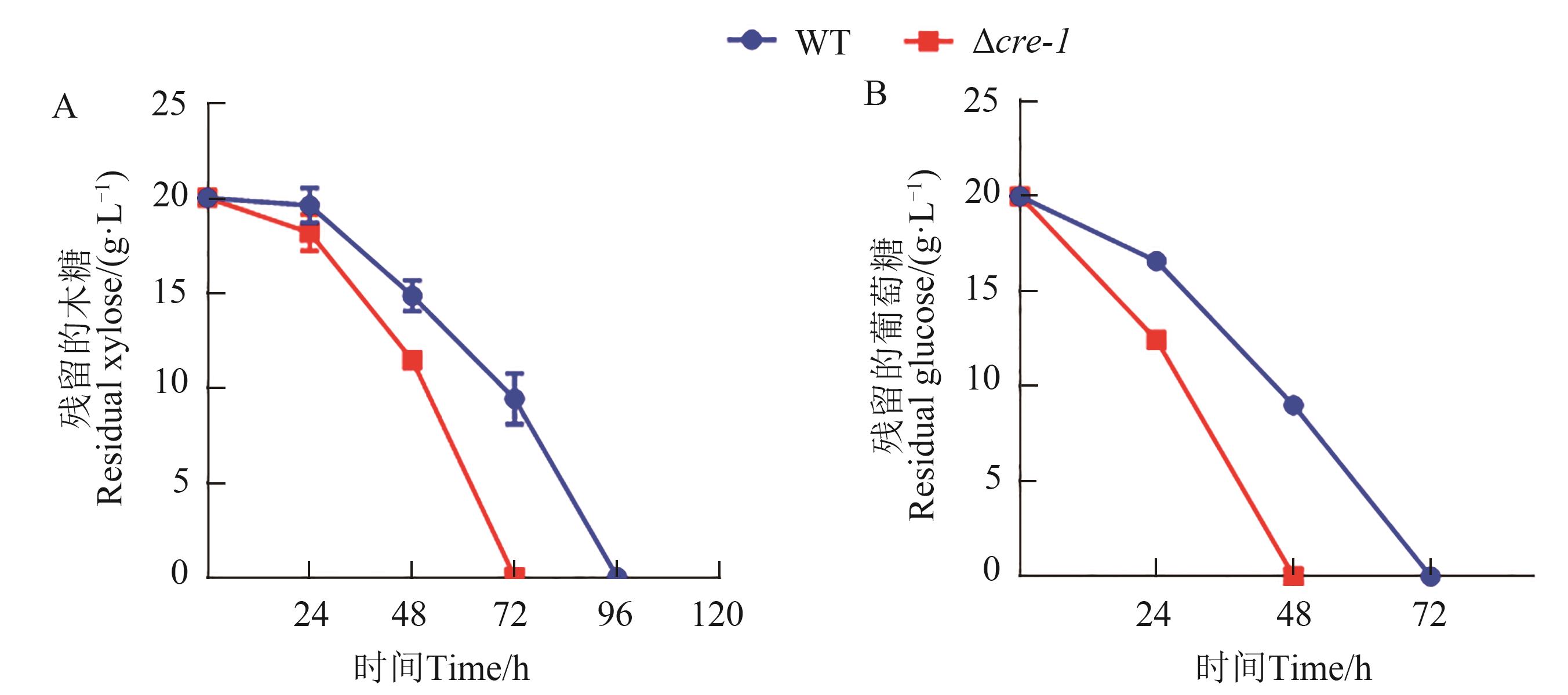
图3 嗜热毁丝霉突变体Δcre-1表型分析A:嗜热毁丝霉野生型菌株和突变体Δcre-1的木糖利用;B:嗜热毁丝霉野生型菌株和突变体Δcre-1的葡萄糖利用
Fig. 3 Phenotypic analysis of M. thermophila strain Δcre-1A: Xylose utilization of M. thermophila strains WT and Δcre-1; B: Glucose utilization of M. thermophila strains WT and Δcre-1
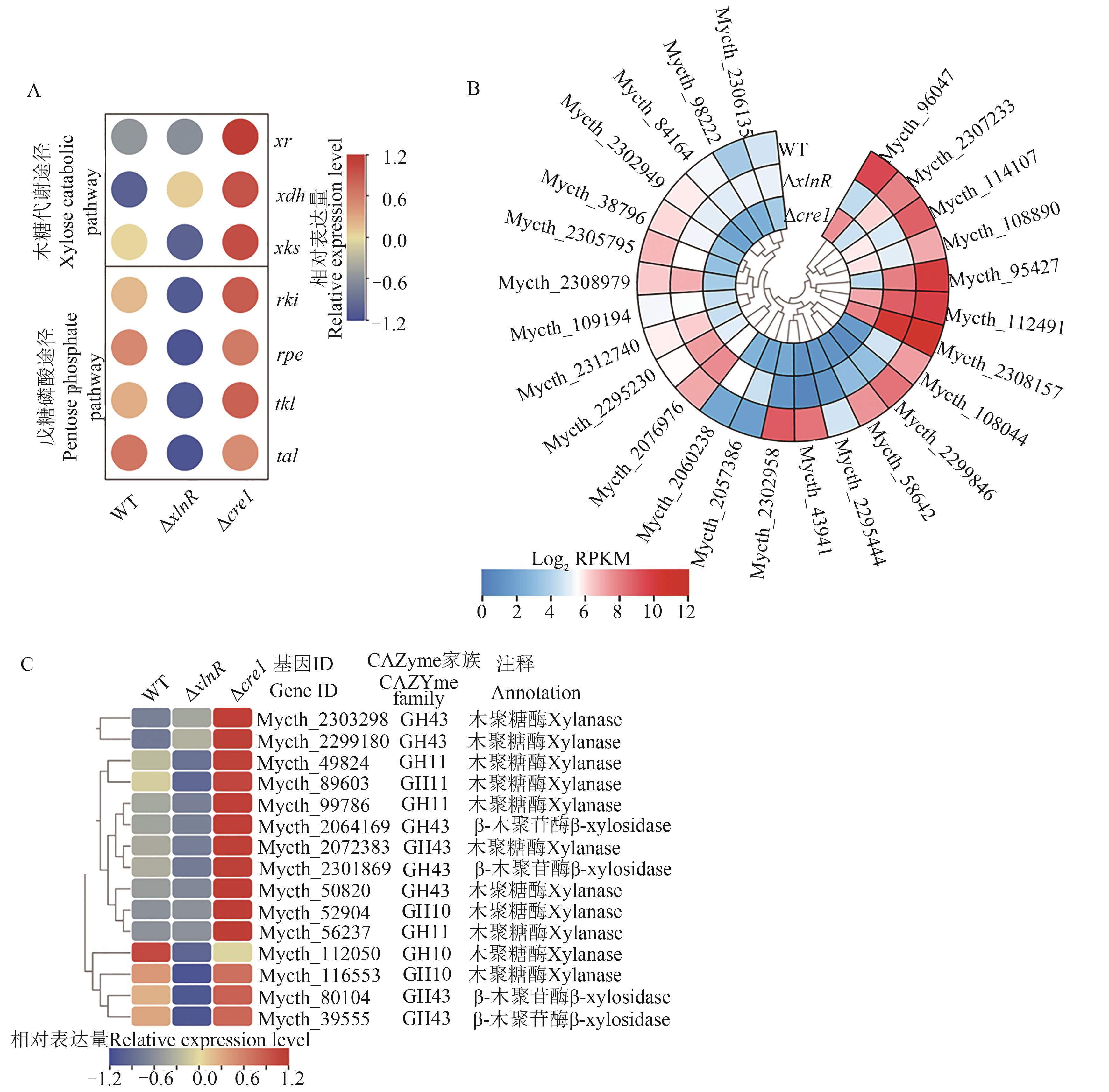
图4 嗜热毁丝霉野生型菌株和突变体Δcre-1、ΔxlnR在木糖诱导条件下的转录组分析A: 嗜热毁丝霉野生型菌株和突变体Δcre-1、ΔxlnR在木糖诱导条件下的木糖代谢途径及磷酸戊糖途径相关基因表达量聚类分析;B:糖转运蛋白基因表达水平聚类分析;C:木聚糖酶基因及β-木糖苷酶基因表达水平聚类分析。xr—木糖还原酶基因;xdh—木酮糖还原酶基因;xks—木酮糖激酶基因;rki—核糖5-磷酸异构酶基因;rpe—磷酸核酮糖3-差向异构酶基因;tkl—转酮醇酶基因;tal—转醛酶基因
Fig. 4 Transcriptome analysis of M. thermophila strains WT, Δcre-1, and ΔxlnR mutation induced by xyloseA: Heatmap analysis of expression levels of genes involved in xylose catabolism and pentose phosphate pathway in strains WT, ΔxlnR, and Δcre-1 under xylose condition; B; Heatmap analysis of expression levels of putative sugar transporter genes; C: Heatmap analysis of expression levels of genes. xr—Xylose reductase gene; xdh—Xylitol dehydrogenase gene; xks—Xylulose kinase gene; rki—Ribose 5-phosphate isomerase gene; rpe—Ribulose phosphate 3-epimerase gene; tkl—Transketolase gene; tal—Transaldolase gene

图5 嗜热毁丝霉中Cre-1对转录激活因子XlnR的调控功能分析A:EMSA分析;B:在木糖条件下,基因xlnR 在野生型菌株和突变体Δcre-1 的表达水平;C:转录因子Cre-1和XlnR之间,以及二者对木糖代谢的调控关系
Fig. 5 Analysis of regulatory function of Cre-1 on transcriptional activator XlnR in M. thermophilaA: Analysis of EMSA; B: Expression level of xlnR in the wild-type strain and mutant Δcre-1 under xylose condition; C: Relationship between transcription factor Cre-1 and XlnR, and their regulation in xylose metabolism
| 1 | TAKKELLAPATI S, LI T, GONZALEZ M A. An overview of biorefinery derived platform chemicals from a cellulose and hemicellulose biorefinery [J]. Clean Technol. Environ. Policy, 2018, 20(7):1615-1630. |
| 2 | ALI S S, NUGENT B, MULLINS E, et al.. Fungal-mediated consolidated bioprocessing: the potential of Fusarium oxysporum for the lignocellulosic ethanol industry [J/OL]. AMB Express, 2016, 6(1):13 [2023-02-10]. . |
| 3 | ZNAMEROSKI E A, CORADETTI S T, ROCHE C M, et al.. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins [J]. Proc. Natl. Acad. Sci. USA, 2012, 109(16):6012-6017. |
| 4 | BALAT M. Production of bioethanol from lignocellulosic materials via the biochemical pathway: a review [J]. Energy Convers. Manage., 2011, 52(2):858-875. |
| 5 | VITIKAINEN M, ARVAS M, PAKULA T, et al.. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties [J/OL]. BMC Genomics, 2010, 11(1):441 [2023-02-10]. . |
| 6 | ALI S S, NUGENT B, MULLINS E, et al.. Insights from the fungus Fusarium oxysporum point to high affinity glucose transporters as targets for enhancing ethanol production from lignocellulose [J/OL]. PLoS One, 2013, 8(1):e54701 [2023-02-10]. . |
| 7 | LIU G, QU Y. Engineering of filamentous fungi for efficient conversion of lignocellulose: tools, recent advances and prospects [J]. Biotechnol. Adv., 2019, 37(4):519-529. |
| 8 | VISSER H, JOOSTEN V, PUNTPETER J, et al.. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1 [J]. Ind. Biotechnol., 2011, 7:214-223. |
| 9 | DRUZHININA I S, KUBICEK C P. Genetic engineering of Trichoderma reesei cellulases and their production [J]. Microb. Biotechnol., 2017, 10(6):1485-1499. |
| 10 | LI N, LIU Y, LIU D, et al.. MtTRC-1, a novel transcription factor, regulates cellulase production via directly modulating the genes expression of the Mthac-1 and Mtcbh-1 in Myceliophthora thermophila [J/OL]. Appl. Environ. Microbiol., 2022, 88(19):e0126322 [2023-02-10]. . |
| 11 | HECTOR R E, QURESHI N, RHUGHES S, et al.. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption [J]. Appl. Microbiol. Biotechnol., 2008, 80(4):675-684. |
| 12 | WU V W, THIEME N, HUBERMAN L B, et al.. The regulatory and transcriptional landscape associated with carbon utilization in a filamentous fungus [J]. Proc. Natl. Acad. Sci. USA, 2020, 117(11):6003-6013. |
| 13 | DENG H, BAI Y, FAN T P, et al.. Advanced strategy for metabolite exploration in filamentous fungi [J]. Crit. Rev. Biotechnol., 2020, 40(2):180-198. |
| 14 | ADNAN M, ZHENG W, ISLAM W, et al.. Carbon catabolite repression in filamentous fungi [J/OL]. Int. J. Mol. Sci., 2017, 19(1):48 [2023-02-10]. . |
| 15 | TAMAYO E N, VILLANUEVA A, HASPER A A, et al.. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans [J]. Fungal Genet. Biol., 2008, 45(6):984-993. |
| 16 | ANTONIETO A C, DOS SANTOS CASTRO L, SILVA-ROCHA R, et al.. Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis [J]. Fungal Genet. Biol., 2014, 73:93-103. |
| 17 | PORTNOY T, MARGEOT A, LINKE R, et al.. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei:a master regulator of carbon assimilation [J/OL]. BMC Genomics, 2011, 12:269 [2023-02-10]. . |
| 18 | FURUKAWA T, SHIDA Y, KITAGAMI N, et al.. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei [J]. Fungal Genet. Biol., 2009, 46(8):564-574. |
| 19 | PORTNOY T, MARGEOT A, SEIDL-SEIBOTH V, et al.. Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase [J]. Eukaryot. Cell, 2011, 10(2):262-271. |
| 20 | DOS SANTOS GOMES A C, FALKOSKI D, BATTAGLIA E, et al.. Myceliophthora thermophila Xyr1 is predominantly involved in xylan degradation and xylose catabolism [J/OL]. Biotechnol. Biofuels, 2019, 12:220 [2023-02-10]. . |
| 21 | CRAIG J P, CORADETTI S T, STARR T L, et al.. Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1 [J/OL]. Mbio, 2015, 6(5):e01452-15 [2023-02-10]. . |
| 22 | HASPER A A, TRINDADE L M, VAN DER VEEN D, et al.. Functional analysis of the transcriptional activator XlnR from Aspergillus niger [J]. Microbiology, 2004, 150(Pt5):1367-1375. |
| 23 | HASPER A A, VISSER J, DE GRAAFF L H. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression [J]. Mol. Microbiol., 2000, 36(1):193-200. |
| 24 | MACH-AIGNER R, PUCHER E, STEIGER G, et al.. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina [J]. Appl. Environ. Microbiol., 2008,74(21):6554-6562. |
| 25 | MACH-AIGNER A R, OMONY J, JOVANOVIC B, et al.. D-Xylose concentration-dependent hydrolase expression profiles and the function of CreA and XlnR in Aspergillus niger [J]. Appl. Environ. Microbiol., 2012, 78(9):3145-3155. |
| 26 | BERKA R M, GRIGORIEV I V, OTILLAR R, et al.. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris [J]. Nat. Biotechnol., 2011, 29(10):922-927. |
| 27 | XU J, LI J, LIN L, et al.. Development of genetic tools for Myceliophthora thermophila [J/OL]. BMC Biotechnol., 2015, 15:35 [2023-02-10]. . |
| 28 | SONG R, ZHAI Q, SUN L, et al.. CRISPR/Cas9 genome editing technology in filamentous fungi:progress and perspective [J]. Appl. Microbiol. Biotechnol., 2019, 103(17):6919-6932. |
| 29 | SWIAT M A, DASHKO S, DEN RIDDER M, et al.. FnCpf1:a novel and efficient genome editing tool for Saccharomyces cerevisiae [J]. Nucleic Acids Res., 2017, 45(21):12585-12598. |
| 30 | PATEL H, RAWAT S. Thermophilic fungi: diversity, physiology, genetics, and applications [J]. New Future Dev. Microb. Biotechnol. Bioeng., 2021, 13(6):69-93. |
| 31 | LIU Q, GAO R, LI J, et al.. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering [J/OL]. Biotechnol. Biofuels, 2017, 10:1 [2023-02-10]. . |
| 32 | LI J, GU S, ZHAO Z, et al.. Dissecting cellobiose metabolic pathway and its application in biorefinery through consolidated bioprocessing in Myceliophthora thermophila [J/OL].Fungal Biol. Biotechnol., 2019, 6:21 [2023-02-10]. . |
| 33 | XU Q, SINGH A, HIMMEL M E. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose [J]. Curr. Opin. Biotechnol., 2009, 20(3):364-371. |
| 34 | WANG B, LI J, GAO J, et al.. Identification and characterization of the glucose dual-affinity transport system in Neurospora crassa:pleiotropic roles in nutrient transport, signaling, and carbon catabolite repression [J/OL]. Biotechnol. Biofuels, 2017, 10:17 [2023-02-10]. . |
| 35 | SUN J, TIAN C, DIAMOND S, et al.. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa [J]. Eukaryot. Cell, 2012, 11(4):482-493. |
| 36 | SUN J, GLASS N L. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa [J/OL]. PLoS One, 2011, 6(9):e25654 [2023-02-10]. . |
| 37 | FURUKAWA T, SHIDA Y, KITAGAMI N, et al.. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei [J]. Fungal Genet. Biol., 2009, 46(8):564-574. |
| 38 | LI J, LIN L, SUN T, et al.. Direct production of commodity chemicals from lignocellulose using Myceliophthora thermophila [J]. Metab. Eng., 2020, 61:416-426. |
| 39 | WEINHANDL K, WINKLER M, GLIEDER A, et al.. Carbon source dependent promoters in yeasts [J/OL]. Microb. Cell Fact., 2014, 13:5[2023-02-10].. |
| [1] | 李丹丹1,2,汪海2*,朱莉2,黄大昉1,2*,郎志宏2. 玉米虫害诱导基因启动子在拟南芥中的活性分析[J]. , 2015, 17(4): 23-29. |
| [2] | 赵俊星1, 岳万福2. 表观遗传调控骨骼肌发育的研究进展(英文)[J]. , 2014, 16(3): 42-47. |
| [3] | 尚立国,弓湃,战嵛华,邓志平,燕永亮*. 固氮施氏假单胞菌全局性调控因子RsmA的进化分析及表达特性研究[J]. , 2014, 16(3): 62-69. |
| [4] | 吴港1,2,王金辉1,2,张维2,林敏2,陈明2*,张云华1,2*. 细菌转录调控蛋白CarD的研究进展[J]. , 2014, 16(3): 48-52. |
| [5] | 周庆新1,2,戴炳业3,陈蕾蕾1,刘孝永1,裘纪莹1,陈相艳1*. 瑞氏木霉中β-葡萄糖苷酶基因功能研究进展[J]. , 2014, 16(2): 74-78. |
| [6] | 张健飞,权瑞党,黄荣峰. EAR转录抑制子结构及功能的研究[J]. , 2011, 13(4): 53-57. |
| [7] | 张融雪,张治礼,张执金,黄荣峰. 植物低温信号的感知、转导与转录调控[J]. , 2009, 11(3): 5-11. |
| [8] | 李成霞 敖光明. 植物TFⅢA类型锌指蛋白的研究进展[J]. , 2000, 2(6): 9-12. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号