




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (7): 43-53.DOI: 10.13304/j.nykjdb.2021.1090
收稿日期:2021-12-23
接受日期:2022-05-06
出版日期:2023-07-15
发布日期:2023-08-25
通讯作者:
蔡小艳
作者简介:贾晶莹 E-mail:jiajingying176@163.com;
基金资助:
Jingying JIA( ), Yahui LI, Bingzhe FU, Yun MA, Xiaoyan CAI(
), Yahui LI, Bingzhe FU, Yun MA, Xiaoyan CAI( )
)
Received:2021-12-23
Accepted:2022-05-06
Online:2023-07-15
Published:2023-08-25
Contact:
Xiaoyan CAI
摘要:
为探明宁夏苜蓿耐旱新品系‘新盐52号’(XY52)的miRNA表达谱,并筛选有跨界研究价值的microRNA(miR),以‘中苜一号’(ZM1)为参照,对ZM1和XY52进行高通量miRs组学测序和分析(RNA-seq);通过生物信息学技术筛选差异表达miRs及预测靶基因,并对预测靶基因进行GO和KEGG富集分析;然后运用RT-qPCR技术对10个miRs进行检测和分析。结果表明,①成功构建了2种苜蓿miR表达谱,ZM1和XY52中分别检测到656和703个miRs,其中已知miRs分别为433和480个,新预测miRs数量均为233个;②2种苜蓿中共检测到21个差异表达miRs,其中novel-miR54、miR156f和miR166a表达量较高,并且novel-miR54在XY52中表达量极显著高于ZM1(P<0.01),miR156f在ZM1中的表达量极显著高于XY52(P<0.01);③5个已知有跨界调控功能miRs中,miR166a在ZM1和XY52中均为表达水平最高的miR;品种之间,miR166a在XY52中的表达量较ZM1极显著上调(P<0.01);④KEGG通路和GO功能富集分析发现差异表达的21个miRs共预测得到623个靶基因,主要与RNA转运、ABC转运蛋白和泛素介导的蛋白水解等信号通路有密切关系。以上结果为解析苜蓿品种miRs差异表达及研究苜蓿源miRs调控奶牛体内基因奠定初步基础。
中图分类号:
贾晶莹, 李雅辉, 伏兵哲, 马云, 蔡小艳. 苜蓿miRs表达谱分析及跨界潜力miRs初步筛选[J]. 中国农业科技导报, 2023, 25(7): 43-53.
Jingying JIA, Yahui LI, Bingzhe FU, Yun MA, Xiaoyan CAI. Analysis on miRs Expression Profiles of Alfalfa and Screening of Trans-border Potential miRs[J]. Journal of Agricultural Science and Technology, 2023, 25(7): 43-53.
miRs名称 miRs name | 引物序列 Primer sequence (5’-3’) |
|---|---|
| 5s | F:GCAGACGAAGTCCTTGTGTTG R:AGTGCAGGGTCCGAGGTATT |
| miR156b-5p | F:CGCAGACAGTTGACAGAAGAGA |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGCTCAC | |
| miR166a | F:CACAGTTCGGACCAGGCTT |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGGAATG | |
| miR167a | F:AAGCTTTGAAGCTGCCAGC |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAGATCAT | |
| miR168a | F:CATGTGTCGCTTGGTGCAG |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCCCGAC | |
| miR319a-3p | F:CGAACGTTTTGGACTGAAGG |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGAGCTC |
表1 引物信息
Table 1 Primer information
miRs名称 miRs name | 引物序列 Primer sequence (5’-3’) |
|---|---|
| 5s | F:GCAGACGAAGTCCTTGTGTTG R:AGTGCAGGGTCCGAGGTATT |
| miR156b-5p | F:CGCAGACAGTTGACAGAAGAGA |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGCTCAC | |
| miR166a | F:CACAGTTCGGACCAGGCTT |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGGAATG | |
| miR167a | F:AAGCTTTGAAGCTGCCAGC |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAGATCAT | |
| miR168a | F:CATGTGTCGCTTGGTGCAG |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCCCGAC | |
| miR319a-3p | F:CGAACGTTTTGGACTGAAGG |
| R:AGTGCAGGGTCCGAGGTATT | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGGAGCTC |
| 组分Component | 体积Volume/μL |
|---|---|
预混液 TB Green Premix Ex Taq Ⅱ (Tli RNaseH Plus) (2×) | 10.0 |
PCR上游引物 PCR forward primer (10 μmol·L-1) | 0.4 |
PCR下游引物 PCR reverse primer (10 μmol·L-1) | 0.4 |
| DNA (100 ng·μL-1) | 2.0 |
| 灭菌水dd H2O | 7.2 |
| 总计Total | 20.0 |
表2 荧光定量反应体系
Table 2 Fluorescence quantitative reaction system
| 组分Component | 体积Volume/μL |
|---|---|
预混液 TB Green Premix Ex Taq Ⅱ (Tli RNaseH Plus) (2×) | 10.0 |
PCR上游引物 PCR forward primer (10 μmol·L-1) | 0.4 |
PCR下游引物 PCR reverse primer (10 μmol·L-1) | 0.4 |
| DNA (100 ng·μL-1) | 2.0 |
| 灭菌水dd H2O | 7.2 |
| 总计Total | 20.0 |
miRs名称 miRs name | 引物序列 Primer sequence (5’-3’) |
|---|---|
| miR5754 | F:TATTGCACTCATCTTCCATGGC |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCATG | |
| miR156f | F:CCGTTGACAGAAGATAGAGAGCAC |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGCTC | |
| miR5743a | F:TGAGAACTGTTTTCCGCACCTT |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGGTG | |
| Novel-miR54 | F:CCAAGTCCTTGTGTTGCATCTC |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGATG | |
| Novel-miR158 | F:GCGCAAAGGATCATTGGATAAGTTc |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAACTT |
表1 引物信息 (续表Continued)
Table 1 Primer information
miRs名称 miRs name | 引物序列 Primer sequence (5’-3’) |
|---|---|
| miR5754 | F:TATTGCACTCATCTTCCATGGC |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCATG | |
| miR156f | F:CCGTTGACAGAAGATAGAGAGCAC |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGCTC | |
| miR5743a | F:TGAGAACTGTTTTCCGCACCTT |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGGTG | |
| Novel-miR54 | F:CCAAGTCCTTGTGTTGCATCTC |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGATG | |
| Novel-miR158 | F:GCGCAAAGGATCATTGGATAAGTTc |
| R:ATCCAGTGCAGGGTCCGAGG | |
| RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAACTT |
| 品种Variety | miRs数量Number of miRs | 优势表达miRNA(miRs TPM占总TPM百分比) Advantage expressed known miRs (miRs TPM/all miRs TPM) | |
|---|---|---|---|
| 已知Known | 新预测Novel | ||
中苜1号 Zhongmu 1 | 433 | 223 | mtr-miR5213-5p(25.82%), mtr-miR159a(22.40%), mtr-miR396a-5p(11.72%), mtr-miR166e-3p(5.22%) |
新盐52号 Xinyan 52 | 480 | 223 | mtr-miR159a(27.27%), mtr-miR5213-5p(20.07%), mtr-miR396a-5p(14.19%), mtr-miR166e-3p(3.89%) |
表3 中苜1号和新盐52号中优势表达的miRs
Table 3 Advantage expression miRs in Zhongmu 1 andXinyan 52
| 品种Variety | miRs数量Number of miRs | 优势表达miRNA(miRs TPM占总TPM百分比) Advantage expressed known miRs (miRs TPM/all miRs TPM) | |
|---|---|---|---|
| 已知Known | 新预测Novel | ||
中苜1号 Zhongmu 1 | 433 | 223 | mtr-miR5213-5p(25.82%), mtr-miR159a(22.40%), mtr-miR396a-5p(11.72%), mtr-miR166e-3p(5.22%) |
新盐52号 Xinyan 52 | 480 | 223 | mtr-miR159a(27.27%), mtr-miR5213-5p(20.07%), mtr-miR396a-5p(14.19%), mtr-miR166e-3p(3.89%) |
| miRs名称miRs name | 序列Sequence(5’-3’) | TPM值TPM value | |
|---|---|---|---|
| 中苜1号Zhongmu1 | 新盐52号Xinyan52 | ||
| miR5743a | TGAGAACTGTTTTCCGCACCTT | 6 019.360 539 | 61.234 256 |
| miR5743b | TGAGAACTGTTTTCCGCACCTT | 6 019.360 539 | 61.234 256 |
| novel-miR158 | AAAGGAUCAUUGGAUAAGUUC | 1 034.191 978 | 57.554 978 |
| miR5754 | TATTGCACTCATCTTCCATGGC | 904.998 534 | 6.803 806 |
| novel-miR54 | AAGUCCUUGUGUUGCAUCUC | 645.517 630 | 1 877.094 533 |
| miR156f | TTGACAGAAGATAGAGAGCAC | 163.659 584 | 10.655 880 |
| miR156e | TTGACAGAAGATAGAGAGCAC | 163.659 584 | 10.655 880 |
| miR156h-5p | TTGACAGAAGATAGAGAGCAC | 163.659 584 | 10.655 880 |
表4 差异表达miRs序列
Table 4 miRs sequence of differentially expressed
| miRs名称miRs name | 序列Sequence(5’-3’) | TPM值TPM value | |
|---|---|---|---|
| 中苜1号Zhongmu1 | 新盐52号Xinyan52 | ||
| miR5743a | TGAGAACTGTTTTCCGCACCTT | 6 019.360 539 | 61.234 256 |
| miR5743b | TGAGAACTGTTTTCCGCACCTT | 6 019.360 539 | 61.234 256 |
| novel-miR158 | AAAGGAUCAUUGGAUAAGUUC | 1 034.191 978 | 57.554 978 |
| miR5754 | TATTGCACTCATCTTCCATGGC | 904.998 534 | 6.803 806 |
| novel-miR54 | AAGUCCUUGUGUUGCAUCUC | 645.517 630 | 1 877.094 533 |
| miR156f | TTGACAGAAGATAGAGAGCAC | 163.659 584 | 10.655 880 |
| miR156e | TTGACAGAAGATAGAGAGCAC | 163.659 584 | 10.655 880 |
| miR156h-5p | TTGACAGAAGATAGAGAGCAC | 163.659 584 | 10.655 880 |
| GO编码 GO ID | 富集层面Enrichment level | 功能Function | 差异表达miR靶基因富集到该通路的数量(百分比/%) Number of differential expressed miR target genes enriched into pathway (Percentage/%) | 全部miR靶基因富集到该通路的数量(百分比/%) Number of all miR target genes enriched into the pathway (Percentage/%) |
|---|---|---|---|---|
| GO:0006468 | 生物过程 Biological process | 蛋白质磷酸化Protein phosphorylation | 36(7.16) | 467(6.88) |
| GO:0055114 | 氧化还原过程Oxidation-reduction process | 29(5.77) | 440(6.49) | |
| GO:0006952 | 防御反应Defense response | 19(3.78) | 683(10.07) | |
| GO:0006508 | 蛋白水解作用Proteolysis | 18(3.58) | 169(2.49) | |
| GO:0006355 | 转录调控,DNA模板化 Regulation of transcription, DNA-templated | 17(3.38) | 360(5.31) | |
| GO:0005975 | 碳水化合物代谢过程 Carbohydrate metabolic process | 15(2.98) | 116(1.71) | |
| GO:0055085 | 跨膜运输Transmembrane transport | 13(2.58) | 126(1.86) |
表5 miRs靶基因GO功能富集分析
Table 5 GO functional enrichment analysis of miRs target genes
| GO编码 GO ID | 富集层面Enrichment level | 功能Function | 差异表达miR靶基因富集到该通路的数量(百分比/%) Number of differential expressed miR target genes enriched into pathway (Percentage/%) | 全部miR靶基因富集到该通路的数量(百分比/%) Number of all miR target genes enriched into the pathway (Percentage/%) |
|---|---|---|---|---|
| GO:0006468 | 生物过程 Biological process | 蛋白质磷酸化Protein phosphorylation | 36(7.16) | 467(6.88) |
| GO:0055114 | 氧化还原过程Oxidation-reduction process | 29(5.77) | 440(6.49) | |
| GO:0006952 | 防御反应Defense response | 19(3.78) | 683(10.07) | |
| GO:0006508 | 蛋白水解作用Proteolysis | 18(3.58) | 169(2.49) | |
| GO:0006355 | 转录调控,DNA模板化 Regulation of transcription, DNA-templated | 17(3.38) | 360(5.31) | |
| GO:0005975 | 碳水化合物代谢过程 Carbohydrate metabolic process | 15(2.98) | 116(1.71) | |
| GO:0055085 | 跨膜运输Transmembrane transport | 13(2.58) | 126(1.86) |
| GO编码 GO ID | 富集层面Enrichment level | 功能Function | 差异表达miR靶基因富集到该通路的数量(百分比/%) Number of differential expressed miR target genes enriched into pathway (Percentage/%) | 全部miR靶基因富集到该通路的数量(百分比/%) Number of all miR target genes enriched into the pathway (Percentage/%) |
|---|---|---|---|---|
| GO:0043161 | 蛋白酶体介导的泛素依赖性蛋白分解代谢过程Proteasome-mediated ubiquitin-dependent protein catabolic process | 12(2.39) | 26(0.38) | |
| GO:0016021 | 细胞组分 Cellular component | 膜的组成部分 Integral component of membrane | 160(31.81) | 2 454(36.18) |
| GO:0005634 | 核Nucleus | 85(16.90) | 591(8.71) | |
| GO:0005886 | 质膜Plasma membrane | 24(4.77) | 294(4.33) | |
| GO:0009506 | 胞间连丝Plasmodesma | 16(3.18) | 122(1.80) | |
| GO:0005829 | 胞浆Cytosol | 13(2.58) | 159(2.34) | |
| GO:0005622 | 胞内Intracellular | 12(2.39) | 102(1.50) | |
| GO:0005737 | 细胞质Cytoplasm | 12(2.39) | 255(3.76) | |
| GO:0005524 | 分子功能Molecular function | ATP结合ATP binding | 97(19.28) | 1 227(18.09) |
| GO:0003677 | DNA结合DNA binding | 84(16.70) | 502(7.40) | |
| GO:0008270 | 锌离子结合Zinc ion binding | 34(6.76) | 317(4.67) | |
| GO:0004674 | 蛋白质丝氨酸/苏氨酸激酶活性 Protein serine/threonine kinase activity | 34(6.76) | 394(5.81) | |
| GO:0043531 | ADP结合ADP binding | 14(2.78) | 658(9.70) | |
| GO:0046872 | 金属离子结合Metal ion binding | 14(2.78) | 267(3.94) | |
| GO:0016887 | ATP酶活性ATPase activity | 11(2.19) | 44(0.65) |
表5 miRs靶基因GO功能富集分析 (续表Continued)
Table 5 GO functional enrichment analysis of miRs target genes
| GO编码 GO ID | 富集层面Enrichment level | 功能Function | 差异表达miR靶基因富集到该通路的数量(百分比/%) Number of differential expressed miR target genes enriched into pathway (Percentage/%) | 全部miR靶基因富集到该通路的数量(百分比/%) Number of all miR target genes enriched into the pathway (Percentage/%) |
|---|---|---|---|---|
| GO:0043161 | 蛋白酶体介导的泛素依赖性蛋白分解代谢过程Proteasome-mediated ubiquitin-dependent protein catabolic process | 12(2.39) | 26(0.38) | |
| GO:0016021 | 细胞组分 Cellular component | 膜的组成部分 Integral component of membrane | 160(31.81) | 2 454(36.18) |
| GO:0005634 | 核Nucleus | 85(16.90) | 591(8.71) | |
| GO:0005886 | 质膜Plasma membrane | 24(4.77) | 294(4.33) | |
| GO:0009506 | 胞间连丝Plasmodesma | 16(3.18) | 122(1.80) | |
| GO:0005829 | 胞浆Cytosol | 13(2.58) | 159(2.34) | |
| GO:0005622 | 胞内Intracellular | 12(2.39) | 102(1.50) | |
| GO:0005737 | 细胞质Cytoplasm | 12(2.39) | 255(3.76) | |
| GO:0005524 | 分子功能Molecular function | ATP结合ATP binding | 97(19.28) | 1 227(18.09) |
| GO:0003677 | DNA结合DNA binding | 84(16.70) | 502(7.40) | |
| GO:0008270 | 锌离子结合Zinc ion binding | 34(6.76) | 317(4.67) | |
| GO:0004674 | 蛋白质丝氨酸/苏氨酸激酶活性 Protein serine/threonine kinase activity | 34(6.76) | 394(5.81) | |
| GO:0043531 | ADP结合ADP binding | 14(2.78) | 658(9.70) | |
| GO:0046872 | 金属离子结合Metal ion binding | 14(2.78) | 267(3.94) | |
| GO:0016887 | ATP酶活性ATPase activity | 11(2.19) | 44(0.65) |
富集通路 Enrichment Pathway | 富集通路类型 Enrichment pathway type | 富集基因数量(百分比/%) Enrichment gene number (Percentage/%) |
|---|---|---|
| 内吞作用Endocytosis | 细胞过程Cellular processes | 6(6.74) |
| ABC转运蛋白ABC transporters | 环境信息处理 Environmental information processing | 5(5.62) |
植物激素信号转运体 Plant hormone signal transporters | 3(3.37) | |
| RNA转运RNA Transport | 遗传信息处理 Genetic information processing | 6(6.74) |
泛素介导的蛋白质水解作用 Ubiquitin mediated proteolysis | 5(5.62) | |
| 蛋白酶体Proteasome | 4(4.49) | |
| RNA聚合酶RNA polymerase | 4(4.49) | |
| 核糖体Ribosome | 3(3.37) | |
氨基酸的生物合成 Biosynthesis of amino acids | 新陈代谢 Metabolism | 4(4.49) |
氨基糖和核苷酸糖的代谢 Amino sugar and nucleotide sugar metabolism | 4(4.49) | |
淀粉和蔗糖代谢 Starch and sucrose metabolism | 4(4.49) | |
GPI锚生物合成 Glycosyl phosphatidylinositol(GPI)-anchor biosynthesis | 3(3.37) | |
谷胱甘肽代谢 Glutathione metabolism | 3(3.37) | |
植物-病原互作 Plant-pathogen interaction | 有机系统 Organismal system | 3(3.37) |
表6 差异表达miRs靶基因KEGG通路富集分析
Table 6 Target gene KEGG pathway enrichment of differential expressed miRs
富集通路 Enrichment Pathway | 富集通路类型 Enrichment pathway type | 富集基因数量(百分比/%) Enrichment gene number (Percentage/%) |
|---|---|---|
| 内吞作用Endocytosis | 细胞过程Cellular processes | 6(6.74) |
| ABC转运蛋白ABC transporters | 环境信息处理 Environmental information processing | 5(5.62) |
植物激素信号转运体 Plant hormone signal transporters | 3(3.37) | |
| RNA转运RNA Transport | 遗传信息处理 Genetic information processing | 6(6.74) |
泛素介导的蛋白质水解作用 Ubiquitin mediated proteolysis | 5(5.62) | |
| 蛋白酶体Proteasome | 4(4.49) | |
| RNA聚合酶RNA polymerase | 4(4.49) | |
| 核糖体Ribosome | 3(3.37) | |
氨基酸的生物合成 Biosynthesis of amino acids | 新陈代谢 Metabolism | 4(4.49) |
氨基糖和核苷酸糖的代谢 Amino sugar and nucleotide sugar metabolism | 4(4.49) | |
淀粉和蔗糖代谢 Starch and sucrose metabolism | 4(4.49) | |
GPI锚生物合成 Glycosyl phosphatidylinositol(GPI)-anchor biosynthesis | 3(3.37) | |
谷胱甘肽代谢 Glutathione metabolism | 3(3.37) | |
植物-病原互作 Plant-pathogen interaction | 有机系统 Organismal system | 3(3.37) |
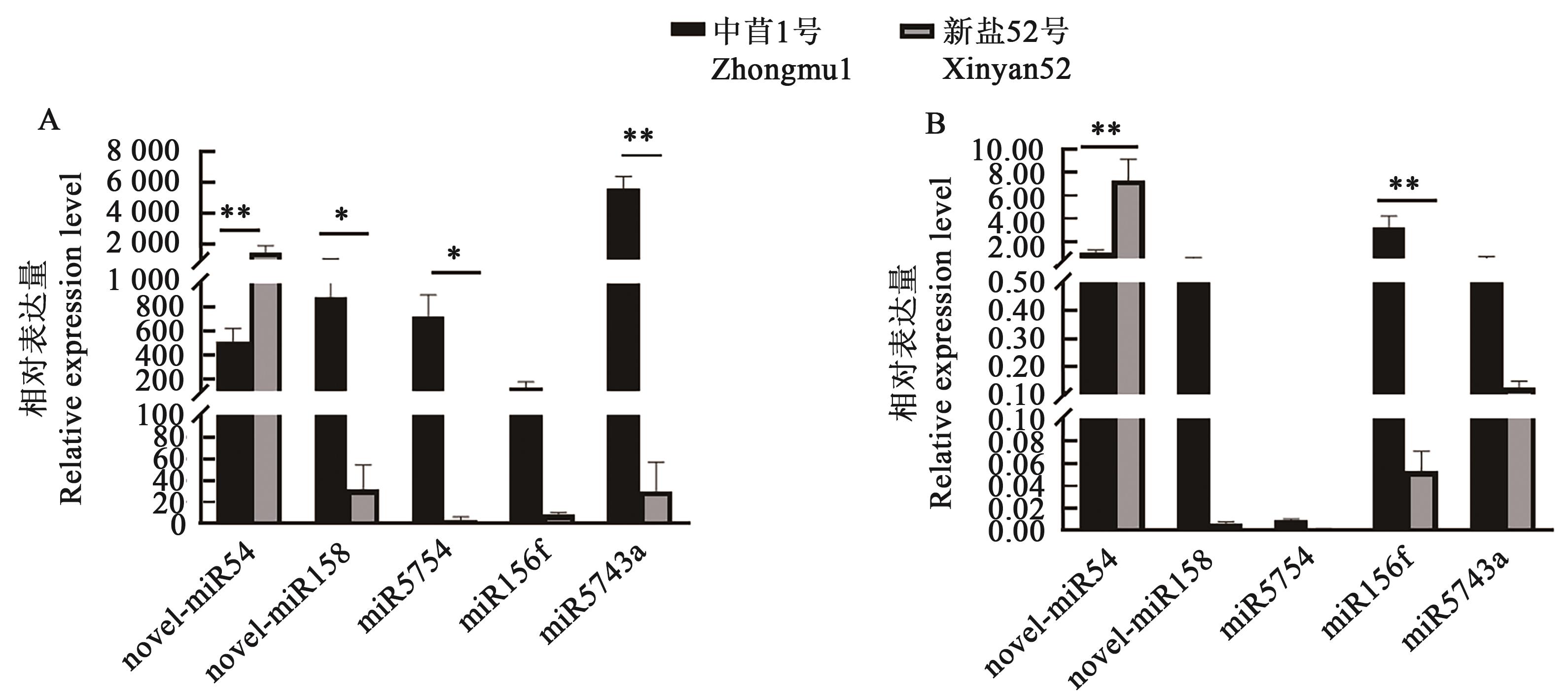
图4 差异表达miRs的相对表达量A:基于RNA-Seq;B:基于RT-qPCR;*和**分别表示不同品种间在P<0.05和P<0.01水平差异显著
Fig. 4 Relative expression level of differentially expressed miRsA: Base on RNA-Seq; B: Bsae on RT-qPCR;* and ** indicate signifiant differences between different varieties at P<0.05 and P<0.01 levels, respectively
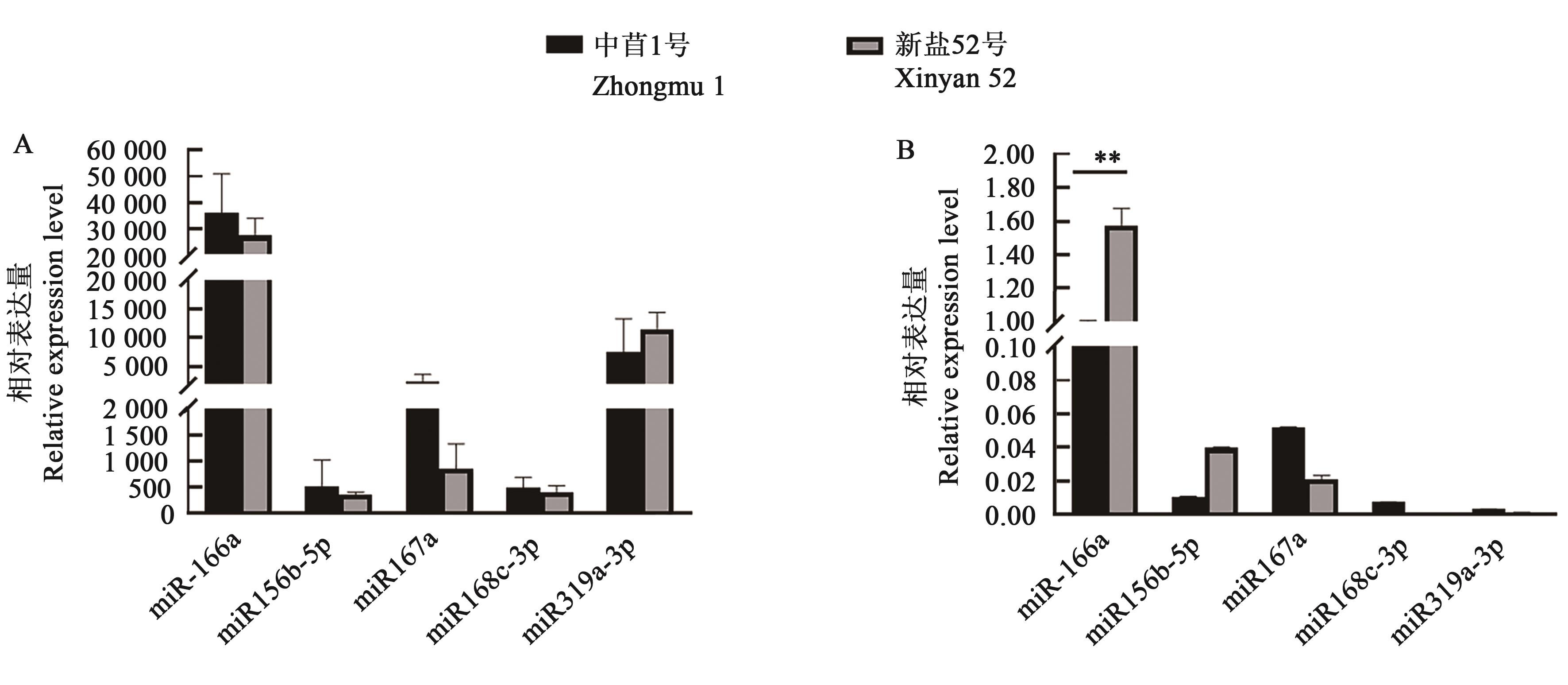
图5 跨界调控miRs的相对表达量A:基于RNA-Seq;B:基于RT-qPCR;**表示不同品种间在P<0.01水平差异显著
Fig. 5 Relative expression level of cross-kingdom regulated miRsA: Base on RNA-Seq; B: Bsae on RT-qPCR; ** indicates significant differences between different varieties at P<0.01 level
| 1 | AMBROS V, BARTEL B, BARTEL D P, et al.. A uniform system for microRNA annotation [J] . RNA (Cambridge), 2003, 9(3):277-279. |
| 2 | CUI J, YOU C, CHEN X. The evolution of microRNAs in plants [J]. Curr. Opin. Plant Biol., 2017, 35(1):61-67. |
| 3 | FILIPOWICA W, BHATTACHARYYA S N, SOENBERG N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? [J]. Nat. Rev. Genet., 2008, 9(2):102-114. |
| 4 | ZHANG L, HOU D, CHEN X, et al.. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA [J]. Cell Res., 2012, 22(1):107-126. |
| 5 | CHIN A R, FONG M Y, SOMLO G, et al..Cross-kingdom inhibition of breast cancer growth by plant miR159 [J]. Cell Res., 2016, 26(2):217-228. |
| 6 | ZHOU L K, ZHOU Z, JIANG X M, et al.. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients [J/OL]. Cell Discov., 2020, 6:54 [2021-11-10]. . |
| 7 | ZHOU Z, LI X, LIU J, et al.. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses [J]. Cell Res., 2015, 25(1):39-49. |
| 8 | LUO Y, WANG P, WANG X, et al.. Detection of dietetically absorbed maize-derived microRNAs in pigs [J/OL]. Sci. Rep.Uk., 2017, 7(1):645 [2021-11-10]. . |
| 9 | MARZANO F, CARATOZZOLO M F, CONSIGLIO A, et al.. Plant miRNAs reduce cancer cell proliferation by targeting MALAT1 and NEAT1: a beneficial cross-Kingdom interaction [J/OL]. Front. Genet., 2020, 11:552490 [2021-11-10]. . |
| 10 | DAVALOS A, PINILLA L, LOPEZ D L H M C, et al.. Dietary microRNAs and cancer: a new therapeutic approach? [J]. Seminars Cancer Biol., 2021, 73(1):19-29. |
| 11 | WANG D, LIANG G, WANG B, et al.. Systematic microRNAome profiling reveals the roles of microRNAs in milk protein metabolism and quality: insights on low-quality forage utilization [J/OL]. Sci. Rep., 2016,6: 21194 [2021-11-10]. . |
| 12 | 李跃.紫花苜蓿对干旱的响应:形态、生理以及miRNA组学研究[D].北京:中国农业科学院,2017. |
| LI Y. Drought response of alfalfa: morphologic, physiological, and miRNA omics studies [D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. | |
| 13 | 何芳芳.植物MIR156a通过下调JAM-A抑制内皮细胞对单核细胞的黏附[D].南京:南京大学,2019. |
| HE F F. Plant MIR156a inhibits adhension of monocyte to endothelial cells by down-regulating JAM-A [D]. Nanjing: Nanjing University, 2019. | |
| 14 | 彭朦媛.人参水煎剂及其microRNA对气虚疲劳小鼠的干预和跨界调控作用研究[D].广州:广东药科大学,2020. |
| PENG M Y. Intervention of ginseng decoction and its microRNAs on qi-deficient fatigue mice and study on cross-kingdom regulation of ginseng microRNAs [D]. Guangzhou: Guangdong Pharmaceutical University, 2020. | |
| 15 | LUKASIK A, ZIELENKIEWICZ P. In silico identification of plant miRNAs in mammalian breast milk exosomes—a small step forward? [J/OL]. PloS One, 2014, 9:e99963 [2021-11-10]. . |
| 16 | 母治平.猪体内食物源性植物miRNAs的鉴定[D].雅安:四川农业大学,2013. |
| MU Z P. Identification of food-derived exogenous plant miRNAs in swine [D]. Ya’an: Sichuan Agricultural University, 2013. | |
| 17 | ARSHAD M, FEYISSA B A, AMYOT L, et al.. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13 [J]. Plant Sci., 2017, 258(1):122-136. |
| 18 | 孙铭优,吴照晨,王斌,等. ABC转运蛋白及其相关的多药抗性研究现状[J].植物保护学报,2022, 49(1): 374-382. |
| SUN M Y, WU Z C, WANG B, et al.. Research status of ABC transporter and its related multidurg resistance [J]. J. Plant Prot., 2022, 49(1):374-382. | |
| 19 | LOCHER K P. Mechanistic diversity in ATP-binding cassette (ABC) transporters [J]. Nat. Structural Mol. Biol., 2016, 23(6):487-493. |
| 20 | HIGGINS C F. ABC transporters: from microorganisms to man [J]. Annual Rev. Cell Biol., 1992, 8(1):67-113. |
| 21 | 王依纯,李佳赟,马进.南方型紫花苜蓿(Medicago sativa‘Millenium’)耐盐相关miRNAs及靶基因预测[J].分子植物育种,2019,17(24):8072-8081. |
| WANG Y C, LI J Y, MA J. Identification and analysis of microRNAs and targets involved in salt stress responses in alfalfa (Medicago sativa‘Millenium’) [J]. Mole. Plant Breeding, 2019, 17(24):8072-8081. | |
| 22 | KAMMES K L, HEEMINK G B, ALBRENCHT K A, et al.. Utilization of kura clover-reed canarygrass silage versus alfalfa silage by lactating dairy cows [J]. J. Dairy Sci., 2008, 91(8):3138-3144. |
| 23 | SUN H Z, WANG D M, WANG B, et al.. Metabolomics of four biofluids from dairy cows: potential biomarkers for milk production and quality [J]. J. Proteome Res., 2015,14(2): 1287-1298. |
| 24 | WANG B, MAO S Y, YANG H J, et al.. Effects of alfalfa and cereal straw as a forage source on nutrient digestibility and lactation performance in lactating dairy cows [J]. J. Dairy Sci., 2014, 97(12):7706-7715. |
| 25 | 王莹,宋响文. miRNAs在不同物种进化中序列保守性的研究[J].辽宁师范大学学报(自然科学版),2009,32(3):351-355. |
| WANG Y, SONG X W. Conservation of miRNAs sequences in the process of evolution of different species[J]. J. Liaoning Norm. Univ. (Nat. Sci.), 2009, 32(3):351-355. | |
| 26 | 王鹏俊.猪体内玉米miRNAs的检测及其潜在靶基因的预测和鉴定[D].雅安:四川农业大学,2017. |
| WANG P J. Detection of maize-derived microRNAs and prediction and validation of their potential target genes in pigs [D]. Ya’an: Sichuan Agricultural University, 2017. | |
| 27 | 贾凌.桑树miRNA的鉴定及其在蚕桑互作中的功能研究[D].重庆:西南大学,2015. |
| JIA L. Identification of mulberry miRNA and its function in silkworm mulberry interaction [D]. Chongqing: Southwest University, 2015. | |
| 28 | LUKASIK A, BRZOZOWSKA I, ZIZELENKIEWICZ U, et al.. Detection of plant miRNAs abundance in human breast milk [J/OL]. Int. J. Mol. Sci., 2017, 19(1):37 [2021-11-10].. |
| [1] | 赵柏霞, 闫建芳. 高通量技术分析‘砂蜜豆’甜樱桃不同组织内生细菌多样性[J]. 中国农业科技导报, 2023, 25(3): 66-77. |
| [2] | 闫宁, 战宇, 苗馨月, 王二刚, 陈长宝, 李琼. 强还原土壤灭菌处理对人参连作土壤细菌群落结构及土壤酶活的影响[J]. 中国农业科技导报, 2022, 24(6): 133-144. |
| [3] | 李洁, 林莹, 徐美玉, 王飞, 徐凌川. 泰山白首乌根际土壤真菌多样性分析[J]. 中国农业科技导报, 2022, 24(6): 70-81. |
| [4] | 赵宏岩, 谭君伟, 张杰, 陈浩楠, 王春旭, 赵地, 李海鹏, 朱李霞, 韩毅强. 小豆和绿豆茎基感病部位真菌群落结构研究[J]. 中国农业科技导报, 2022, 24(5): 129-136. |
| [5] | 魏艳晨, 陈吉祥, 王永刚, 孟彤彤, 韩亚龙, 李美. 荒漠植物珍珠猪毛菜根际土壤细菌多样性与土壤理化性质相关性分析[J]. 中国农业科技导报, 2022, 24(5): 209-217. |
| [6] | 范鹤龄, 朱清, 孙雪冰, 张丽, 李长江, 陈萍, 黄小龙, 张荣萍. 不同农用酵素的微生物多样性和群落结构[J]. 中国农业科技导报, 2022, 24(11): 179-189. |
| [7] | 苏雨萌§,张旭婷§,特日格乐,田敏,尚晓蕊,李国婧,王瑞刚*. 高通量测序鉴定中间锦鸡儿干旱条件下的microRNA[J]. 中国农业科技导报, 2021, 23(3): 51-57. |
| [8] | 刘璐1,名晓东1,张晓艳2,郝俊杰2,付丽平1,王乾坤1,吕鑫1,陈旺1,刘全兰1*. 高通量测序分析蚕豆种子内生细菌的多样性[J]. 中国农业科技导报, 2021, 23(2): 73-80. |
| [9] | 杨晶晶,张青青*,吐尔逊娜依·热依木,阿马努拉·依明尼亚孜,雪热提江·麦提努日. 游牧和定居对伊犁绢蒿荒漠草地土壤真菌群落多样性的影响[J]. 中国农业科技导报, 2020, 22(7): 166-173. |
| [10] | 史芳芳,李向泉*. 葡萄根际土壤真菌群落多样性分析[J]. 中国农业科技导报, 2019, 21(7): 47-58. |
| [11] | 张艺洁,邵惠芳*,张珂,贾宏昉,黄五星,韩丹. 基于高通量测序研究施肥对连作植烟土壤环境及微生物的影响[J]. 中国农业科技导报, 2018, 20(5): 16-25. |
| [12] | 曲梦楠1,2§,蒋炳军2§,刘薇2,毛婷婷2,马立明2,林抗雪2,韩天富1,2*. 大豆分子育种研究新进展[J]. , 2014, 16(3): 8-13. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号