




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (2): 48-59.DOI: 10.13304/j.nykjdb.2021.0699
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Man ZHANG( ), Zhicheng WANG, Zhengwen LIU, Guoning WANG, Xingfen WANG, Yan ZHANG(
), Zhicheng WANG, Zhengwen LIU, Guoning WANG, Xingfen WANG, Yan ZHANG( )
)
Received:2021-08-14
Accepted:2021-09-22
Online:2023-02-15
Published:2023-05-17
Contact:
Yan ZHANG
张曼( ), 王志城, 刘正文, 王国宁, 王省芬, 张艳(
), 王志城, 刘正文, 王国宁, 王省芬, 张艳( )
)
通讯作者:
张艳
作者简介:张曼 E-mail:zhangman920601@163.com;
基金资助:CLC Number:
Man ZHANG, Zhicheng WANG, Zhengwen LIU, Guoning WANG, Xingfen WANG, Yan ZHANG. Genome-Wide Identification and Analysis of BGLU Genes Family in Gossypium hirsutum[J]. Journal of Agricultural Science and Technology, 2023, 25(2): 48-59.
张曼, 王志城, 刘正文, 王国宁, 王省芬, 张艳. 陆地棉BGLU基因家族成员的全基因组鉴定与表达分析[J]. 中国农业科技导报, 2023, 25(2): 48-59.
引物名称 Primer name | 引物序列 Primer sequence(3’-5’) |
|---|---|
| GhUBQ14-F | CAACGCTCCATCTTGTCCTT |
| GhUBQ14-R | TGATCGTCTTTCCCGTAAGC |
| GhBGLU9-RT-F | GGCGGTGATGGACACTGGA |
| GhBGLU9-RT-R | GCCACCCTTATTAGCCATCC |
| GhBGLU19-RT-F | CGTAGAACATGATGATTCCAACACT |
| GhBGLU19-RT-R | CCACCTTCGTTCACACCTCCTT |
| GhBGLU05-RT-F | GTTTTATTTTCGGTGCCGGAT |
| GhBGLU05-RT-R | GATGTTGTCTAACGAGAATGTCCC |
| GhBGLU23-RT-F | CGAGGGAGCAGTGAAGGAGGAT |
| GhBGLU23-RT-R | TATCTTCAACATAGCGGTGGTACT |
| GhBGLU26-RT-F | AAGATGGAAAAGGCCTAAGCAAC |
| GhBGLU26-RT-R | GCATCATCTCCATTCTCATTGTTTT |
| GhBGLU34-RT-F | TGTGTTCTATCTGTTGTTCCTTTGG |
| GhBGLU34-RT-R | CTGTATCTCCATTGCTTCCATCCAT |
| GhBGLU52-RT-F | CTGAAACAAAGCCTGGGTAATGGTA |
| GhBGLU52-RT-R | CTTCATACTGGTAAGCAGAGGAGGC |
| GhBGLU35-RT-F | GTGAATGAAGGAAACAAGGGAGATA |
| GhBGLU35-RT-R | AAGCATCCATTCCCAGATCTTTCAT |
| GhBGLU14-RT-F | GTTCTTCTTCTTACTCAACTTCATTTC |
| GhBGLU14-RT-R | TTCATACTGGTAAGCGGAAGAGGC |
Table 1 Primer sequences for qRT?PCR in this study
引物名称 Primer name | 引物序列 Primer sequence(3’-5’) |
|---|---|
| GhUBQ14-F | CAACGCTCCATCTTGTCCTT |
| GhUBQ14-R | TGATCGTCTTTCCCGTAAGC |
| GhBGLU9-RT-F | GGCGGTGATGGACACTGGA |
| GhBGLU9-RT-R | GCCACCCTTATTAGCCATCC |
| GhBGLU19-RT-F | CGTAGAACATGATGATTCCAACACT |
| GhBGLU19-RT-R | CCACCTTCGTTCACACCTCCTT |
| GhBGLU05-RT-F | GTTTTATTTTCGGTGCCGGAT |
| GhBGLU05-RT-R | GATGTTGTCTAACGAGAATGTCCC |
| GhBGLU23-RT-F | CGAGGGAGCAGTGAAGGAGGAT |
| GhBGLU23-RT-R | TATCTTCAACATAGCGGTGGTACT |
| GhBGLU26-RT-F | AAGATGGAAAAGGCCTAAGCAAC |
| GhBGLU26-RT-R | GCATCATCTCCATTCTCATTGTTTT |
| GhBGLU34-RT-F | TGTGTTCTATCTGTTGTTCCTTTGG |
| GhBGLU34-RT-R | CTGTATCTCCATTGCTTCCATCCAT |
| GhBGLU52-RT-F | CTGAAACAAAGCCTGGGTAATGGTA |
| GhBGLU52-RT-R | CTTCATACTGGTAAGCAGAGGAGGC |
| GhBGLU35-RT-F | GTGAATGAAGGAAACAAGGGAGATA |
| GhBGLU35-RT-R | AAGCATCCATTCCCAGATCTTTCAT |
| GhBGLU14-RT-F | GTTCTTCTTCTTACTCAACTTCATTTC |
| GhBGLU14-RT-R | TTCATACTGGTAAGCGGAAGAGGC |
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU01 | Ghir_A01G020950 | 359 | 42.213 | 8.740 | 35~36 | 细胞质膜Plasma membrane |
| GhBGLU02 | Ghir_A01G021100 | 495 | 57.410 | 6.123 | 23~24 | 液泡Vacuole |
| GhBGLU03 | Ghir_A01G021370 | 502 | 58.129 | 7.219 | 23~24 | 液泡Vacuole |
| GhBGLU04 | Ghir_A02G012630 | 390 | 44.336 | 5.309 | 38~39 | 细胞质Cytoplasmic |
| GhBGLU05 | Ghir_A05G007460 | 476 | 54.519 | 5.022 | 21~22 | 液泡Vacuole |
| GhBGLU06 | Ghir_A06G012310 | 491 | 55.875 | 9.314 | 23~24 | 溶酶体Lysosomal |
| GhBGLU07 | Ghir_A06G015860 | 527 | 59.958 | 6.555 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU08 | Ghir_A06G015870 | 494 | 56.087 | 6.519 | 21~22 | 液泡Vacuole |
| GhBGLU09 | Ghir_A07G008330 | 331 | 37.350 | 9.361 | 19~20 | 胞外Extracellular |
| GhBGLU10 | Ghir_A07G008340 | 497 | 56.916 | 8.338 | 19~20 | 液泡Vacuole |
| GhBGLU11 | Ghir_A08G024630 | 520 | 58.768 | 7.434 | 28~29 | 细胞质Cytoplasmic |
| GhBGLU12 | Ghir_A08G025080 | 371 | 41.575 | 6.883 | 26~27 | 细胞质膜Plasma membrane |
Table 2 Characteristics of 53 BGLU proteins in G. hirsutum
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU01 | Ghir_A01G020950 | 359 | 42.213 | 8.740 | 35~36 | 细胞质膜Plasma membrane |
| GhBGLU02 | Ghir_A01G021100 | 495 | 57.410 | 6.123 | 23~24 | 液泡Vacuole |
| GhBGLU03 | Ghir_A01G021370 | 502 | 58.129 | 7.219 | 23~24 | 液泡Vacuole |
| GhBGLU04 | Ghir_A02G012630 | 390 | 44.336 | 5.309 | 38~39 | 细胞质Cytoplasmic |
| GhBGLU05 | Ghir_A05G007460 | 476 | 54.519 | 5.022 | 21~22 | 液泡Vacuole |
| GhBGLU06 | Ghir_A06G012310 | 491 | 55.875 | 9.314 | 23~24 | 溶酶体Lysosomal |
| GhBGLU07 | Ghir_A06G015860 | 527 | 59.958 | 6.555 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU08 | Ghir_A06G015870 | 494 | 56.087 | 6.519 | 21~22 | 液泡Vacuole |
| GhBGLU09 | Ghir_A07G008330 | 331 | 37.350 | 9.361 | 19~20 | 胞外Extracellular |
| GhBGLU10 | Ghir_A07G008340 | 497 | 56.916 | 8.338 | 19~20 | 液泡Vacuole |
| GhBGLU11 | Ghir_A08G024630 | 520 | 58.768 | 7.434 | 28~29 | 细胞质Cytoplasmic |
| GhBGLU12 | Ghir_A08G025080 | 371 | 41.575 | 6.883 | 26~27 | 细胞质膜Plasma membrane |
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU13 | Ghir_A09G005920 | 506 | 58.313 | 9.323 | 20~21 | 溶酶体Lysosomal |
| GhBGLU14 | Ghir_A10G001000 | 536 | 61.583 | 6.510 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU15 | Ghir_A10G001010 | 500 | 57.229 | 7.590 | 27~28 | 液泡Vacuole |
| GhBGLU16 | Ghir_A10G001020 | 536 | 61.339 | 6.976 | 24~25 | 溶酶体Lysosomal |
| GhBGLU17 | Ghir_A11G032270 | 514 | 58.455 | 6.337 | 26~27 | 细胞质Cytoplasmic |
| GhBGLU18 | Ghir_A11G033190 | 199 | 23.264 | 4.694 | 40~41 | 细胞质Cytoplasmic |
| GhBGLU19 | Ghir_A11G033200 | 195 | 22.294 | 6.799 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU20 | Ghir_A12G017270 | 132 | 15.366 | 7.963 | 19~20 | 胞外Extracellular |
| GhBGLU21 | Ghir_A12G026530 | 482 | 55.028 | 9.470 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU22 | Ghir_A13G008250 | 498 | 56.644 | 5.970 | 23~24 | 液泡Vacuole |
| GhBGLU23 | Ghir_A13G008260 | 136 | 15.630 | 9.499 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU24 | Ghir_A13G008270 | 504 | 56.842 | 7.635 | 24~25 | 液泡Vacuole |
| GhBGLU25 | Ghir_A13G014930 | 383 | 43.308 | 4.834 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU26 | Ghir_A13G015150 | 546 | 62.303 | 6.561 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU27 | Ghir_D01G022500 | 445 | 51.828 | 8.334 | 18~19 | 细胞质膜Plasma membrane |
| GhBGLU28 | Ghir_D01G022510 | 500 | 57.851 | 7.275 | 19~20 | 液泡Vacuole |
| GhBGLU29 | Ghir_D01G022660 | 499 | 58.085 | 6.203 | 23~24 | 液泡Vacuole |
| GhBGLU30 | Ghir_D01G022670 | 511 | 59.569 | 6.400 | 23~24 | 液泡Vacuole |
| GhBGLU31 | Ghir_D03G006350 | 328 | 37.578 | 6.139 | 27~28 | 细胞质Cytoplasmic |
| GhBGLU32 | Ghir_D05G007520 | 482 | 55.193 | 5.549 | 21~22 | 液泡Vacuole |
| GhBGLU33 | Ghir_D06G012190 | 437 | 50.036 | 9.662 | 21~22 | 溶酶体Lysosomal |
| GhBGLU34 | Ghir_D06G016620 | 483 | 54.692 | 6.788 | 16~17 | 细胞质膜Plasma membrane |
| GhBGLU35 | Ghir_D06G016640 | 534 | 60.754 | 6.782 | 21~22 | 细胞质膜Plasma membrane |
| GhBGLU36 | Ghir_D07G008440 | 139 | 15.520 | 8.918 | 19~20 | 胞外Extracellular |
| GhBGLU37 | Ghir_D07G008450 | 479 | 54.695 | 8.085 | 19~20 | 溶酶体Lysosomal |
| GhBGLU38 | Ghir_D08G025520 | 514 | 57.962 | 6.515 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU39 | Ghir_D08G025990 | 463 | 52.616 | 7.511 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU40 | Ghir_D09G005490 | 341 | 39.515 | 9.728 | 24~25 | 胞外Extracellular |
| GhBGLU41 | Ghir_D10G001780 | 466 | 53.328 | 5.882 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU42 | Ghir_D10G001790 | 509 | 58.366 | 7.255 | 27~28 | 细胞质膜Plasma membrane |
| GhBGLU43 | Ghir_D10G001800 | 518 | 58.984 | 6.416 | 24~25 | 溶酶体Lysosomal |
| GhBGLU44 | Ghir_D11G032750 | 514 | 58.322 | 6.206 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU45 | Ghir_D11G033820 | 490 | 56.008 | 5.179 | 38~29 | 细胞质Cytoplasmic |
| GhBGLU46 | Ghir_D12G017490 | 121 | 13.931 | 7.500 | 47~48 | 胞外Extracellular |
| GhBGLU47 | Ghir_D12G026540 | 482 | 54.786 | 9.368 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU48 | Ghir_D12G026620 | 472 | 53.808 | 9.573 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU49 | Ghir_D13G008490 | 504 | 56.847 | 6.680 | 24~25 | 液泡Vacuole |
| GhBGLU50 | Ghir_D13G008500 | 205 | 23.526 | 8.424 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU51 | Ghir_D13G008510 | 498 | 56.553 | 5.218 | 23~24 | 液泡Vacuole |
| GhBGLU52 | Ghir_D13G015630 | 488 | 55.822 | 8.565 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU53 | Ghir_D13G015870 | 546 | 61.929 | 6.658 | 26~27 | 细胞质膜Plasma membrane |
Table 2 Characteristics of 53 BGLU proteins in G. hirsutum
基因名称 Gene name | 基因ID号 Gene ID | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| GhBGLU13 | Ghir_A09G005920 | 506 | 58.313 | 9.323 | 20~21 | 溶酶体Lysosomal |
| GhBGLU14 | Ghir_A10G001000 | 536 | 61.583 | 6.510 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU15 | Ghir_A10G001010 | 500 | 57.229 | 7.590 | 27~28 | 液泡Vacuole |
| GhBGLU16 | Ghir_A10G001020 | 536 | 61.339 | 6.976 | 24~25 | 溶酶体Lysosomal |
| GhBGLU17 | Ghir_A11G032270 | 514 | 58.455 | 6.337 | 26~27 | 细胞质Cytoplasmic |
| GhBGLU18 | Ghir_A11G033190 | 199 | 23.264 | 4.694 | 40~41 | 细胞质Cytoplasmic |
| GhBGLU19 | Ghir_A11G033200 | 195 | 22.294 | 6.799 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU20 | Ghir_A12G017270 | 132 | 15.366 | 7.963 | 19~20 | 胞外Extracellular |
| GhBGLU21 | Ghir_A12G026530 | 482 | 55.028 | 9.470 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU22 | Ghir_A13G008250 | 498 | 56.644 | 5.970 | 23~24 | 液泡Vacuole |
| GhBGLU23 | Ghir_A13G008260 | 136 | 15.630 | 9.499 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU24 | Ghir_A13G008270 | 504 | 56.842 | 7.635 | 24~25 | 液泡Vacuole |
| GhBGLU25 | Ghir_A13G014930 | 383 | 43.308 | 4.834 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU26 | Ghir_A13G015150 | 546 | 62.303 | 6.561 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU27 | Ghir_D01G022500 | 445 | 51.828 | 8.334 | 18~19 | 细胞质膜Plasma membrane |
| GhBGLU28 | Ghir_D01G022510 | 500 | 57.851 | 7.275 | 19~20 | 液泡Vacuole |
| GhBGLU29 | Ghir_D01G022660 | 499 | 58.085 | 6.203 | 23~24 | 液泡Vacuole |
| GhBGLU30 | Ghir_D01G022670 | 511 | 59.569 | 6.400 | 23~24 | 液泡Vacuole |
| GhBGLU31 | Ghir_D03G006350 | 328 | 37.578 | 6.139 | 27~28 | 细胞质Cytoplasmic |
| GhBGLU32 | Ghir_D05G007520 | 482 | 55.193 | 5.549 | 21~22 | 液泡Vacuole |
| GhBGLU33 | Ghir_D06G012190 | 437 | 50.036 | 9.662 | 21~22 | 溶酶体Lysosomal |
| GhBGLU34 | Ghir_D06G016620 | 483 | 54.692 | 6.788 | 16~17 | 细胞质膜Plasma membrane |
| GhBGLU35 | Ghir_D06G016640 | 534 | 60.754 | 6.782 | 21~22 | 细胞质膜Plasma membrane |
| GhBGLU36 | Ghir_D07G008440 | 139 | 15.520 | 8.918 | 19~20 | 胞外Extracellular |
| GhBGLU37 | Ghir_D07G008450 | 479 | 54.695 | 8.085 | 19~20 | 溶酶体Lysosomal |
| GhBGLU38 | Ghir_D08G025520 | 514 | 57.962 | 6.515 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU39 | Ghir_D08G025990 | 463 | 52.616 | 7.511 | 26~27 | 细胞质膜Plasma membrane |
| GhBGLU40 | Ghir_D09G005490 | 341 | 39.515 | 9.728 | 24~25 | 胞外Extracellular |
| GhBGLU41 | Ghir_D10G001780 | 466 | 53.328 | 5.882 | 29~30 | 细胞质膜Plasma membrane |
| GhBGLU42 | Ghir_D10G001790 | 509 | 58.366 | 7.255 | 27~28 | 细胞质膜Plasma membrane |
| GhBGLU43 | Ghir_D10G001800 | 518 | 58.984 | 6.416 | 24~25 | 溶酶体Lysosomal |
| GhBGLU44 | Ghir_D11G032750 | 514 | 58.322 | 6.206 | 28~29 | 细胞质膜Plasma membrane |
| GhBGLU45 | Ghir_D11G033820 | 490 | 56.008 | 5.179 | 38~29 | 细胞质Cytoplasmic |
| GhBGLU46 | Ghir_D12G017490 | 121 | 13.931 | 7.500 | 47~48 | 胞外Extracellular |
| GhBGLU47 | Ghir_D12G026540 | 482 | 54.786 | 9.368 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU48 | Ghir_D12G026620 | 472 | 53.808 | 9.573 | 23~24 | 细胞质膜Plasma membrane |
| GhBGLU49 | Ghir_D13G008490 | 504 | 56.847 | 6.680 | 24~25 | 液泡Vacuole |
| GhBGLU50 | Ghir_D13G008500 | 205 | 23.526 | 8.424 | 37~38 | 细胞质Cytoplasmic |
| GhBGLU51 | Ghir_D13G008510 | 498 | 56.553 | 5.218 | 23~24 | 液泡Vacuole |
| GhBGLU52 | Ghir_D13G015630 | 488 | 55.822 | 8.565 | 22~23 | 细胞质膜Plasma membrane |
| GhBGLU53 | Ghir_D13G015870 | 546 | 61.929 | 6.658 | 26~27 | 细胞质膜Plasma membrane |
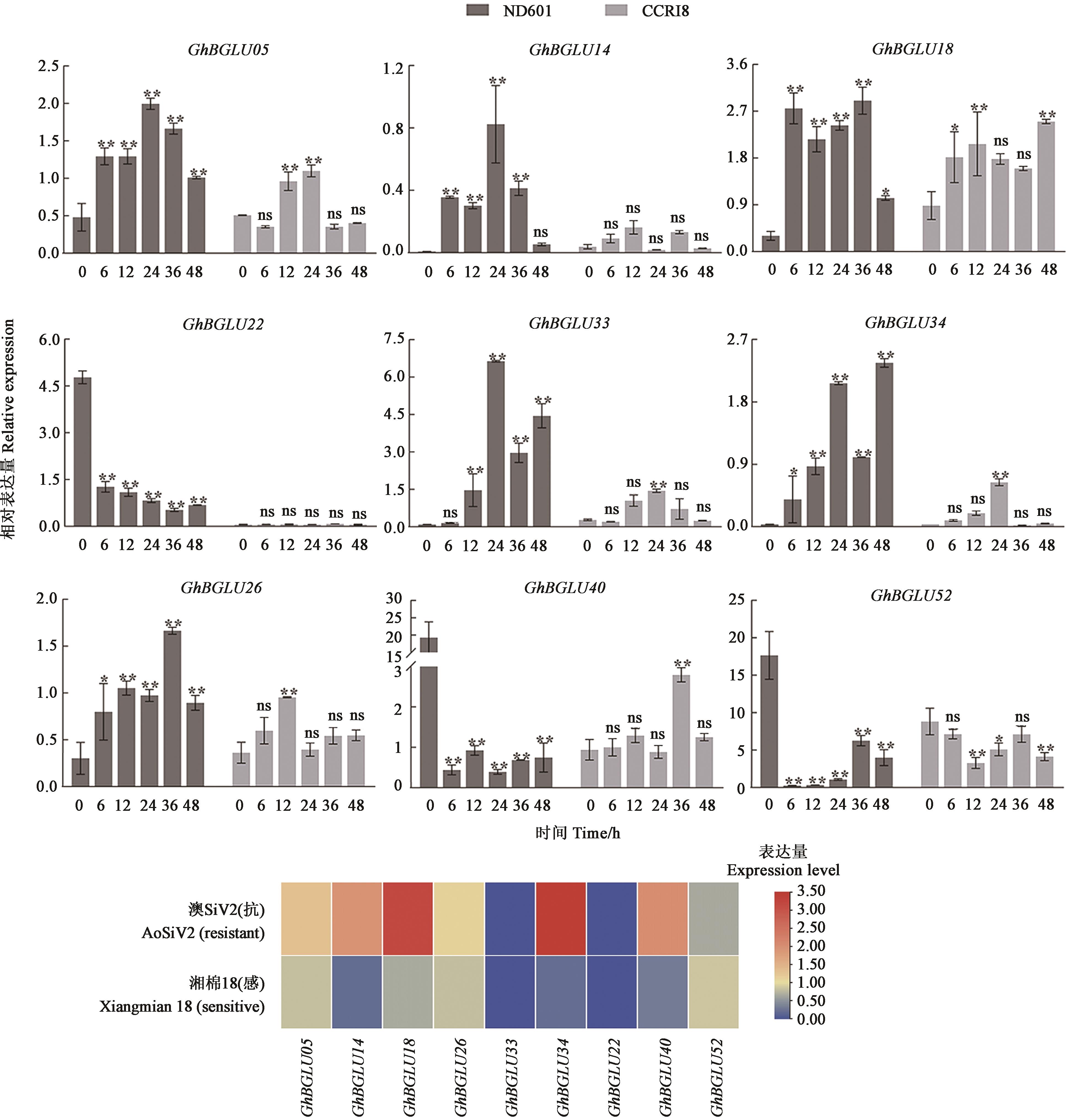
Fig. 7 Analysis of the expression of 9 GhBGLU genes under the stress of V. dahliaeNote: ** and * indicate significant differences at P<0.01 and P<0.05 levels, respectively.
| 1 | WANG K, WANG Z, LI F, et al.. The draft genome of a diploid cotton Gossypium raimondii [J]. Nat. Genet., 2012, 44(10): 1098-1103. |
| 2 | LI F G, FAN G Y, WANG K B, et al.. Genome sequence of the cultivated cotton Gossypium arboreum [J]. Nat. Genet., 2014, 46(6): 567-572. |
| 3 | PATERSON A H, WENDEL J F, GUNDLACH H, et al.. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers [J]. Nature, 2012, 492(7429): 423-427. |
| 4 | HU Y, CHEN J, FANG L, et al.. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton [J]. Nat. Genet., 2019: 51(54): 739-748. |
| 5 | 王晨,李家儒.植物β-葡萄糖苷酶的研究进展[J].生物资源, 2021, 43(2): 101-109. |
| WANG C, LI J. Research progress of plant β⁃glucosidase [J]. Biotic. Resour., 2021, 43(2): 101-109. | |
| 6 | HENRISSAT B A. A classification of glycosyl hydrolases based on amino acid sequence similarities [J]. Biochem. J., 1992, 280(2): 309-316. |
| 7 | DUROUX L, DELMOTTE F M, LANCELIN J M, et al.. Insight into naphthoquinone metabolism: β-glucosidase-catalysed hydrolysis of hydrojuglone β-D-glucopyranoside [J]. Biochem. J., 1998, 333(2): 275-283. |
| 8 | DHARMAWARDHANA D. A beta-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin [J]. Plant Physiol., 1995, 107(2): 331-339. |
| 9 | BRZOBOHATY B, MOORE I, KRISTOFFERSEN P, et al.. Release of active cytokinin by a beta-glucosidase localized to the maize root meristem [J]. Science, 1993, 262(5136): 1051-1054. |
| 10 | SUN H, XUE Y, LIN Y, et al.. Enhanced catalytic efficiency in quercetin-4'-glucoside hydrolysis of Thermotoga maritima β-glucosidase a by site-directed mutagenesis [J]. J. Agric. Food Chem., 2014, 62(28):6763-6770. |
| 11 | SAMPEDRO J, VALDIVIA E R, FRAGA P, et al.. Soluble and membrane-bound b-glucosidases are involved in trimming the xyloglucan backbone1 [J]. Plant Physiol., 2017, 173: 1017-1030. |
| 12 | LUIS L E-T W, CHEN, MARCELLA L, et al.. Arabidopsis thaliana β-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides science direct [J]. Phytochemistry, 2006, 67(15): 1651-1660. |
| 13 | WANG C, CHEN S, DONG Y, et al.. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice [J]. New Phytol., 2020, 226(4): 1042-1054. |
| 14 | WANG H, ZHANG Y, ZHAO Y, et al.. Genome-wide comparative analysis of the β-glucosidase family in five rosaceae species and their potential role on lignification of stone cells in Chinese white pear [J/OL]. Peer J., 2019, 7(3):46971 [2021-12-07] . |
| 15 | KONG W L, ZHONG H, DENG X X, et al.. Evolutionary analysis of GH3 genes in six oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica [J]. Plants, 2019: 8(2):30. |
| 16 | QANMBER G, DAOQIAN Y U, JIE L I, et al.. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses [J]. J. Cotton Res., 2018, 1(1): 3-19. |
| 17 | YANG, XIU, XU, et al.. Genome-wide identification of OSCA gene family and their potential function in the regulation of dehydration and salt stress in Gossypium hirsutum [J]. J. Cotton Res., 2019, 2(02): 14-31. |
| 18 | XU Q F, DUNBRACK RL. Assignment of protein sequences to existing domain and family classification systems: Pfam and the PDB [J]. Bioinformatics, 2012, 28(21): 2763-2772. |
| 19 | LARKIN M. Clustal W and Clustal X v. 2.0 [J]. Bioinformatics, 2007, 23: 2947-2948. |
| 20 | CHENG H Q, HAN L B, YANG C L, et al.. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection [J]. J. Exp. Bot., 2016(6): 1935-1950. |
| 21 | ZHAO Y L, ZHOU T T, GUO H S, et al.. Hyphopodium-specific VdNoxB/VdPls1-dependent ROS-Ca2+ signaling is required for plant infection by Verticillium dahliae [J/OL]. PLoS Pathog., 2016, 12(7): e1005793 [2021-09-10]. . |
| 22 | YANG C L, LIANG S, WANG H Y, et al.. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae [J]. Mol. Plant, 2015, 8(3): 399-411. |
| 23 | WANG G, WANG X, ZHANG Y, et al.. Dynamic characteristics and functional analysis provide new insights into long non-coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum [J/OL]. BMC Plant Biol., 2021, 21(1):8 [2021-09-10]. . |
| 24 | LIVAK K J, SCHIMTTGEN T D. Analysis of relative gene expression data using real-time quantitative and the 2-ΔΔ CT method [J]. Methods, 2001(25):492-408. |
| 25 | DONG O X, METEIGNIER L V, PLOURDE M B, et al.. Arabidopsis TAF15b localizes to RNA processing bodies and contributes to snc1-mediated autoimmunity [J]. Mol. Plant Microbe Interact., 2015, 29(4):247-257. |
| 26 | ROEPKE J, BOZZO G G. Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides [J]. Phytochemistry, 2015, 109(1): 14-24. |
| 27 | NAKAZAKI A, YAMADA K, KUNIEDA T, et al.. Biogenesis of leaf endoplasmic reticulum body is regulated by both jasmonate-dependent and independent pathways [J]. Plant Signal Behav., 2019, 14(8): 1-3. |
| 28 | XU Z Y, LEE K H, DONG T, et al.. A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis [J]. Plant Cell, 2012, 24(5): 2184-2199. |
| 29 | 柯丹霞,刘永辉,张静静,等.大豆BGLU基因家族全基因组鉴定与表达分析[J].信阳师范学院学报, 2019, 32(3): 1003-0972. |
| KE D X, LIU Y H, ZHANG J J, et al.. Genome-wide identification and expression analysis of BGLU family genes in soybean [J]. J. Xinyang Norm.Univ., 2019, 32(3): 1003-0972. | |
| 30 | OPASSIRI R, POMTHONG B, ONKOKSOONG T, et al.. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase [J/OL]. BMC Plant Biol., 2006, 6(1):33 [2021-09-10]. . |
| 31 | CHANG H C, TSAI M C, WU S S, et al.. Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana [J/OL]. Bot. Stud., 2019, 60(1): 16 [2021-09-10]. . |
| 32 | YANG J F, MA L, JIANG W B, et al.. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula [J]. Plant Physiol. Biochem., 2020, 158: 21-33. |
| 33 | GOMEZ A G, CENICEROS E A, CASADOS-V L E, et al.. Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73) [J]. Plant Mol. Biol., 2011, 77(1-2): 159-183. |
| 34 | KWAK G Y, GOO E, JEONG H, et al.. Adverse effects of adaptive mutation to survive static culture conditions on successful fitness of the rice pathogen Burkholderia glumae in a host [J/OL]. PLoS ONE, 2020, 15(8): e0238151 [2021-09-10]. . |
| 35 | CHAPELLE A, MORREEL K, VANHOLME R, et al.. Impact of the absence of stem-specific β-glucosidases on lignin and monolignols [J]. Plant Physiol., 2012, 160(3): 1204-1217. |
| 36 | ZHANG Z Q, LI Z M, STASWICKPAUL E, et al.. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction [J]. Plant Physiol., 2007, 145(2): 450-464. |
| 37 | ISHIHARA H, TOHGE T V, PRISCA F, et al.. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6 [J]. J. Exp. Bot. 2016, 67(5): 1505-1517. |
| 38 | HASS C, LOHRMANN J, ALBRECHT V, et al.. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis [J]. Embo. J., 2014, 23(16): 3290-3302. |
| 39 | BOLLER T. Antimicrobial functions of the plant hydrolases, chitinase and -1,3-glucanase [J]. Plant and Soil., 1984, 2: 391-400. |
| 40 | DUMAS G E, SLEZACK S, DASSI B, et al.. Plant hydrolytic enzymes (chitinases and beta-1, 3-glucanases) in root reactions to pathogenic and symbiotic microorganisms [J]. Plant Soil, 1996, 185(2): 211-221. |
| 41 | 王妮妍,蒋德安.茉莉酸及其甲酯与植物诱导抗病性[J].植物生理学报, 2002, 38(3): 279-284. |
| WANG N Y, JIANG D A. The role of jasmonic acid and methyl jasmonate in plant induced disease resistance [J]. Plant Physiol. Commun., 2002, 38(3): 279-284. |
| [1] | Meili LI, Junji SU, Yonglin YANG, Jianghong QIN, Xianxian LI, Delong YANG, Qi MA, Caixiang WANG. Identification of COI Family Genes and Their Expression in Gossypium hirsutum L. Under Drought and Salt Stress [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 63-74. |
| [2] | Shaojing MO, Zhicheng WANG, Xingfen WANG, Zhengwen LIU, Liqiang WU, Guiyin ZHANG, Zhiying MA, Yan ZHANG, Huijun DUAN. Genome-wide Identification of GELP Family Genes in Cotton and Expression Analysis Under Stress [J]. Journal of Agricultural Science and Technology, 2022, 24(2): 93-103. |
| [3] | SU Yue§, LIU Juanjuan§, WAN Bin, ZHANG Pengju, CHEN Zhenggen, SU Junji, WANG Caixiang. Chloroplast Genome Structure Characteristic and Phylogenetic Analysis of Mulgedium tataricum [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 33-42. |
| [4] | GE Chuan1, YANG Rong2, LI Liujun2, ZHANG Jiancheng2, ZHENG Xingwei2*. Genome-Wide Identification and Characterization of the YABBY Family Genes of Wheat (Trticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2019, 21(8): 11-18. |
| [5] | SONG Peiyong, ZHENG Yaqiang, LI Bin, XIAO Zhongjiu. Studies on Antimicrobial Activity of Actinomycetes Isolated from Soils of Chishui River Basin [J]. , 2013, 15(1): 136-143. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号