




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (7): 77-86.DOI: 10.13304/j.nykjdb.2022.0170
收稿日期:2022-03-07
接受日期:2022-04-20
出版日期:2023-07-15
发布日期:2023-08-25
通讯作者:
张传博
作者简介:王云胜 E-mail:1710818668@qq.com;
基金资助:
Yunsheng WANG( ), Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG(
), Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG( )
)
Received:2022-03-07
Accepted:2022-04-20
Online:2023-07-15
Published:2023-08-25
Contact:
Chuanbo ZHANG
摘要:
冠突散囊菌是茯砖茶加工过程中的优势益生真菌,其产生的胞外酶和丰富的次生代谢产物影响成品茶的品质和口感。为探究veA对冠突散囊菌次级代谢的调控机制,使用无缝克隆及农杆菌转化技术构建了冠突散囊菌veA基因过表达突变株,使用高效液相色谱技术(high performance liquid chromatography,HPLC)和转录组测序初步解析过表达veA对冠突散囊菌次级代谢相关基因表达的影响。结果表明,与野生型冠突散囊菌相比,突变株发酵产物中多种化合物含量具有明显变化,物质吸收峰1的相对峰面积提高了4.09倍,且检测到3种新的物质吸收峰。转录组测序结果表明,与野生型相比,突变株的基因表达谱具有较大差异,共检测出737个显著差异表达基因(differentially expressed genes,DEGs),其中,282个基因显著上调表达,455个基因显著下调表达。DEGs主要富集在氨基酸代谢、蛋白酶和萜类化合物合成等通路。使用antiSMASH软件注释到545个次级代谢产物合成基因,10.64%的基因表达存在显著差异,其中,聚戊烯基合成酶基因、异戊二烯基二磷酸δ-异构酶基因和法尼基焦磷酸合成酶基因均显著上调表达。上述结果表明VeA能够激活一些次级代谢相关基因的表达,为冠突散囊菌的发酵应用和新颖次级代谢产物的挖掘提供了新的见解和方法。
中图分类号:
王云胜, 陈银翠, 程在, 张锦, 张传博. 过表达veA基因对冠突散囊菌次级代谢的影响[J]. 中国农业科技导报, 2023, 25(7): 77-86.
Yunsheng WANG, Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG. Effects of Overexpression of veA Gene on Secondary Metabolism of Eurotium cristatus[J]. Journal of Agricultural Science and Technology, 2023, 25(7): 77-86.
引物 Primer | 序列 Sequence (5’-3’) |
|---|---|
| veAF | AGACATCACCATGGTAGATCTATGGCGACGCGAATGCCC |
| veAR | GGGGAAATTCGAGCTGGTCACCTTAACTAGAAAAAGCCGGCGG |
| GAPDHF | CTGCCGTATCGAGAAGGGTG |
| GAPDHR | GATGAAGTTGGGGTTGAGGG |
| QveAF | CGACATCACCTTTGCCTAC |
| QveAR | TCGGGAAAGATGAAGTAGC |
表 1 本研究使用的引物
Table 1 Primers used in this study
引物 Primer | 序列 Sequence (5’-3’) |
|---|---|
| veAF | AGACATCACCATGGTAGATCTATGGCGACGCGAATGCCC |
| veAR | GGGGAAATTCGAGCTGGTCACCTTAACTAGAAAAAGCCGGCGG |
| GAPDHF | CTGCCGTATCGAGAAGGGTG |
| GAPDHR | GATGAAGTTGGGGTTGAGGG |
| QveAF | CGACATCACCTTTGCCTAC |
| QveAR | TCGGGAAAGATGAAGTAGC |
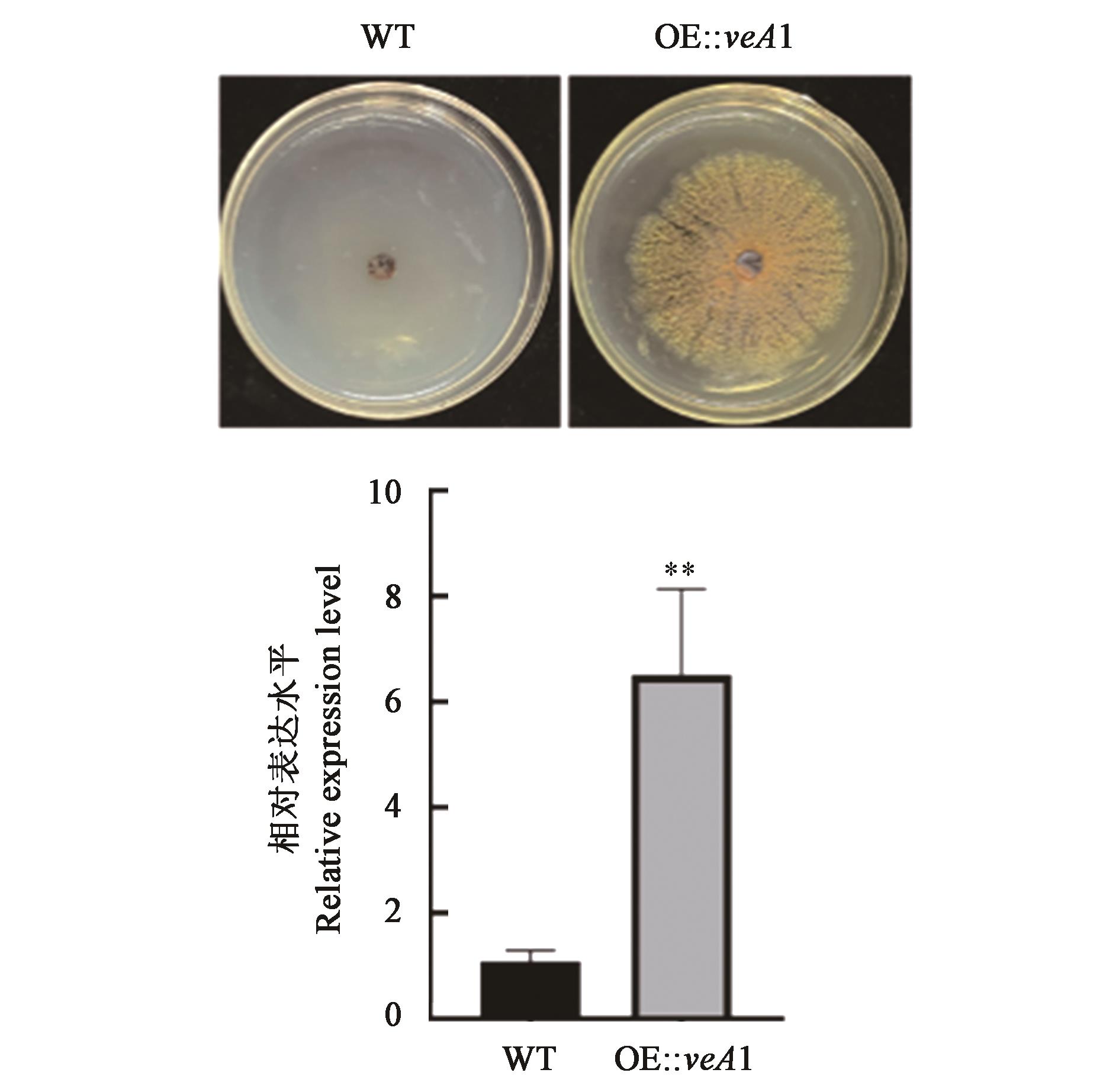
图1 veA过表达突变株的鉴定注:**表示两组间差异在P<0.01水平显著。
Fig. 1 Screening of veA overexpression mutantNote: ** indicated significant difference between the two groups at P<0.01 level.
样品 Sample | 原始数据 Raw reads | 质控数据 Clean reads | 错配率 Error rate/% | Q20/% | Q30/% |
|---|---|---|---|---|---|
| OE::veA1_1 | 53 215 236 | 52 880 844 | 0.023 9 | 98.44 | 95.28 |
| OE::veA1_2 | 58 759 294 | 58 355 148 | 0.024 0 | 98.4 | 95.21 |
| OE::veA1_3 | 56 979 242 | 56 535 564 | 0.024 1 | 98.36 | 95.11 |
| WT1 | 52 014 002 | 51 659 080 | 0.024 0 | 98.41 | 95.24 |
| WT2 | 57 594 222 | 57 246 210 | 0.023 9 | 98.43 | 95.26 |
| WT3 | 58 672 768 | 58 282 756 | 0.023 9 | 98.42 | 95.25 |
表2 转录组测序数据统计
Table 2 Transcriptome sequencing data statistics
样品 Sample | 原始数据 Raw reads | 质控数据 Clean reads | 错配率 Error rate/% | Q20/% | Q30/% |
|---|---|---|---|---|---|
| OE::veA1_1 | 53 215 236 | 52 880 844 | 0.023 9 | 98.44 | 95.28 |
| OE::veA1_2 | 58 759 294 | 58 355 148 | 0.024 0 | 98.4 | 95.21 |
| OE::veA1_3 | 56 979 242 | 56 535 564 | 0.024 1 | 98.36 | 95.11 |
| WT1 | 52 014 002 | 51 659 080 | 0.024 0 | 98.41 | 95.24 |
| WT2 | 57 594 222 | 57 246 210 | 0.023 9 | 98.43 | 95.26 |
| WT3 | 58 672 768 | 58 282 756 | 0.023 9 | 98.42 | 95.25 |
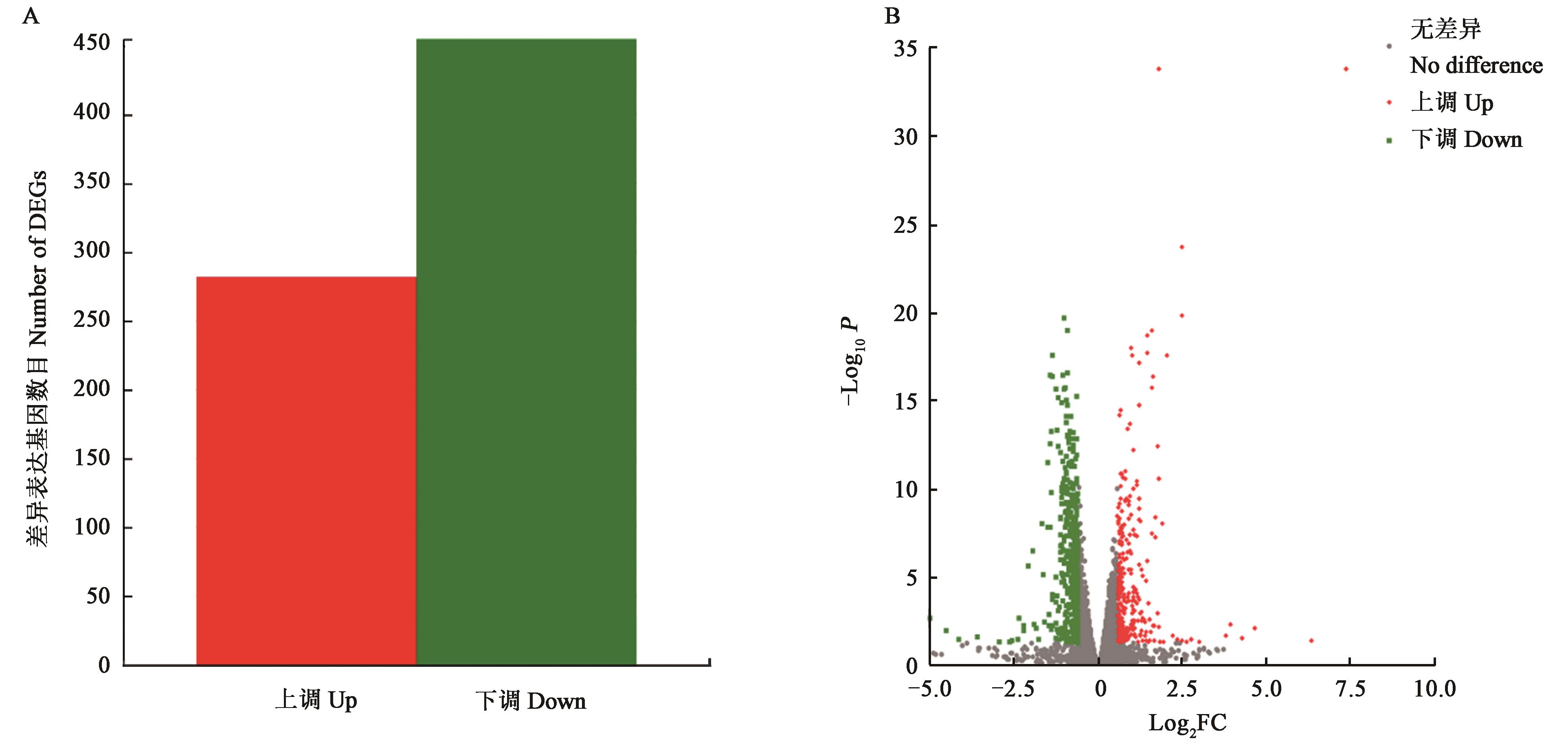
图3 野生型与过表达突变株的差异表达基因A:差异表达基因柱状统计图;B:差异表达基因火山图
Fig. 3 Differentially expressed genes between WT and OE::veA1A: Histogram of differentially expressed genes; B: Volcano map of differentially expressed genes
基因编号 Gene ID | 功能注释 Function annotation | 变化倍数 Fold change | 调控 Regulate |
|---|---|---|---|
| SI65_05759 | 氧化还原酶 Flavoprotein-ubiquinone oxidoreductase | 1.01ns | 上调Up |
| SI65_05760 | 反转录酶 Reverse transcriptase | 0.61ns | 下调Down |
| SI65_05761 | TATA框结合蛋白 TATA-box-binding protein | 0.72ns | 下调Down |
| SI65_05762 | L-甲基哌啶氧化酶 L-pipecolate oxidase | 1.50* | 上调Up |
| SI65_05763 | 甲基转移酶 Methyltransferase | 1.05ns | 上调Up |
| SI65_05764 | 假设蛋白 Hypothetical protein | 1.45ns | 上调Up |
| SI65_05765 | C6指域转录因子 C6 finger domain transcription factor | 1.25ns | 上调Up |
| SI65_05766 | NADH-生化色素b5还原酶 NADH-cytochrome b5 reductase | 1.27ns | 上调 Up |
| SI65_05767 | 甲基转移酶 Methyltransferase | 1.72* | 上调Up |
| SI65_05768 | 羧基酸合成酶 Carboxylic acid synthase | 2.02* | 上调Up |
| SI65_05769 | 金属化合物-β-内酰胺酶 Metallo-beta-lactamase | 2.31* | 上调Up |
| SI65_05770 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 1.92ns | 上调Up |
| SI65_05771 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 2.47* | 上调 Up |
| SI65_05772 | 氧化还原酶 Oxidoreductase | 2.67* | Up 上调 |
| SI65_05773 | 乙酰基转移酶 Acetyltransferase | 2.74* | Up 上调 |
| SI65_05774 | 假定蛋白 Hypothetical protein | NA | NA |
| SI65_05775 | 假定蛋白 Hypothetical protein | 0.55* | Down 下调 |
| SI65_05776 | 转录因子 Transcription factor | 0.64* | Down 下调 |
表3 次级代谢产物基因簇32中基因差异表达及功能注释
Table 3 Gene expression and functional annotation in secondary metabolite gene cluster 32
基因编号 Gene ID | 功能注释 Function annotation | 变化倍数 Fold change | 调控 Regulate |
|---|---|---|---|
| SI65_05759 | 氧化还原酶 Flavoprotein-ubiquinone oxidoreductase | 1.01ns | 上调Up |
| SI65_05760 | 反转录酶 Reverse transcriptase | 0.61ns | 下调Down |
| SI65_05761 | TATA框结合蛋白 TATA-box-binding protein | 0.72ns | 下调Down |
| SI65_05762 | L-甲基哌啶氧化酶 L-pipecolate oxidase | 1.50* | 上调Up |
| SI65_05763 | 甲基转移酶 Methyltransferase | 1.05ns | 上调Up |
| SI65_05764 | 假设蛋白 Hypothetical protein | 1.45ns | 上调Up |
| SI65_05765 | C6指域转录因子 C6 finger domain transcription factor | 1.25ns | 上调Up |
| SI65_05766 | NADH-生化色素b5还原酶 NADH-cytochrome b5 reductase | 1.27ns | 上调 Up |
| SI65_05767 | 甲基转移酶 Methyltransferase | 1.72* | 上调Up |
| SI65_05768 | 羧基酸合成酶 Carboxylic acid synthase | 2.02* | 上调Up |
| SI65_05769 | 金属化合物-β-内酰胺酶 Metallo-beta-lactamase | 2.31* | 上调Up |
| SI65_05770 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 1.92ns | 上调Up |
| SI65_05771 | 细胞色素P450单加氧酶 Cytochrome P450 monooxygenase | 2.47* | 上调 Up |
| SI65_05772 | 氧化还原酶 Oxidoreductase | 2.67* | Up 上调 |
| SI65_05773 | 乙酰基转移酶 Acetyltransferase | 2.74* | Up 上调 |
| SI65_05774 | 假定蛋白 Hypothetical protein | NA | NA |
| SI65_05775 | 假定蛋白 Hypothetical protein | 0.55* | Down 下调 |
| SI65_05776 | 转录因子 Transcription factor | 0.64* | Down 下调 |
| 1 | BOK J, HOFFMEISTER D, MAGGIO-HALL L, et al.. Genomic mining for Aspergillus natural products [J]. Chem. Biol., 2006, 13(1):7-31. |
| 2 | RAJA H A, MILLER A N, PERACE C J, et al.. Fungal identification using molecular tools: a primer for the natural products research community [J]. J. Nat. Prod., 2017, 80(3):756-770. |
| 3 | ZHANG X J, ZHU Y Y, BAO L F, et al.. Putative methyltransferase laea and transcription factor CREA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression [J]. Fungal. Genet. Biol., 2016, 94:32-46. |
| 4 | BAYRAM O, BRAUS G H. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins [J]. FEMS Microbiol. Lett., 2012, 36(1):1-24. |
| 5 | KELLER N. Fungal secondary metabolism: regulation, function and drug discovery [J]. Nat. Rev. Microbiol., 2019, 17(3):167-180. |
| 6 | CHIANG Y M, OAKLEY C E, AHUJA M E, et al.. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus Nidulans [J]. J. Am. Chem. Soc., 2013, 135(20):7720-7731. |
| 7 | YAEGASHI J, OAKLEY B, WANG C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans [J]. J. Appl. Microbiol., 2014, 41(2):433-442. |
| 8 | SARIKAYA-BATRAM Ö, PALMER JM, KELLER N, et al.. One juliet and four romeos: vea and its methyltransferases [J/OL]. Front. Microbiol., 2015, 6:1 [2022-02-10]. . |
| 9 | KATO N, BROOKS W, CALVO A. The expression of sterigmatocystin and penicillin genes in Aspergillus Nidulans is controlled by vea, a gene required for sexual development [J]. Eukaryot. Cell, 2003, 2(6):1178-1186. |
| 10 | MARUI J, OHASHI-KUNIHIRO S, ANDO T, et al.. Penicillin biosynthesis in Aspergillus Oryzae and its overproduction by genetic engineering [J]. J. Biosci. Bioeng., 2010, 110(1):8-11. |
| 11 | 曹双,郑跃亮,张虎,等.过表达veA对橘青霉美伐他汀生物合成的影响[J].西南师范大学学报(自然科学版),2016,41(6):67-73. |
| CAO S, ZHENG Y L, ZHANG H, et al.. On investigation of veA gene in regulating mevastatin biosynthesis, conidia development in Penicillium citrinum [J]. J. Southwest China Normal Univ. (Nat. Sci.), 2016, 41(6):67-73. | |
| 12 | 张月,崔旋旋,刘英学,等.茯砖茶中冠突散囊菌的分离鉴定及其发酵工艺和生物活性研究[J].食品与发酵工业,2020,418(22):202-207. |
| ZHANG Y, CUI X X, LIU Y X, et al.. Isolation, identification, fermentation technology and bioactivity of Eurotium cristatum in Fuzhuan brick tea [J]. Food. Ferment. Ind., 2020, 46(22):202-207. | |
| 13 | AN T T, CHEN M X, ZU Z Q, et al.. Untargeted and targeted metabolomics reveal changes in the chemical constituents of instant dark tea during liquid-state fermentation by Eurotium cristatum [J]. Food. Res. Int., 2021, 148:110-125. |
| 14 | 陈敏,谢发,游玲,等.冠突散囊菌发酵苦丁茶工艺研究[J].食品与发酵工业, 2020,402(6):224-228. |
| CHNEG M, XIE F, YOU L, et al.. Fermentation of ligustrum robustum by Eurotium cristatum [J]. Food. Ferment. Ind., 2020, 46(6):224-228. | |
| 15 | GE Y Y, WANG Y C, LIU Y X, et al.. Comparative genomic and transcriptomic analyses of the Fuzhuan brick tea-fermentation fungus Aspergillus cristatus [J/OL]. BMC Genomics, 2016, 17(1): 428 [2022-02-10]. . |
| 16 | 周国庆.冠突曲霉LaeA基因功能的研究[D].贵阳:贵州大学,2018. |
| ZHOU G Q. Study on the function of LaeA gene in Aspergillus cristatum [D]. Guiyang: Guizhou University, 2018. | |
| 17 | TAN Y M, WANG H, WANG Y P, et al.. The role of the vea gene in adjusting developmental balance and environmental stress response in Aspergillus cristatus [J]. Fungal. Biol., 2018, 122(10):952-964. |
| 18 | WANG Y P, TAN Y M, WANG Y C, et al.. Role of Acndta in cleistothecium formation, osmotic stress response, pigmentation and carbon metabolism of Aspergillus cristatus [J]. Fungal. Biol., 2021, 125(10):749-763. |
| 19 | 刘逸梅.冠突曲霉钙信号调控基因Fig的功能研究[D].贵阳:贵州大学, 2019. |
| LIU YM. Study on the function of Fig gene regulating calcium signal in Aspergillus cristatum [D]. Guiyang: Guizhou University, 2019. | |
| 20 | 朱思远,徐岩,喻晓蔚.农杆菌介导转化黑曲霉条件优化及脂肪酶表达[J].食品与生物技术学报,2020,39(5):51-58. |
| ZHU S Y, XU Y, YU X Y. Optimization of agrobacterium tumefaciens-mediated transformation of Aspergillus niger and expression of lipase [J]. J. Food Sci. Biotechnol., 2020, 39(5):51-58. | |
| 21 | 邵蕾,谭玉梅,任春光,等.转录因子ACZ影响冠突曲霉色素及产孢数量的研究[J].基因组学与应用生物学,2019,38(5):2041-2048. |
| SHAO L, TAN Y M, REN C G, et al.. Study on the transcription factor ACZ affecting the number of spores and pigmentation of Asperillus cristatus [J]. Genom. Appl. Biol., 2019, 38(5):2041-2048. | |
| 22 | 向婷,于凤明,杨正军,等.冠突曲霉preA基因的功能分析[J].基因组学与应用生物学,2020,39(7):3070-3078. |
| XIANG T, YU F M, YANG Z J, et al.. Functional analysis on the preA gene in Aspergillus cristatus [J]. Genom. Appl. Biol., 2020, 39(7):3070-3078. | |
| 23 | 杜静.冠突散囊菌对中药材三七成分的转化及机理研究[D].贵阳:贵州师范大学,2020. |
| DU J. Biotransformation of bioactive compounds in Pannax notoginseng fermented by Eurotium cristatum and its mechanism [D]. Guiyang: Guizhou Normal University, 2020. | |
| 24 | 王琪琪.黑茶中散囊属真菌及其对茶叶品质提升研究[D].贵阳:贵州师范大学,2021. |
| WANG Q Q. The diversity of Eurotium sp . in dark tea and its effect on the quality improvement [D]. Guiyang: Guizhou Normal University, 2021. | |
| 25 | REN X X, WANG Y C, LIU Y X, et al.. Comparative transcriptome analysis of the calcium signaling and expression analysis of sodium/calcium exchanger in Aspergillus cristatus [J]. J. Basic. Microbiol., 2018, 58(1):76-87. |
| 26 | WANG D Y, TONG S M, GUAN Y Y, et al.. The velvet protein vea functions in asexual cycle, stress tolerance and transcriptional regulation of Beauveria bassiana [J]. Fungal. Genet. Biol., 2019, 127:1-11. |
| 27 | LIU H, SANG S L, WANG H, et al.. Vea comparative proteomic analysis reveals the regulatory network of the gene during asexual and sexual spore development of Aspergillus cristatus [J/OL]. Biosci. Rep., 2018, 38(4):67 [2022-02-10]. . |
| 28 | 张玉婷,李伟国,梁冬梅,等. P450s在萜类合成方面的合成生物学研究进展[J].中国生物工程杂志,2020,40(8):84-96. |
| ZHANG Y T, LI W G, LIANG D M, et al.. Research progress in synthetic biology of P450s in terpenoid synthesis [J]. Chin. J. Biotechnol., 2020, 40(8):84-96. | |
| 29 | CHANG M, KEASLING J. Production of isoprenoid pharmaceuticals by engineered microbes [J]. Nat. Chem. Biol., 2006, 2(12):674-681. |
| 30 | CARY J W, HAN Z, YIN Y, et al.. Transcriptome analysis of Aspergillus flavus reveals vea-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster [J]. Eukaryot. Cell., 2015, 14(10):983-997. |
| 31 | 张梦薇.过表达laeA和veA基因对黑曲霉生长和代谢的影响[D].天津:天津科技大学,2020. |
| ZHANG M W. Effects of overexpression of laeA and veA genes on growth and metabolism of Aspergillus niger [D]. Tianjin: Tianjin University of Science and Technology, 2020. | |
| 32 | KONG W P, HUANG C S, SHI J, et al.. Recycling of Chinese herb residues by endophytic and probiotic fungus Aspergillus cristatus Cb10002 for the production of medicinal valuable anthraquinones [J/OL]. Microb. Cell. Fact., 2019, 18(1):9 [2022-02-10]. . |
| [1] | 马蓝, 彭晴, 徐小轻, 杨硕, 张宇微, 田丹丹, 施琳波, 石波, 乔宇. 大肠杆菌O157∶H7生物被膜状态下基因表达分析[J]. 中国农业科技导报, 2023, 25(6): 71-88. |
| [2] | 周雨青, 杨永飞, 葛常伟, 沈倩, 张思平, 刘绍东, 马慧娟, 陈静, 刘瑞华, 李士丛, 赵新华, 李存东, 庞朝友. 基于WGCNA的棉花子叶抗冷相关共表达模块鉴定[J]. 中国农业科技导报, 2022, 24(4): 52-62. |
| [3] | 李舒欣, 张浩, 郑厚胜, 郑培和, 逄世峰, 许世泉. 转录组分析二马牙和长脖类型林下参表型差异[J]. 中国农业科技导报, 2021, 23(9): 56-68. |
| [4] | 刘源, 张秀妍, 徐妙云, 郑红艳, 邹俊杰, 张兰, 王磊. 水稻干旱胁迫的small RNA转录组分析[J]. 中国农业科技导报, 2021, 23(6): 23-32. |
| [5] | 张文云1,张建诚2,姚景珍2*. 氮胁迫下小麦叶片转录组分析[J]. 中国农业科技导报, 2020, 22(11): 26-34. |
| [6] | 陈海龙1,蒋丽华1,陈帅1,张晓戈1,朱年青1,韦平和1*,周长林2. Adk1过表达和柠檬酸钠补料促进酵母S-腺苷甲硫氨酸的合成[J]. 中国农业科技导报, 2020, 22(10): 69-76. |
| [7] | 陈诚1,刘晓飞2,李强3,王剑1,伏荣桃1,张鸿1,卢代华1*. 美国大灵芝(Ganoderma oregonense)转录组SSR位点的生物信息学分析[J]. 中国农业科技导报, 2018, 20(7): 48-55. |
| [8] | 王林1,杨立均2,赵佳佳2,黄海棠2,许自成1*. 组学分析技术在烟草研究中的应用进展[J]. 中国农业科技导报, 2018, 20(7): 56-62. |
| [9] | 黄娟,邓娇,陈庆富*. 荞麦根的转录组学分析及黄酮合成基因的鉴定[J]. 中国农业科技导报, 2017, 19(2): 9-19. |
| [10] | 岳春江1§,陈川川1§,郭凤仙1,李华1,孙洪波1,裴丹宁1,马晓清1,陈富欣1,杨获莉1,李琴1,刘越1,2*. 蒙药冷蒿转录组SSR信息分析[J]. 中国农业科技导报, 2016, 18(6): 31-43. |
| [11] | 黎瑞源1,潘凡2,陈庆富2,石桃雄2*. 苦荞转录组EST-SSR发掘及多态性分析[J]. , 2015, 17(4): 42-52. |
| [12] | 苏世友,滕超,张维,陈明*. 细菌Ⅲ型聚酮合酶研究进展[J]. , 2013, 15(6): 119-129. |
| [13] | 王辰,赵为民,牟玉莲*. 调控动物表型microRNA在分子育种中新的重要标记[J]. , 2013, 15(3): 108-112. |
| [14] | 李晓晖,李鑫鑫,张维,燕永亮,陈明,陆伟. 宏转录组学在微生物生态学研究中的应用[J]. , 2011, 13(4): 58-65. |
| [15] | 张松焕,李春奇,郭惠明,裴熙祥,程红梅. 过量表达紫茎泽兰类黄酮-3’羟化酶基因对转基因烟草POD、PAL活性的影响[J]. , 2009, 11(3): 98-101. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号