




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (8): 239-249.DOI: 10.13304/j.nykjdb.2025.0124
• INNOVATIVE METHODS AND TECHNOLOGIES • Previous Articles
Yanqin MA1,2( ), Yujie ZHOU1,2, Haicheng LONG1,2, Ju LI1,2, Haie WANG1,2, Wei CHANG3, Zhi LI1,2, Jian ZHONG1,2, Mingjun MIAO1,2, Liang YANG1,2(
), Yujie ZHOU1,2, Haicheng LONG1,2, Ju LI1,2, Haie WANG1,2, Wei CHANG3, Zhi LI1,2, Jian ZHONG1,2, Mingjun MIAO1,2, Liang YANG1,2( )
)
Received:2025-02-26
Accepted:2025-06-23
Online:2025-08-15
Published:2025-08-26
Contact:
Liang YANG
马燕勤1,2( ), 周玉洁1,2, 龙海成1,2, 李菊1,2, 王海娥1,2, 常伟3, 李志1,2, 钟建1,2, 苗明军1,2, 杨亮1,2(
), 周玉洁1,2, 龙海成1,2, 李菊1,2, 王海娥1,2, 常伟3, 李志1,2, 钟建1,2, 苗明军1,2, 杨亮1,2( )
)
通讯作者:
杨亮
作者简介:马燕勤 E-mail:dora0514@sina.cn;
基金资助:CLC Number:
Yanqin MA, Yujie ZHOU, Haicheng LONG, Ju LI, Haie WANG, Wei CHANG, Zhi LI, Jian ZHONG, Mingjun MIAO, Liang YANG. Construction of TRV-mediated VIGS System in Brassica rapa subsp. chinensis and Brassica juncea[J]. Journal of Agricultural Science and Technology, 2025, 27(8): 239-249.
马燕勤, 周玉洁, 龙海成, 李菊, 王海娥, 常伟, 李志, 钟建, 苗明军, 杨亮. TRV介导的上海青和芥菜VIGS体系的构建[J]. 中国农业科技导报, 2025, 27(8): 239-249.

Fig. 1 Structure of TRV-based VIGS vectorNote: Replicase—RNA-dependent RNA polymerase; 16 KD—16 kD cysteine-rich protein; MP—Move protein; CP—Coat protein; LB and RB—left and right border of T-DNA; Rz—Ribozyme; MCS—Multiple clone site.
引物名称 Primer name | 序列 Sequence (5’-3’) | 备注 Reference |
|---|---|---|
| BrPDS-F | ATGGTTGTGTTTGGGAATGT | 扩增BrPDS开放阅读框序列 |
| BrPDS-R | TCATGTTGATACAGTTGTCTC | Amplify BrPDS open reading frame |
| BjuPDS-g-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-g 开放阅读框序列 |
| BjuPDS-g-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BjuPDS-c-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-c 开放阅读框序列 |
| BjuPDS-c-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BrPDS-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BrPDS沉默表达载体 Construct TRV2-BrPDS vector |
| BrPDS-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-g-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BjuPDS-g沉默表达载体 Construct TRV2-BjuPDS-g vector |
| BjuPDS-g-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-c-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2- BjuPDS-c沉默表达载体 Construct TRV2-BjuPDS-c vector |
| BjuPDS-c-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| YL192-F | CTTGAAGAAGAAGACTTTCGAAGTCTC | 鉴定TRV1,约900 bp Identify TRV1, 900 bp |
| YL192-R | GTAAAATCATTGATAACAACACAGACAAAC | |
| YL156-F | GGTCAAGGTACGTAGTAGAG | 鉴定TRV2,约390 bp Identify TRV1, 390 bp |
| YL156-R | CGAGAATGTCAATCTCGTAGG | |
| BrPDS-F2 | CCTGATCGCGTGACTGATG | 内源BrPDS表达检测 Detect expression of BrPDS |
| BrPDS-R2 | TGTTCAACAATCGGCATGCA | |
| BrActin-F | GTCTCCATCTCCTGCTCATAGT | 上海青内参基因 |
| BrActin-R | GCTGACCGTATGAGCAAAGA | actin gene of B. rapa subsp. chinensis |
| BjuPDS-g-F2 | CTGATCGCGTGACTGATGAG | 内源BjuPDS-g表达检测 |
| BjuPDS-g-R2 | CCATGTTTCTCCTGAAGAAACC | Detect expression of BjuPDS-g |
| BjuPDS-c-F2 | TATAGCCATGTCAAAGGCGC | 内源BjuPDS-c表达检测 |
| BjuPDS-c-R2 | GCTCAATCTTCCTTATCCTTG | Detect expression of BjuPDS-c |
| BjuActin-R | GCTGACCGTATGAGCAAAGA | 芥菜内参基因 |
| BjuActin-R | GTTGGAAAGTGCTGAGGGAT | actin gene of B. juncea |
Table 1 Primers for vector construction
引物名称 Primer name | 序列 Sequence (5’-3’) | 备注 Reference |
|---|---|---|
| BrPDS-F | ATGGTTGTGTTTGGGAATGT | 扩增BrPDS开放阅读框序列 |
| BrPDS-R | TCATGTTGATACAGTTGTCTC | Amplify BrPDS open reading frame |
| BjuPDS-g-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-g 开放阅读框序列 |
| BjuPDS-g-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BjuPDS-c-F | ATGGTTGTGTTTGGGAATGT | 扩增BjuPDS-c 开放阅读框序列 |
| BjuPDS-c-R | TCATGTTGATACAGTTGTCTC | Amplify BjuPDS-g open reading frame |
| BrPDS-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BrPDS沉默表达载体 Construct TRV2-BrPDS vector |
| BrPDS-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-g-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2-BjuPDS-g沉默表达载体 Construct TRV2-BjuPDS-g vector |
| BjuPDS-g-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| BjuPDS-c-F1 | CTGTGAGTAAGGTTACCGAATTCACCTGATCGCGTGACTGATG | 构建pTRV2- BjuPDS-c沉默表达载体 Construct TRV2-BjuPDS-c vector |
| BjuPDS-c-R1 | GTGAGCTCGGTACCGGATCCAGCGTCTCCTTGGATAGTGG | |
| YL192-F | CTTGAAGAAGAAGACTTTCGAAGTCTC | 鉴定TRV1,约900 bp Identify TRV1, 900 bp |
| YL192-R | GTAAAATCATTGATAACAACACAGACAAAC | |
| YL156-F | GGTCAAGGTACGTAGTAGAG | 鉴定TRV2,约390 bp Identify TRV1, 390 bp |
| YL156-R | CGAGAATGTCAATCTCGTAGG | |
| BrPDS-F2 | CCTGATCGCGTGACTGATG | 内源BrPDS表达检测 Detect expression of BrPDS |
| BrPDS-R2 | TGTTCAACAATCGGCATGCA | |
| BrActin-F | GTCTCCATCTCCTGCTCATAGT | 上海青内参基因 |
| BrActin-R | GCTGACCGTATGAGCAAAGA | actin gene of B. rapa subsp. chinensis |
| BjuPDS-g-F2 | CTGATCGCGTGACTGATGAG | 内源BjuPDS-g表达检测 |
| BjuPDS-g-R2 | CCATGTTTCTCCTGAAGAAACC | Detect expression of BjuPDS-g |
| BjuPDS-c-F2 | TATAGCCATGTCAAAGGCGC | 内源BjuPDS-c表达检测 |
| BjuPDS-c-R2 | GCTCAATCTTCCTTATCCTTG | Detect expression of BjuPDS-c |
| BjuActin-R | GCTGACCGTATGAGCAAAGA | 芥菜内参基因 |
| BjuActin-R | GTTGGAAAGTGCTGAGGGAT | actin gene of B. juncea |
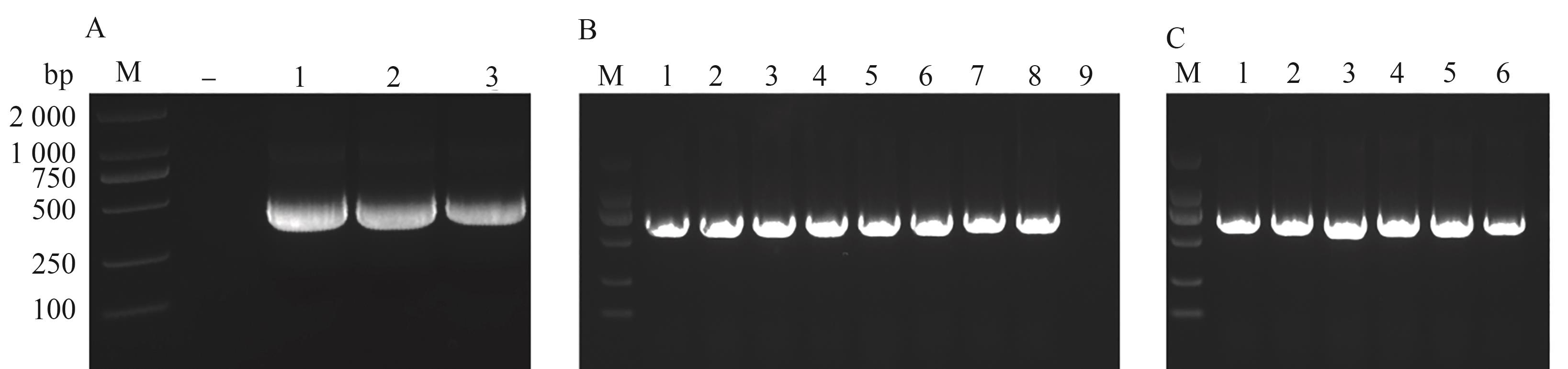
Fig. 3 Construction of VIGS vectorsA: Amplification fragments of Endogenous PDS gene, - indicates negative control; 1~3 are BrPDS, BjuPDS-g and BjuPDS-c gene fragments, respectively; B: PCR detection of recombinant vector-transformed Escherichia coli DH5α colonies, 1~3 are pTRV2-BrPDS DH5αcolonies PCR detection, respectively; 4~6 are pTRV2-BjuPDS-g DH5αcolonies PCR detection, respectively; 7~9 are pTRV2-BjuPDS-c DH5αcolonies PCR detection, respectively; C: PCR detection of recombinant vector-transformed Agrobacterium tumefaciens GV3101 strain colonies, 1~2 are pTRV2-BrPDS GV3101 colonies PCR detection, respectively; 3~4 are pTRV2-BjuPDS-g GV3101 colonies PCR detection, respectively; 5~6 are pTRV2-BjuPDS-c GV3101 colonies PCR detection. M—DL2000 DNA Marker

Fig. 4 Albinism rate of plant under different Agrobacterium levelsNote: Different lowercase letters indicate significant differences between different treatments at P<0.05 level.

Fig. 6 Leaf albinismof B. juncea plant with downregulation of PDSA: B. juncea var. gemmifera with OD600=0.3; B: B. juncea var. gemmifera with OD600=0.5; C: B. juncea var. gemmifera with OD600=0.8; D: B. juncea var. gemmifera with OD600=1.0;E: B. juncea var. capitate with OD600=0.3; F: B. juncea var. capitate with OD600=0.5; G: B. juncea var. capitate with OD600=0.8; H: B. juncea var. capitate with OD600=1.0. Scale bar =1 cm

Fig. 9 Analysis of TRV RNA expression in leafA: B. rapa subsp. Chinensis; B: B. juncea var. gemmifera; C: B. juncea var. capitata. TRV1 is the detection of TRV1 expression in leaves, TRV2 is the detection of TRV1 expression in leaves
| [1] | WATERHOUSE P M, WANG M B, LOUGH T. Gene silencing as an adaptive defence against viruses [J]. Nature, 2001, 411(6839): 834-842. |
| [2] | LIU Y, SUN W, ZENG S, et al.. Virus-induced gene silencing in two novel functional plants, Lycium barbarum L. and Lycium ruthenicum Murr [J]. Sci. Hortic., 2014, 170: 267-274. |
| [3] | ARCE-RODRÍGUEZ M L, OCHOA-ALEJO N. Virus-induced gene silencing (VIGS) in chili pepper (Capsicum spp.) [J]. Methods Mol. Biol., 2020, 2172: 27-38. |
| [4] | BACHAN S, DINESH-KUMAR S P. Tobacco rattle virus (TRV)-based virus-induced gene silencing [J]. Methods Mol. Biol., 2012, 894: 83-92. |
| [5] | RATCLIFF F, MARTIN-HERNANDEZ A M, BAULCOMBE D C. Technical advance. tobacco rattle virus as a vector for analysis of gene function by silencing [J]. Plant J., 2001, 25(2): 237-245. |
| [6] | SINGH A K, GHOSH D, CHAKRABORTY S. Optimization of tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) in tomato [J]. Methods Mol. Biol., 2022, 2408: 133-145. |
| [7] | GOULD B, KRAMER E M. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (Columbine, Ranunculaceae) [J/OL]. Plant Methods, 2007, 3: 6 [2025-01-20]. . |
| [8] | ZHANG J, WANG F, ZHANG C, et al.. A novel VIGS method by agroinoculation of cotton seeds and application for elucidating functions of GhBI-1 in salt-stress response [J]. Plant Cell Rep., 2018, 37(8): 1091-1100. |
| [9] | CAI C, WANG X, ZHANG B, et al.. Tobacco rattle virus-induced gene silencing in cotton [J]. Methods Mol. Biol., 2019, 1902: 105-119. |
| [10] | YAN H, SHI S, MA N, et al.. Graft-accelerated virus-induced gene silencing facilitates functional genomics in rose flowers [J]. J. Integr. Plant Biol., 2018, 60(1): 34-44. |
| [11] | CHEN R, CHEN X, HAGEL J M, et al.. Virus-induced gene silencing to investigate alkaloid biosynthesis in opium poppy [J]. Methods Mol. Biol., 2020, 2172: 75-92. |
| [12] | MA Y Q, LI Q, CHENG H, et al.. Alternative splicing variants of IiSEP3 in Isatis indigotica are involved in floral transition and flower development [J/OL]. Plant Physiol. Biochem., 2024, 216: 109153 [2025-01-20]. . |
| [13] | SHEN Z, SUN J, YAO J, et al.. High rates of virus-induced gene silencing by tobacco rattle virus in Populus [J]. Tree Physiol., 2015, 35(9): 1016-1029. |
| [14] | LI H L, GUO D, WANG Y, et al.. Tobacco rattle virus-induced gene silencing in Hevea brasiliensis Free [J]. Biosci. Biotechnol. Biochem., 2021, 85(3): 562-567. |
| [15] | KOUDOUNAS K, THOMOPOULOU M, ANGELI E, et al.. Virus-induced gene silencing in olive tree (Oleaceae) [J]. Methods Mol. Biol., 2020, 2172: 165-182. |
| [16] | ZHANG Y, NIU N, LI S, et al.. Virus-induced gene silencing (VIGS) in Chinese jujube [J/OL]. Plants (Basel), 2023, 12(11): 2115 [2025-01-20]. . |
| [17] | YU J, YANG X D, WANG Q, et al.. Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector [J]. Biol. Plant., 2018, 62(4): 826-834. |
| [18] | WANG L, WU Y, DU W, et al.. Virus-induced gene silencing (VIGS) analysis shows involvement of the LsSTPK gene in lettuce (Lactuca sativa L.) in high temperature-induced bolting [J/OL]. Plant Signal. Behav., 2021, 16(7): 1913845 [2025-01-20]. . |
| [19] | LI G, LI Y, YAO X, et al.. Establishment of a virus-induced gene-silencing (VIGS) system in tea plant and its use in the functional analysis of CsTCS1 [J/OL]. Int. J. Mol. Sci., 2022, 24(1): 392 [2025-01-20]. . |
| [20] | PARK H, KREUNEN S S, CUTTRISS A J, et al.. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis [J]. Plant Cell, 2002, 14(2): 321-332. |
| [21] | ISAACSON T, RONEN G, ZAMIR D, et al.. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants [J]. Plant Cell, 2002, 14(2): 333-342. |
| [22] | KANT R, DASGUPTA I. Phenotyping of VIGS-mediated gene silencing in rice using a vector derived from a DNA virus [J]. Plant Cell Rep., 2017, 36(7): 1159-1170. |
| [23] | ZHANG J, YU D, ZHANG Y, et al.. Vacuum and co-cultivation agroinfiltration of (germinated) seeds results in tobacco rattle virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize [J/OL]. Front. Plant Sci., 2017, 8: 393 [2025-01-20]. . |
| [24] | YAMAGISHI N, YOSHIKAWA N. Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with apple latent spherical virus vectors [J]. Plant Mol. Biol., 2009, 71(1-2): 15-24. |
| [25] | ROMERO I, TIKUNOV Y, BOVY A. Virus-induced gene silencing in detached tomatoes and biochemical effects of phytoene desaturase gene silencing [J]. J. Plant Physiol., 2011, 168(10): 1129-1135. |
| [26] | C-MRYU, ANAND A, KANG L, et al.. Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species [J]. Plant J., 2004, 40(2): 322-331. |
| [27] | SENTHIL-KUMAR M, MYSORE K S. Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato [J]. Plant Biotechnol. J., 2011, 9(7): 797-806. |
| [28] | BURCH-SMITH T M, ANDERSON J C, MARTIN G B, et al.. Applications and advantages of virus-induced gene silencing for gene function studies in plants [J]. Plant J., 2004, 39(5): 734-746. |
| [29] | BECKER A, LANGE M. VIGS: genomics goes functional [J]. Trends Plant Sci., 2010, 15(1): 1-4. |
| [30] | ZHANG F, WEN Y, GUO X. CRISPR/Cas9 for genome editing: progress, implications and challenges [J]. Huuman Mol. Genet., 2014, 23(r1): R40-R46. |
| [1] | Huiting WENG, Haiyang LIU, Huiming GUO, Hongmei CHENG, Jun LI, Chao ZHANG, Xiaofeng SU. Preliminary Function Analysis of GhERF020 Gene in Response to Verticillium Wilt in Cotton [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 112-121. |
| [2] | Man ZHANG, Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG. Cloning and Functional Analysis of GhNAC1 in Upland Cotton Involved in Verticillium Wilt Resistance [J]. Journal of Agricultural Science and Technology, 2023, 25(10): 35-44. |
| [3] | Mengyuan HAO, Qi HANG, Gongyao SHI. Application and Prospect of Virus-induced Gene Silencing in Crop Gene Function Research [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 1-13. |
| [4] | Qinqin WANG, Xiugui CHEN, Xuke LU, Shuai WANG, Yuexin ZHANG, Yapeng FAN, Quanjia CHEN, Wuwei YE. Bioinformatics Analysis and Functional Verification of GhPKE1 inUpland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 38-45. |
| [5] | XU Mengjun, GAO Tian, WANG Pengfei, LI Gezi, KANG Guozhang*. Function of Calcium Dependent Protein Kinase 34 in Grain Starch Synthesis of Wheat (Triticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2019, 21(2): 26-33. |
| [6] | YANG Xiaomin, RUI Cun, ZHANG Yuexin, WANG Delong, WANG Junjuan, LU Xuke, CHEN Xiugui, GUO Lixue, WANG Shuai, CHEN Chao, YE Wuwei*. Cloning and Stress Resistance Analysis of Cotton DNA Methyltransferase GhDMT9 Gene [J]. Journal of Agricultural Science and Technology, 2019, 21(10): 12-19. |
| [7] | JIN Qiaochun1, YU Fang2, YU Zongxia1*, FENG Baomin1*. Functional Characterization of PatPTS Gene Using Virus Induced Gene Silencing (VIGS) System in Patchouli (Pogostemon cablin) [J]. Journal of Agricultural Science and Technology, 2018, 20(3): 39-45. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号