




















Journal of Agricultural Science and Technology ›› 2022, Vol. 24 ›› Issue (12): 78-89.DOI: 10.13304/j.nykjdb.2022.1030
• INNOVATIVE TECHNOLOGY • Previous Articles Next Articles
Lei YAN1( ), Jinshan ZHANG2, Jiankang ZHU1,3(
), Jinshan ZHANG2, Jiankang ZHU1,3( ), Lanqin XIA1(
), Lanqin XIA1( )
)
Received:2022-10-25
Accepted:2022-12-02
Online:2022-12-15
Published:2023-02-06
Contact:
Jiankang ZHU,Lanqin XIA
通讯作者:
朱健康,夏兰琴
作者简介:闫磊 E-mail:yanlei@caas.cn
CLC Number:
Lei YAN, Jinshan ZHANG, Jiankang ZHU, Lanqin XIA. Genome Editing and Its Applications in Crop Improvement[J]. Journal of Agricultural Science and Technology, 2022, 24(12): 78-89.
闫磊, 张金山, 朱健康, 夏兰琴. 基因编辑技术及其在农作物中的应用进展[J]. 中国农业科技导报, 2022, 24(12): 78-89.
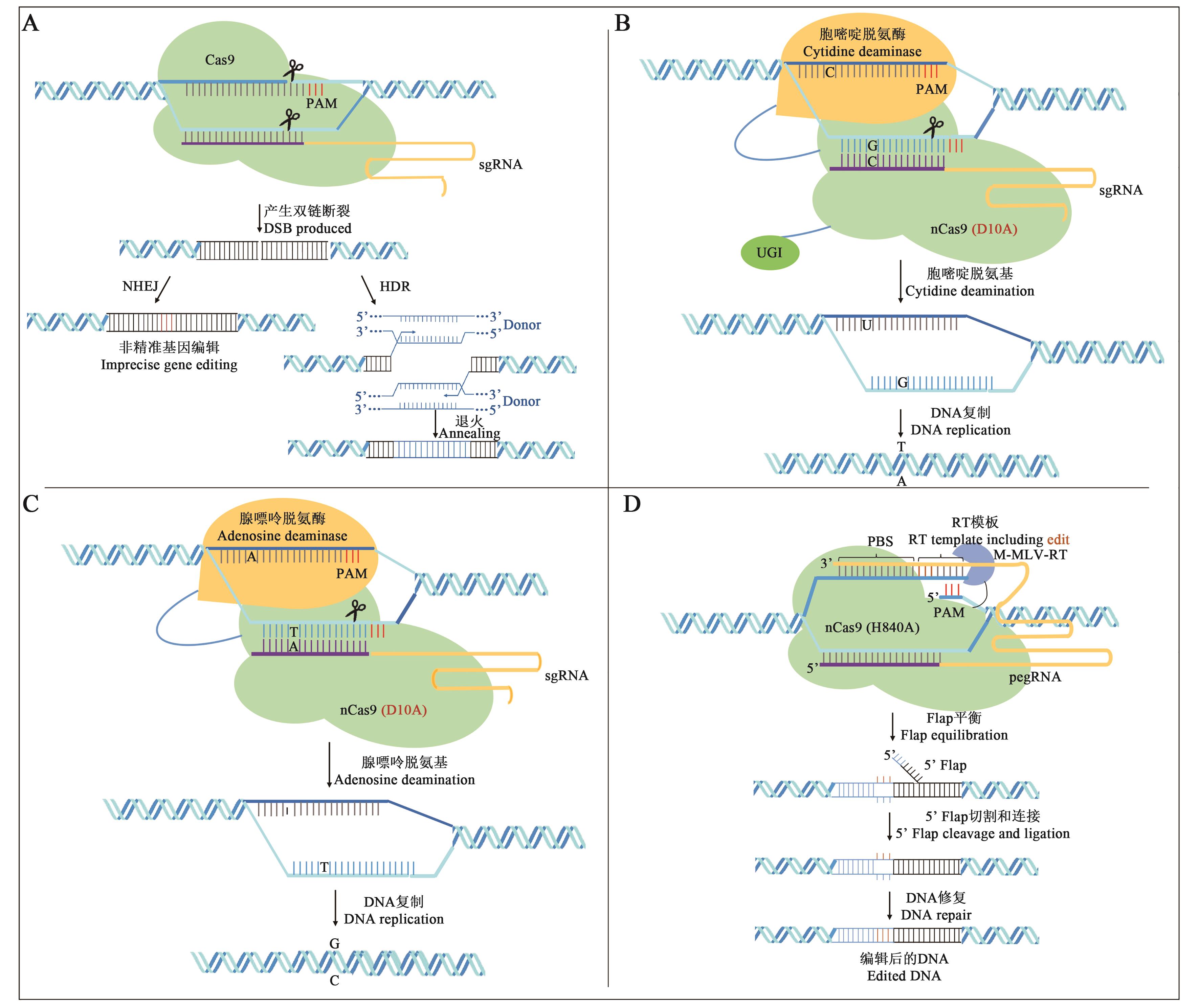
Fig. 1 CRISPR/CAS-mediated gene editing technology[12]A: NHEJ and HDR repairing pathway; B: CBE base editing system; C: ABE base editing system; D: Prime editing system
| 1 | BAILEY-SERRES J, PARKER J E, AINSWORTH E A, et al.. Schroeder genetic strategies for improving crop yields [J]. Nature, 2019, 575(3):109-118. |
| 2 | LI S, XIA L. Precise gene replacement in plants through CRISPR/Cas genome editing technology: current status and future perspectives [J]. aBIOTECH, 2020, 1(1):58-73. |
| 3 | VOYTAS DF, GAO C. Precision genome engineering and agriculture: opportunities and regulatory challenges [J/OL]. PLoS Biol., 2014, 12, e1001877 [2022-09-20]. . |
| 4 | SHAN Q, BALTES N J, ATKINS P, et al.. ZFN, TALEN and CRISPR-Cas9 mediated homology directed gene insertion in Arabidopsis: a disconnect between somatic and germinal cells [J]. J. Genet. Genomics, 2018, 45:681-684. |
| 5 | WOO J W, KIM J, KWON S I, et al.. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins [J]. Nat. Biotechnol., 2015, 33:1162-1164. |
| 6 | CHRISTIAN M, CERMAK T, DOYLE E L, et al.. Targeting DNA double-strand breaks with TAL effector nucleases [J]. Genetics, 2010, 186(2):757-761. |
| 7 | SUN Y, ZHANG X, WU C, et al.. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of Acetolactate synthase [J]. Mol. Plant, 2016, 9(4):628-631. |
| 8 | HUI S, LI H, MAWIA A M, et al.. Production of aromatic three-line hybrid rice using novel alleles of BADH2 [J]. Plant Biotechnol. J., 2022, 20(1):59-74. |
| 9 | ZHOU J, XIN X, HE Y, et al.. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties [J]. Plant Cell Reports, 2019, 38(4):475-485. |
| 10 | TIAN J, WANG C, XIA J, et al.. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields [J]. Science, 2019, 365(6454):658-664. |
| 11 | OLIVA R, JI C, ATIENZA-GRANDE G, et al.. Broad-spectrum resistance to bacterial blight in rice using genome editing [J]. Nat. Biotechnol., 2019, 37(11):1344-1350. |
| 12 | LI J, JIAO G, SUN Y, et al.. Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9 [J]. Plant Biotechnol. J., 2021, 19(5):937-951. |
| 13 | CONG L, RAN F A, COX D, et al.. Multiplex genome engineering using CRISPR/Cas systems [J]. Science, 2013, 339:819-823. |
| 14 | ZETSCHE B, GOOTENBERG-JONATHAN S, ABUDAYYEH O O, et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system [J]. Cell, 2015, 163:759-771. |
| 15 | HIOM K. Coping with DNA double strand breaks [J]. DNA Repair, 2010, 9:1256-1263. |
| 16 | PUCHTA H, FAUSER F. Synthetic nucleases for genome engineering in plants: prospects for a bright future [J]. Plant J., 2014, 78:727-741. |
| 17 | LI S, GAO F, XIE K, et al.. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice [J]. Plant Biotechnol. J., 2016, 14:2134-2146. |
| 18 | ZHENG X, YANG S, ZHANG D, et al.. Effective screen of CRISPR/Cas9-induced mutants in rice by single-strand conformation polymorphism [J]. Plant Cell Rep., 2016, 35(7):1545-1554. |
| 19 | LIU J, CHEN J, ZHENG X, et al.. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice [J/OL]. Nat. Plants, 2017, 3:17043 [2022-09-20]. . |
| 20 | LU K, WU B, WANG J, et al.. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice [J]. Plant Biotechnol. J., 2018, 16:1710-1722. |
| 21 | MIAO C, XIAO L, HUA K, et al.. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity [J]. Proceed. Nat. Academy Sci., 2018, 115:6058-6063. |
| 22 | ZHANG J, ZHOU Z, BAI J, et al.. Disruption of MIR396e and MIR396f improves rice yield under nitrogen-deficient conditions [J]. Nat. Sci. Review., 2020, 7(1):102-112. |
| 23 | ZHANG Y, LIANG Z, ZONG Y, et al.. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA [J/OL]. Nat. Comm., 2016, 7:12617 [2022-09-20]. . |
| 24 | ZHANG Y, LI D, ZHANG D, et al.. Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits [J]. Plant J., 2018, 94:857-866. |
| 25 | WANG W, SIMMONDS J, PAN Q, et al.. Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat [J]. Theor. Appl. Genet., 2018, 131(11):2463-2475. |
| 26 | XU R, YANG Y, QIN R, LI H, QIU C, et al.. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice [J]. J. Genet. Genomics, 2016, 43:529-532. |
| 27 | ZHANG J, ZHANG H, LI S, et al.. Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9 [J]. J. Int. Plant Biol., 2021, 63(9):1649-1663. |
| 28 | DONG L, QI X, ZHU J, et al.. Super sweet and waxy: meeting the diverse demands for specialty maize by genome editing [J]. Plant Biotechnol. J., 2019, 17(10):1853-1855. |
| 29 | SANCHEZ L S, GIL-HUMANES J, OZUNA CV, et al.. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9 [J]. Plant Biotechnol. J., 2018, 16:902-910. |
| 30 | WANG J, KUANG H, ZHANG Z, et al.. Generation of seed lipoxygenase-free soybean using crispr-cas9 [J]. Crop J., 2019, 8(3):432-439. |
| 31 | CHEN Y, FU M, LI H, et al.. High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system [J]. Plant Biotechnol. J., 2021, 19(3):424-426. |
| 32 | WANG Y, CHENG X, SHAN Q, et al.. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew [J]. Nat. Biotechnol., 2014, 32:947-951. |
| 33 | WANG N, TANG C, FAN X, et al.. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi [J]. Cell, 2022, 185(16):2961-2974. |
| 34 | PENG A, CHEN S, LEI T, et al.. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus [J]. Plant Biotechnol. J., 2017, 15(12):1509-1519. |
| 35 | HAO Z, TIAN J, FANG H, et al.. A VQ-motif-containing protein fine-tunes rice immunity and growth by a hierarchical regulatory mechanism [J/OL]. Cell Rep., 2022, 40(7):111235 [2022-09-20]. . |
| 36 | KELLIHER T, STARR D, RICHBOURG L, et al.. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction [J]. Nature. 2017, 542(7639):105-109. |
| 37 | LIU C, ZHONG Y, QI X, et al.. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat [J]. Plant Biotechnol. J., 2020, 18(2):316-318. |
| 38 | LI Y, LIN Z, YUE Y, et al.. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize [J]. Nat. Plants, 2021, 7(12):1579-1588. |
| 39 | ZHONG Y, LIU C, QI X, et al.. Mutation of ZmDMP enhances haploid induction in maize [J]. Nat. Plants, 2019, 5(6):575-580. |
| 40 | LYU J, YU K, WEI J, et al.. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3 [J]. Nat. Biotechnol., 2020, 38(12):1397-1401. |
| 41 | QI X, ZHANG C, ZHU J, et al.. Genome editing enables next-generation hybrid seed production technology [J]. Mol. Plant, 2020, 13(9):1262-1269. |
| 42 | KHANDAY I, SKINNER D, YANG B, et al.. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds [J]. Nature, 2018, 565(7737):91-95. |
| 43 | WANG C, LIU Q, SHEN Y, et al.. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes [J]. Nat. Biotechnol., 2019, 37(1):283-286. |
| 44 | LUO J, LI S, XU J, et al.. Pyramiding favorable alleles in an elite wheat variety in one generation by CRISPR-Cas9-mediated multiplex gene editing [J]. Mol. Plant, 2021, 14(6):847-850. |
| 45 | LI T, YANG X, YU Y, et al.. Domestication of wild tomato is accelerated by genome editing [J/OL]. Nat. Biotechnol., 2018, 10:4273 [2022-09-20]. . |
| 46 | YU H, LIN T, MENG X, et al.. A route to de novo domestication of wild allotetraploid rice [J]. Cell, 2021, 184(5):1156-1170. |
| 47 | KOMOR A C, KIM Y B, PACKER M S, et al.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage [J]. Nature. 2016, 533(7603):420-424. |
| 48 | GAUDELLI N M, KOMOR A C, REES H A, et al.. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage [J]. Nature, 2017, 551:464-471. |
| 49 | BHARAT S S, LI S, LI J . et al .. Base editing in plants: current status and challenges [J]. Crop J., 2020, 8(3):384-395. |
| 50 | HUA K, TAO X, YUAN F K, et al.. Precise A·T to G·C base editing in the rice genome [J]. Mol. Plant, 2018, 11:627-630. |
| 51 | LI C, ZONG Y, WANG Y, et al.. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion [J/OL]. Genome Biol., 2018, 19:59 [2022-09-20]. . |
| 52 | KURT, IC, ZHOU R, IYER S, et al.. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells [J]. Nat. Biotechnol., 2021, 39(1):41-46. |
| 53 | HESS G T, TYCKO J, YAO D, et al.. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes [J]. Mol. Cell., 2017, 68:26-43. |
| 54 | ZONG Y, WANG Y, LI C, et al.. Precise base editing in rice, wheat and maize with a Cas9-cytidined lieaminase fusion [J]. Nat. Biotechnol., 2017, 35(5):438-440. |
| 55 | KIM K, RYU S M, KIM S T, et al.. Highly efficient RNA-guided base editing in mouse embryos [J]. Nat. Biotechnol., 2017, 35(5):435-437. |
| 56 | NISHIDA K, ARAZOE T, YACHIE N, et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems [J/OL]. Science, 2016, 353(6305): aaf8729 [2022-09-20]. . |
| 57 | D’ADDA D I, FAGAGNA F, WELLER G R, et al.. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku [J]. EMBO Rep., 2003, 4(1):47-52. |
| 58 | HUA K, TAO X, HAN P, et al.. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences [J]. Mol. Plant, 2019, 12(7):1003-1014. |
| 59 | REN B, YAN F, KUANG Y, et al.. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant [J]. Mol. Plant, 2018, 11(4):623-626. |
| 60 | REN Q, SRETENOVIC S, LIU S, et al.. PAM-less plant genome editing using a CRISPR-SpRY toolbox [J]. Nat. Plants, 2021, 7(1):25-33. |
| 61 | WALTON R T, CHRISTIE K A, WHITTAKER M N, et al.. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants [J]. Science, 2020, 368(6488):290-296. |
| 62 | LI J, SUN Y, DU J, et al.. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system [J]. Mol. Plant, 2017, 10(3):526-529. |
| 63 | LU Y, ZHU J. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system [J]. Mol. Plant, 2017, 10(3):523-525. |
| 64 | REN B, YAN F, KUANG Y, et al.. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice [J]. Sci. China Life Sci., 2017, 60(5):516-519. |
| 65 | SHIMATANI Z, KASHOJIYA S, TAKAYAMA M, et al.. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion [J]. Nat. Biotechnol., 2017, 35(5):441-443. |
| 66 | TIAN S, JIANG L, CUI X, et al.. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing [J]. Plant Cell Rep., 2018, 37(9):1353-1356. |
| 67 | VEILLET F, PERROT L, CHAUVIN L, et al.. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor [J/OL]. Int. J. Mol. Sci., 2019, 20(2):402 [2022-09-20]. . |
| 68 | QIN L, LI J, WANG Q, et al.. High-efficient and precise base editing of C·G to T·A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system [J]. Plant Biotechnol. J., 2020, 18(1):45-56. |
| 69 | ZHANG R, LIU J, CHAI Z, et al.. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing [J]. Nat. Plants, 2019, 5(5):480-485. |
| 70 | KOBLAN L W, DOMAN J L, WILSON C, et al.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction [J]. Nat. Biotechnol., 2018, 36(9):843-846. |
| 71 | RICHTER M F, ZHAO K T, ETON E, et al.. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity [J]. Nat. Biotechnol., 2020, 38(7):883-891. |
| 72 | TAN J, ZENG D, ZHAO Y, et al.. PhieABEs: a PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants [J]. Plant Biotechnol. J., 2022, 20(5):934-943. |
| 73 | YAN D, REN B, LIU L, et al.. High-efficiency and multiplex adenine base editing in plants using new TadA variants [J]. Mol. Plant, 2021, 14(5):722-731. |
| 74 | YAN F, KUANG Y, REN B, et al.. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice [J]. Mol. Plant, 2018, 11(5):631-634. |
| 75 | KANG B C, YUN J Y, KIM S T, et al.. Precision genome engineering through adenine base editing in plants [J]. Nat. Plants, 2018, 11(4):427-431. |
| 76 | WEI C, WANG C, JIA M, et al.. Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor [J]. J. Int. Plant Biol., 2021, 63(9):1595-1599. |
| 77 | TIAN Y, SHEN R, LI Z, et al.. Efficient C-to-G editing in rice using an optimized base editor [J]. Plant Biotechnol. J., 2022, 20(7):1238-1240. |
| 78 | LI C, ZHANG R, MENG X, et al.. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors [J]. Nat. Biotechnol., 2020;38(7):875-882. |
| 79 | KUANG Y, LI S, REN B, et al.. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms [J]. Mol. Plant, 2020, 13(4):565-572. |
| 80 | WANG H, HE Y, WANG Y, et al.. Base editing-mediated targeted evolution of ACCase for herbicide-resistant rice mutants [J]. J. Int. Plant Biol., 2022, 64(11):2029-2032. |
| 81 | LUO M, GILBERT B, AYLIFFE M . et al .. Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants [J]. Plant Cell Rep., 2016, 35:1439-1450. |
| 82 | LI S, ZHANG Y, XIA L, et al.. CRISPR-Cas12a enables efficient biallelic gene targeting in rice [J]. Plant Biotechnol. J., 2020, 18:1351-1353. |
| 83 | LI S, LI J, HE Y, et al.. Precise gene replacement in rice by RNA transcript-templated homologous recombination [J]. Nat. Biotechnol., 2019, 37(1):445-450. |
| 84 | LI J, SUN Y, DU J, et al.. Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene [J]. J. Int. Plant Biol., 2018, 60(10):536-540. |
| 85 | LI Z, LIU Z B, XING A, et al.. Cas9-guide RNA directed genome editing in soybean [J]. Plant Physiol., 2015, 169(2):960-970. |
| 86 | SVITASHEV S, YOUNG J K, SCHWARTZ C, et al.. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA [J]. Plant Physiol., 2015, 169(2):931-945. |
| 87 | SAUER N J, NARVAEZ-VASQUEZ J, MOZORUK J, et al.. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants [J]. Plant Physiol., 2016, 170(4):1917-1928. |
| 88 | HUMMEL A W, CHAUHAN R D, CERMAK T, et al.. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava [J]. Plant Biotechnol. J., 2017, 16(3):1275-1282. |
| 89 | WANG M, LU Y, BOTELLA J R, et al.. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system [J]. Mol. Plant, 2017, 10(7):1007-1010. |
| 90 | SHI J, GAO H, WANG H, et al.. ARGOS8 variants generated by CRISPRCas9 improve maize grain yield under field drought stress conditions [J]. Plant Biotechnol. J., 2017, 15(3):207-216. |
| 91 | FILLER H S, MELAMED B C, LEVY A A. Targeted recombination between homologous chromosomes for precise breeding in tomato [J/OL]. Nat. Commun., 2017, 8:15605 [2022-09-20]. . |
| 92 | FILLER-HAYUT S, KNIAZEV K, MELAMED-BESSUDO C, et al.. Targeted inter-homologs recombination in Arabidopsis euchromatin and heterochromatin [J/OL]. Int. J. Mol. Sci., 2021, 22(22):12096 [2022-09-20]. . |
| 93 | GIL-HUMANES J, WANG Y, LIANG Z, et al.. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9 [J]. Plant J., 2017, 89(6):1251-1262. |
| 94 | VU T V, SIVANKALYANI V, KIM E J, et al.. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato [J]. Plant Biotechnol. J., 2020, 18(10):2133-2143. |
| 95 | HUANG T K, ARMSTRONG B, SCHINDELE P, et al.. Efficient gene targeting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a [J]. Plant Biotechnol. J., 2021, 19(7):1314-1324. |
| 96 | AIRD E J, LOVENDAHL K N, MARTIN A S T, et al.. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template [J/OL]. Comm. Biol., 2018, 1:54 [2022-09-20]. . |
| 97 | MIKI D, ZHANG W, ZENG W, et al.. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation [J/OL]. Nat. Comm.. 2018, 9:1967 [2022-09-20]. . |
| 98 | LU Y, TIAN Y, SHEN R, et al.. Targeted, efficient sequence insertion and replacement in rice [J]. Nat. Biotechnol., 2020, 38(12):1402-1407. |
| 99 | WILDE J J, AIDA T, DEL ROSARIO R C H, et al.. Efficient embryonic homozygous gene conversion via RAD51-enhanced interhomolog repair [J]. Cell, 2021, 184(12):3267-3280. |
| 100 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al.. Search-and-replace genome editing without double-strand breaks or donor DNA [J]. Nature, 2019, 576(7785):149-157. |
| 101 | LIN Q, ZONG Y, XUE C, et al.. Prime genome editing in rice and wheat [J]. Nat. Biotechnol., 2020, 38(2):582-585. |
| 102 | LI H Y, LI J Y, CHEN J L, et al.. Precise modifications of both exogenous and endogenous genes in rice by prime editing [J]. Mol. Plant, 2020, 13(5):671-674. |
| 103 | TANG X, SRETENOVIC S, REN Q R, et al.. Plant prime editors enable precise gene editing in rice cells [J]. Mol. Plant, 2020, 13(5):667-670. |
| 104 | XU W, ZHANG C W, YANG Y X, et al.. Versatile nucleotides substitution in plant using an improved prime editing system [J]. Mol. Plant, 2020, 13(5):675-678. |
| 105 | XU R F, LI J, LIU X H, et al.. Development of plant prime-editing systems for precise genome editing [J/OL]. Plant Comm., 2020, 1:100043 [2022-09-20]. . |
| 106 | HUA K, JIANG Y W, TAO X P, et al.. Precision genome engineering in rice using prime editing system [J]. Plant Biotechnol. J., 2020, 18:2167-2169. |
| 107 | JIANG Y Y, CHAI Y P, LU M H, et al.. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes of maize [J/OL]. Genome Biol., 2020, 21:257 [2022-09-20]. . |
| 108 | CHOW R D, CHEN J S, SHEN J, et al.. A web tool for the design of prime-editing guide RNAs [J/OL]. Nat. Biomed. Eng., 2020, 5:190 [2022-09-20]. . |
| 109 | KIM H K, YU G, PARK J, et al.. Predicting the efciency of prime editing guide RNAs in human cells [J]. Nat. Biotechnol., 2020, 39(1):198-206. |
| 110 | LIN Q, JIN S, ZONG Y, et al.. High-efficiency prime editing with optimized, paired pegRNAs in plants [J]. Nat. Biotechnol., 2021, 40(2):923-927. |
| 111 | XU W, YANG Y, YANG B, et al.. A design optimized prime editor with expanded scope and capability in plants [J]. Nat. Plants, 2022, 8(1):45-52. |
| 112 | ZOU J, MENG X, LIU Q, et al.. Improving the efficiency of prime editing with epegRNAs and high-temperature treatment in rice [J]. Sci. China Life Sci., 2022, 65(11):2328-2331. |
| 113 | LI H, ZHU Z, LI S, et al.. Multiplex precision gene editing by a surrogate prime editor in rice [J]. Mol. Plant, 2022, 15(7):1077-1080. |
| 114 | CARLSON-STEVERMER J, ABDEEN A A, KOHLENBERG L, et al.. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing [J/OL]. Nat. Comm., 2017, 8:1711 [2022-09-20]. . |
| 115 | WOLTER F, KLEMM J, PUCHTA H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus [J]. Plant J., 2018, 94(2):735-746. |
| 116 | HU J, LI S, LI Z, et al.. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize [J]. Mol. Plant Pathol., 2019, 20(10):1463-1474. |
| 117 | ELLISON E E, NAGALAKSHMI U, GAMO M E, et al.. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs [J]. Nat. Plants, 2020, 6(6):620-624. |
| 118 | MA X, ZHANG X, LIU H, et al.. Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9 [J]. Nat. Plants, 2020, 6(7):773-779. |
| 119 | LI T, HU J, SUN Y, et al.. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture [J]. Mol. Plant, 2021, 14(11):1787-1798. |
| 120 | STAAHL B T, BENEKAREDDY M, COULON-BAINIER C, et al.. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes [J]. Nat. Biotechnol., 2017, 35(5):431-434. |
| 121 | LEE K, CNOBOY M, PARK H M, et al.. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair [J]. Nat. Biomed. Eng., 2017, 1(11):889-901. |
| [1] | Chuang LU, Haitang HU, Yuan QIN, Heju HUAI, Cunjun LI. Delineating Management Zones in Spring Maize Field Based on UAV Multispectral Image [J]. Journal of Agricultural Science and Technology, 2022, 24(9): 106-115. |
| [2] | Gaitian ZHANG, Hui QI, Suoning YANG, Zhiyun CHU, Suxia YUAN, Chun LIU. Isolation and Application of Sepal Colored Protoplast in Hydrangea macrophylla [J]. Journal of Agricultural Science and Technology, 2022, 24(9): 50-57. |
| [3] | Wen XU, Hu LI, Shijiang ZHU, Weiping HE, Yu DU. Applicability of Different Reference Crop Evapotranspiration Calculation Models at Western Region of Hubei Province [J]. Journal of Agricultural Science and Technology, 2022, 24(7): 141-149. |
| [4] | Hai WANG, Jinsheng LAI, Haiyang WANG, Xinhai LI. Bipartite Intelligent Design of Crops—Intelligent Combination of Natural Variation and Intelligent Creation of Artificial Variation [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 1-8. |
| [5] | Li YANG, Li YU, Zhuo SUN, Tongyu ZHANG, Yang ZHANG, Limin YANG. Allelopathic Effects of Organic Acids and Saponins in Ginseng Root Exudates on Pathogenic and Biocontrol Bacteria [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 145-155. |
| [6] | Wei LI, Deli ZHU, Qing WANG, Shaohua ZENG. Research Review on Crop Digital Twin System for Monitoring Growth Status and Environmental Response [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 90-105. |
| [7] | Zunkang CUI, Danyang LI, Xiaoting XU, Junfeng ZHU, Laping WU, Wenge ZUO. Research on the Global Innovation Layout and Competition Situation of Food Crop Bio-breeding Technology: Based on the Perspective of Core Patent Data Mining [J]. Journal of Agricultural Science and Technology, 2022, 24(5): 1-14. |
| [8] | Qionghua LI, Lin ZHANG, Xinru HAN, Lili SONG. Temporal and Spatial Analysis of Total Factor Productivity of Double Cropping Rice in China and Its Countermeasures [J]. Journal of Agricultural Science and Technology, 2022, 24(5): 15-23. |
| [9] | Shanshan WEI, Xingfeng LI, Jing LUO. Proposal Application, Peer Review and Funding of the Basic Agriculture and Crop Science Discipline of National Natural Science Foundation in 2021 [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 1-10. |
| [10] | Xinhai LI, Ming LU, Jun ZHENG, Xiaofeng GU, Lijuan QIU, Yu LI, Youzhi MA, Jianmin WAN. Trends and Strategies for Development of Crop Seed Industry in China [J]. Journal of Agricultural Science and Technology, 2022, 24(12): 1-7. |
| [11] | Peng ZHU, Ke XU, Xitie LING, Jinyan WANG, Yuwen YANG, Baolong ZHANG, Shi QIU. Excavation and Utilization of Herbicide-resistant Rice and Wheat Germplasm Resources Under Rice-Wheat Rotation System [J]. Journal of Agricultural Science and Technology, 2022, 24(12): 142-152. |
| [12] | Beibei FAN, Jin LI, Chen MA. Digital Development of China’s Crop Industry: Achievements, Difficulties and Prospects [J]. Journal of Agricultural Science and Technology, 2022, 24(12): 25-32. |
| [13] | Xianwei SONG, Shanjie TANG, Xiaofeng CAO. Epigenetic Regulation and Crop Breeding [J]. Journal of Agricultural Science and Technology, 2022, 24(12): 33-38. |
| [14] | Jingkun ZHANG, Wenjia LI, Peng ZENG, Xiangbing MENG, Hong YU, Jiayang LI. Innovation and Progresses in de novo Domestication of Crops [J]. Journal of Agricultural Science and Technology, 2022, 24(12): 68-77. |
| [15] | Hua XIANG, Yan YAN, Xiudong WANG. Evaluation of Industrial Chain Integration Driven by Technology Integration of Characteristic Tropical Crops [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 1-12. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号