




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (6): 83-92.DOI: 10.13304/j.nykjdb.2023.0923
梁丽存1( ), 于会民1, 管晓晨2, 黄火清1, 姚斌1, 杨浩萌1(
), 于会民1, 管晓晨2, 黄火清1, 姚斌1, 杨浩萌1( )
)
收稿日期:2023-12-15
接受日期:2024-03-05
出版日期:2025-06-15
发布日期:2025-06-23
通讯作者:
杨浩萌
作者简介:梁丽存 E-mail:lianglicun1022@163.com;
基金资助:
Licun LIANG1( ), Huimin YU1, Xiaochen GUAN2, Huoqing HUANG1, Bin YAO1, Haomeng YANG1(
), Huimin YU1, Xiaochen GUAN2, Huoqing HUANG1, Bin YAO1, Haomeng YANG1( )
)
Received:2023-12-15
Accepted:2024-03-05
Online:2025-06-15
Published:2025-06-23
Contact:
Haomeng YANG
摘要:
塔宾曲霉是一种丝状真菌,在饲料、食品、生物防治、环境修复等领域都有广泛的应用前景。通过形态学观察结合分子检测和Ehrlich试剂染色法,成功地鉴定了1株塔宾曲霉,将其命名为F316。为探索适合F316的基因编辑方法,便于基因工程改造,应用CRISPR/Cas9系统,在F316的孢子色素合成相关基因fwnA的内部插入潮霉素抗性基因,通过孢子颜色统计和抗性基因PCR验证,比较不同基因编辑元件的递送方式对同源重组效率的影响。结果表明,在F316菌株中,表达盒法、自主复制质粒法和核糖核蛋白复合体法3种不同递送方式的同源重组效率分别为70.14%、66.67%和25.70%;另外,供体DNA中同源臂的长度对同源重组效率也有影响,当同源臂长度为2 000 bp时,重组效率最好。
中图分类号:
梁丽存, 于会民, 管晓晨, 黄火清, 姚斌, 杨浩萌. 塔宾曲霉的鉴定及基因编辑方法对其同源重组效率的影响[J]. 中国农业科技导报, 2025, 27(6): 83-92.
Licun LIANG, Huimin YU, Xiaochen GUAN, Huoqing HUANG, Bin YAO, Haomeng YANG. Identification of Aspergillus tubingensis and the Effect of Gene Editing Methods on Its Homologous Recombination Efficiency[J]. Journal of Agricultural Science and Technology, 2025, 27(6): 83-92.
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) | 用途 Usage |
|---|---|---|
| ITS-F | TCCGTAGGTGAACCTGCGC | 扩增ITS序列 |
| ITS-R | TCCT CCGCTTATTGATATGC | Amplification of ITS |
| BenA-F | GGTAACCAAATCGGTGCTGCTTTC | 扩增BenA序列 |
| BenA-R | ACCCTCAGTGTAGTGACCCTTGGC | Amplification of BenA |
| CaM-F | CCGAGTACAAGGAGGCCTTC | 扩增CaM序列 |
| CaM-R | CCGATAGAGGTCATAACGTGG | Amplification of Cam |
| 40-fwn-hph-F | ggtcctccgcctcccagcctacaagtgggatctcaagaactcgacagaagatgatattgaaggagcac | 扩增40 bp同源臂供体DNA |
| 40-fwn-hph-R | ccttgctcaggcagaagttgttggtatagggaatccagtagtatctggaagaggtaaacccgaaacgcg | Amplify 40 bp homologous arm donor DNA |
fwn-up-500-F fwn-dw-500-R | caaccaagctcaaggttccttacgcg tccgcagcagcgcagtcaaag | 扩增500 bp同源臂供体DNA Amplify 500 bp homologous arm donor DNA |
fwn-up-1k-F fwn-dw-1k-R | actctaccgcagttgtttccaattccg gtgataaggcttggaacttgactgcac | 扩增1 kb同源臂供体DNA Amplify 1 kb homologous arm donor DNA |
fwn-up-2k-F fwn-dw-2k-R | cgcagatcgggaacactgcggccgcccactggcaactgtaacacctttgatg cgaaacgcgcgaggcaggcggccgcttcatgtacgggcagttcaatccgtag | 扩增2 kb同源臂供体DNA Amplify 2 kb homologous arm donor DNA |
fwn-dw-F fwn-up-R | ctataccaacaacttctgcctgagcaagg gttcttgagatcccacttgtaggctggg | 扩增上下游同源臂 Amplify upstream and downstream homologous arm |
hph-fu-F hph-fu-R | caggctacaagtgggatctcaagaacgacagaagatgatattgaaggagcactttttg agttgttggtatagggaatccagtagtatctggaagaggtaaacccgaaacg | 潮霉素基因融合 Hygromycin gene fusion |
1K-fwn-F2 1k-fwn-R2 | gctgttggagcgaacctgcaatccctgcg ctcttgctaggagcagcaggggccg | 1 kb同源臂检测引物 1 kb homologous arm detection primers |
500-fwn-F 500-fwn-R2 | cgatgctggccatcaaggcgtccctggc gtgctgttcactgtcaagaataacctcgcg | 500 bp同源臂检测引物 500 bp homologous arm detection primers |
2K-fwn-F 2K-fwn-R | agcgtgctcgtctagtcttgctgctatcc catcataggcgcaaataccacctgcagacc | 2 kb同源臂检测引物 2 kb homologous arm detection primers |
fwn-up-4k-F fwn-dw-4k-R | gggggccctaaatctacatgtgtataaagtgtgcttctcatcgacacggatggggaagg cgaagccccaaatggactgcaggtaatagccggcccacgagccctggcatcaacaatgg | 扩增4 kb同源臂供体DNA Amplify 4 kb homologous arm donor DNA |
HPH-150-R HPH-150-F | gctactgctacaagtggggctgatctgacc cgttggtgtcgatgtcagctccggagttg | 潮霉素基因检测引物 Hygromycin gene detection primers |
5SRNA-F gRNA-R | cacatacgaccacagggtgtg aaaagcaccgactcggtgcc | 扩增gRNA表达盒 Amplify gRNA expression cassette |
Cas9-F Cas9-R | cgagacagcagaatcaccgcccaagttaag attacacttgtattgggatgaattttgtatgcac | 扩增Cas9表达盒 Amplify Cas9 expression cassette |
5s-BglⅡ-F 5s-BglⅡ-R | gcggaacatatactgggcccgggaacacatacgaccacagggtgtgg ctcagcggaaacagctatgaccatgaaaaaaagcaccgactcggtgcc | 扩增gRNA(与Cas9重组) Amplify gRNA(recombination with Cas9) |
Cas9-YZ-F Cas9-YZ-R | ggaacgaactcggttggttgggctg cagaattatcggggttcaggtcaccc | 供体DNA整合检测Detection of donor DNA integration |
fwn gRNA-YZ-F fwn gRNA-YZ-R | cagggtgtggaaaacagggcttccc cggtgccactttttcaagttgata | gRNA整合检测 Detection of gRNA integration |
表1 本研究所用引物 (续表Continued)
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) | 用途 Usage |
|---|---|---|
| ITS-F | TCCGTAGGTGAACCTGCGC | 扩增ITS序列 |
| ITS-R | TCCT CCGCTTATTGATATGC | Amplification of ITS |
| BenA-F | GGTAACCAAATCGGTGCTGCTTTC | 扩增BenA序列 |
| BenA-R | ACCCTCAGTGTAGTGACCCTTGGC | Amplification of BenA |
| CaM-F | CCGAGTACAAGGAGGCCTTC | 扩增CaM序列 |
| CaM-R | CCGATAGAGGTCATAACGTGG | Amplification of Cam |
| 40-fwn-hph-F | ggtcctccgcctcccagcctacaagtgggatctcaagaactcgacagaagatgatattgaaggagcac | 扩增40 bp同源臂供体DNA |
| 40-fwn-hph-R | ccttgctcaggcagaagttgttggtatagggaatccagtagtatctggaagaggtaaacccgaaacgcg | Amplify 40 bp homologous arm donor DNA |
fwn-up-500-F fwn-dw-500-R | caaccaagctcaaggttccttacgcg tccgcagcagcgcagtcaaag | 扩增500 bp同源臂供体DNA Amplify 500 bp homologous arm donor DNA |
fwn-up-1k-F fwn-dw-1k-R | actctaccgcagttgtttccaattccg gtgataaggcttggaacttgactgcac | 扩增1 kb同源臂供体DNA Amplify 1 kb homologous arm donor DNA |
fwn-up-2k-F fwn-dw-2k-R | cgcagatcgggaacactgcggccgcccactggcaactgtaacacctttgatg cgaaacgcgcgaggcaggcggccgcttcatgtacgggcagttcaatccgtag | 扩增2 kb同源臂供体DNA Amplify 2 kb homologous arm donor DNA |
fwn-dw-F fwn-up-R | ctataccaacaacttctgcctgagcaagg gttcttgagatcccacttgtaggctggg | 扩增上下游同源臂 Amplify upstream and downstream homologous arm |
hph-fu-F hph-fu-R | caggctacaagtgggatctcaagaacgacagaagatgatattgaaggagcactttttg agttgttggtatagggaatccagtagtatctggaagaggtaaacccgaaacg | 潮霉素基因融合 Hygromycin gene fusion |
1K-fwn-F2 1k-fwn-R2 | gctgttggagcgaacctgcaatccctgcg ctcttgctaggagcagcaggggccg | 1 kb同源臂检测引物 1 kb homologous arm detection primers |
500-fwn-F 500-fwn-R2 | cgatgctggccatcaaggcgtccctggc gtgctgttcactgtcaagaataacctcgcg | 500 bp同源臂检测引物 500 bp homologous arm detection primers |
2K-fwn-F 2K-fwn-R | agcgtgctcgtctagtcttgctgctatcc catcataggcgcaaataccacctgcagacc | 2 kb同源臂检测引物 2 kb homologous arm detection primers |
fwn-up-4k-F fwn-dw-4k-R | gggggccctaaatctacatgtgtataaagtgtgcttctcatcgacacggatggggaagg cgaagccccaaatggactgcaggtaatagccggcccacgagccctggcatcaacaatgg | 扩增4 kb同源臂供体DNA Amplify 4 kb homologous arm donor DNA |
HPH-150-R HPH-150-F | gctactgctacaagtggggctgatctgacc cgttggtgtcgatgtcagctccggagttg | 潮霉素基因检测引物 Hygromycin gene detection primers |
5SRNA-F gRNA-R | cacatacgaccacagggtgtg aaaagcaccgactcggtgcc | 扩增gRNA表达盒 Amplify gRNA expression cassette |
Cas9-F Cas9-R | cgagacagcagaatcaccgcccaagttaag attacacttgtattgggatgaattttgtatgcac | 扩增Cas9表达盒 Amplify Cas9 expression cassette |
5s-BglⅡ-F 5s-BglⅡ-R | gcggaacatatactgggcccgggaacacatacgaccacagggtgtgg ctcagcggaaacagctatgaccatgaaaaaaagcaccgactcggtgcc | 扩增gRNA(与Cas9重组) Amplify gRNA(recombination with Cas9) |
Cas9-YZ-F Cas9-YZ-R | ggaacgaactcggttggttgggctg cagaattatcggggttcaggtcaccc | 供体DNA整合检测Detection of donor DNA integration |
fwn gRNA-YZ-F fwn gRNA-YZ-R | cagggtgtggaaaacagggcttccc cggtgccactttttcaagttgata | gRNA整合检测 Detection of gRNA integration |
| 名称Name | 序列Sequence(5’-3’) |
|---|---|
| gRNA-fwn | TAATACGACTCACTATAGGAGTGGGATCTCAAGAACTACGTTTTAGAGCTAGAAATAGCAA GTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT |
| 5spro-gRNA-fwn | CACATACGACCACAGGGTGTGGAAAACAGGGCTTCCCGTCCGCTCAGCCGTACTTAA GCCACACGCCGGGAGGTTAGTAGTTGGGTGGGTGACCACCAGCGAATCCCTTCTGTT GTATGAAAGGACGAAACACCAGTGGGATCTCAAGAACTACGTTTTAGAGCTAGAAATA GCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT |
表2 gRNA体外转录模板和体内表达盒的序列信息
Table 2 Sequence information of gRNA both in vitro transcription template and in vivo expression cassette
| 名称Name | 序列Sequence(5’-3’) |
|---|---|
| gRNA-fwn | TAATACGACTCACTATAGGAGTGGGATCTCAAGAACTACGTTTTAGAGCTAGAAATAGCAA GTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT |
| 5spro-gRNA-fwn | CACATACGACCACAGGGTGTGGAAAACAGGGCTTCCCGTCCGCTCAGCCGTACTTAA GCCACACGCCGGGAGGTTAGTAGTTGGGTGGGTGACCACCAGCGAATCCCTTCTGTT GTATGAAAGGACGAAACACCAGTGGGATCTCAAGAACTACGTTTTAGAGCTAGAAATA GCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT |
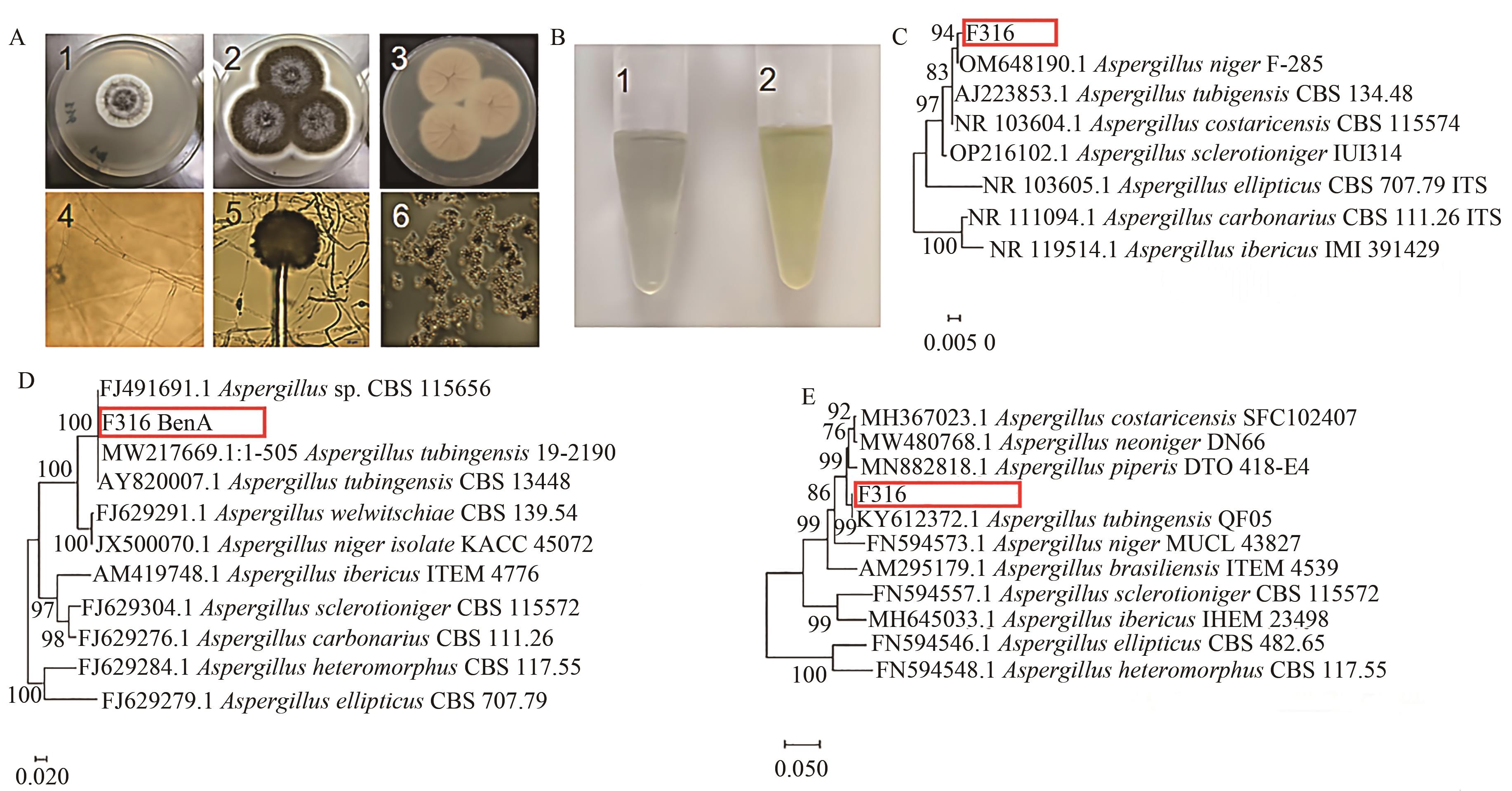
图2 F316菌种的形态、Ehrlich染色和系统进化树A:F316菌株的形态特征,1—菌落正面初期, 2—菌落正面, 3—菌落反面,4—菌丝(标尺:20 μm),5—分生孢子头和梗(标尺:20 μm),6—分生孢子 (标尺:20 μm);B:Ehrlich颜色反应,1—F316,2—黑曲霉;C:基于ITS的菌株系统发育分析;D:基于BenA的菌株系统发育分析;E:基于CaM序列的菌株系统发育分析
Fig. 2 Morphology, Ehrlich staining and phylogenetic analysis of F316 strainA: Morphological characteristics of strain F316, 1— Obverse of young colony, 2—Obverse of Colony, 3—Reverse of colony, 4—Mycelia(bar: 20 μm), 5—Conidiphore (bar: 20 μm), 6—Conidium (bar: 20 μm); B: Ehrlich color reaction, 1—F316, 2—A. tubingensis; C: Phylogenetic analysis based on ITS sequences; D: Phylogenetic analysis based on BenA sequences; E: Phylogenetic analysis based on CaM sequences
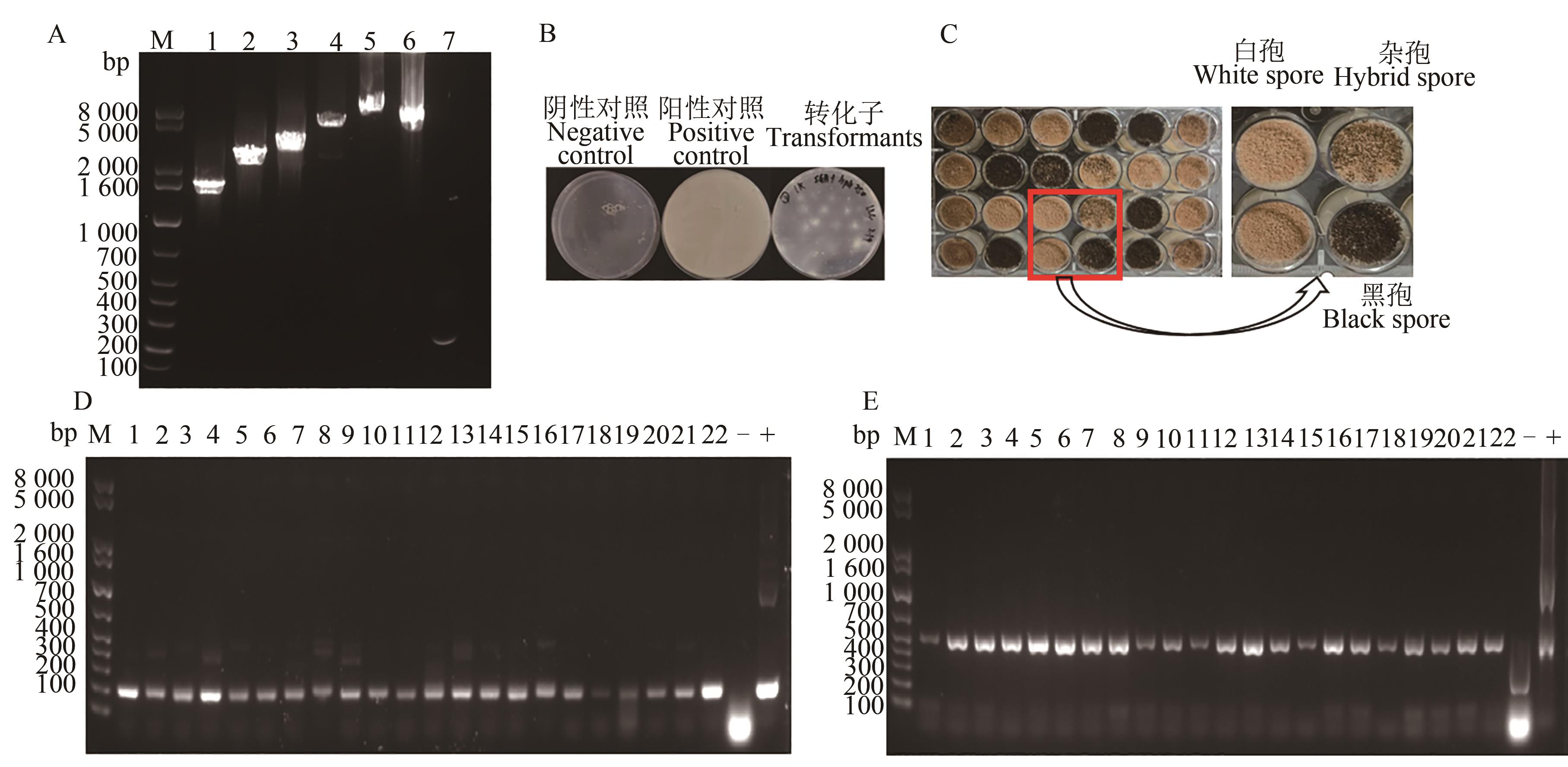
图3 PCR扩增、孢子颜色对比及整合效率验证A:表达原件的扩增,1~5为1.6、2.6、3.6、5.6、9.6 kb的donor DNA,6为cas9 表达盒,7为gRNA表达盒;B:转化子及阴阳对照;C:转化子的生长情况;D和E:gRNA和Cas9表达盒整合进基因组的PCR验证,1~22为随机22个孢子
Fig. 3 PCR amplification, spore color comparison and integration efficiency verificationA: Amplification of expression element, 1~5 indicate 1.6,2.6,3.6,5.6,9.6 kb donor DNA, respectively, 6 indicates cas9 expression cassette, 7 indicates gRNA expression cassette;B: Transformants and negative, positive contrast;C: Growth of transformants; D and E: PCR validation of gRNA and Cas9 cassette integrate into the genome, 1~22 indicate 22 spores selected at random
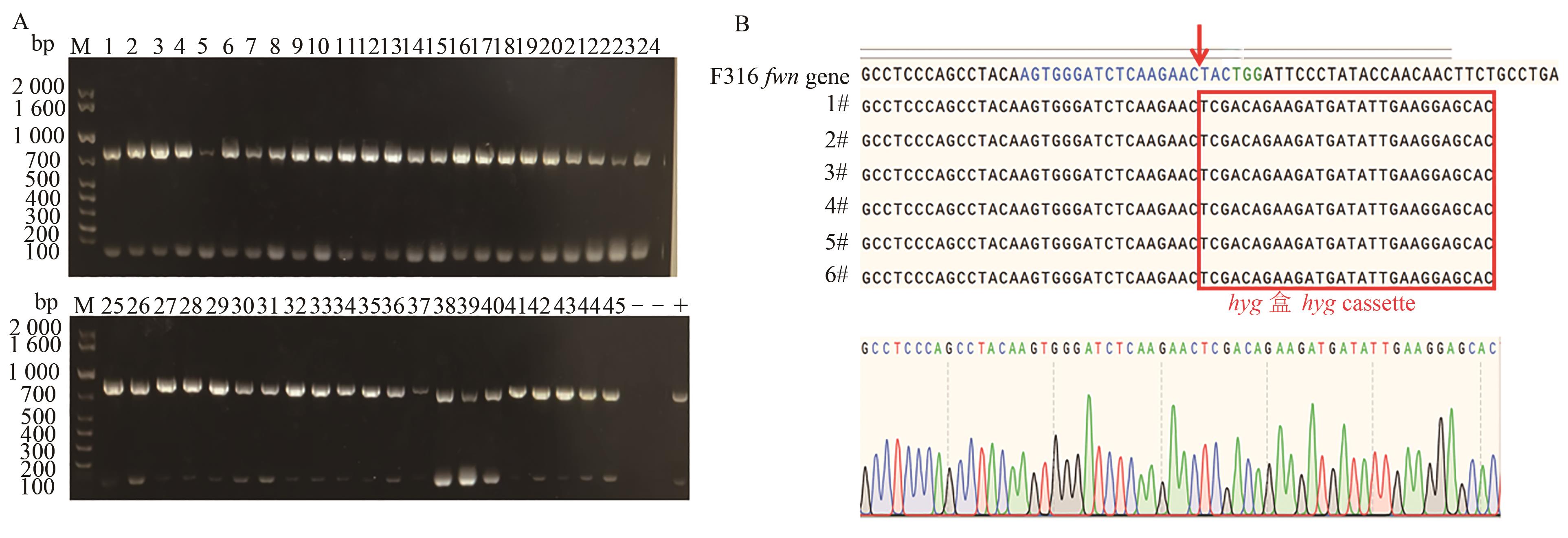
图4 同源重组效率PCR验证及测序结果A:PCR验证,1~24—随机24个孢子;B:测序结果,蓝色碱基序列为gRNA的特异识别序列,1#~6#为基因编辑序列
Fig. 4 Homologous recombination efficiency PCR verification and sequencing resultA: PCR verification, 1~24—24 spores selected at random; B: Sequencing result, the blue base sequence is the specific recognition sequence of gRNA, and 1#~6# are the gene edited sequences
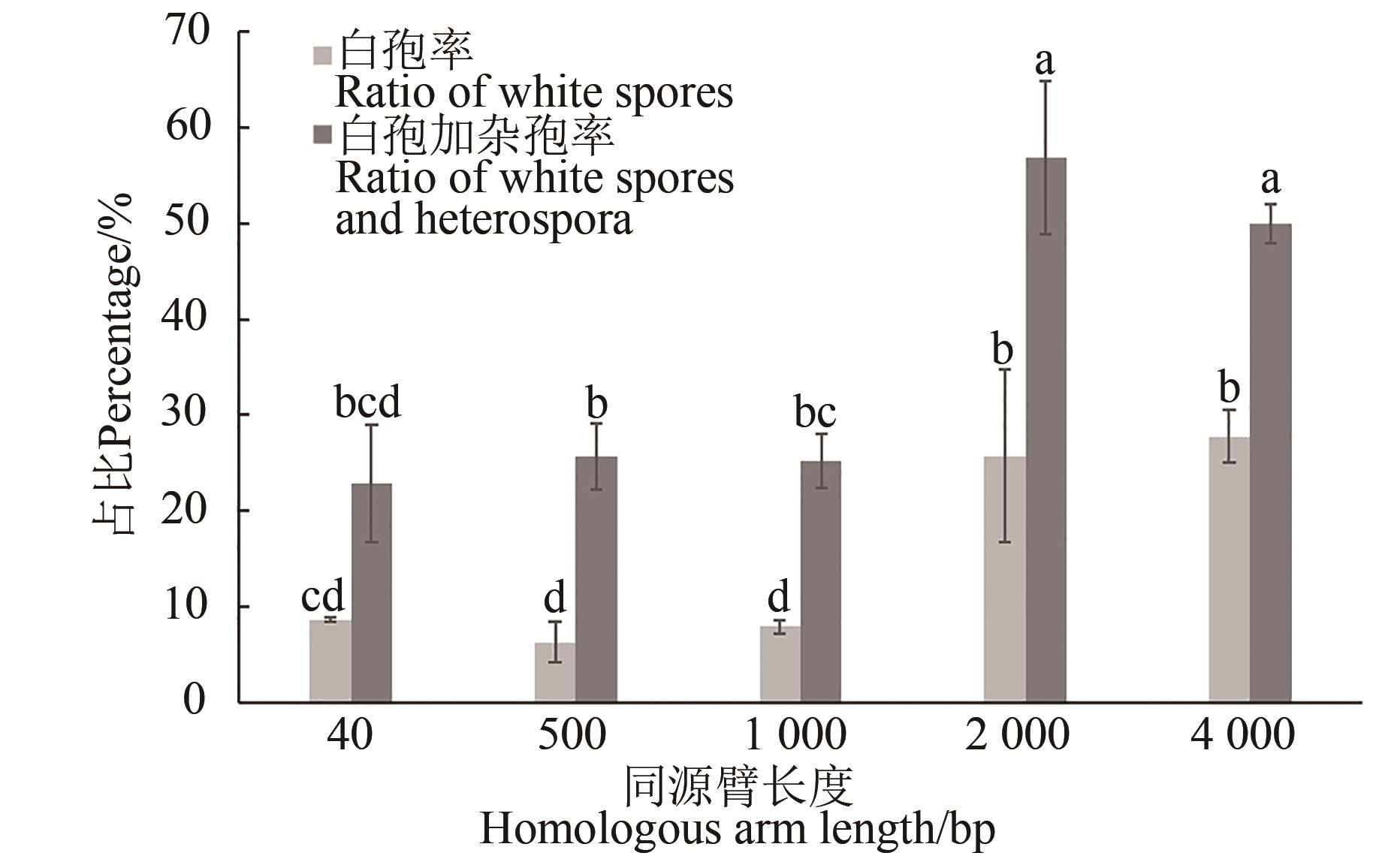
图5 不同长度同源臂的同源重组效率注:不同小写字母表示在P<0.05水平差异显著。
Fig. 5 Homologous recombination efficiency of different lengths of homologous armNote: Different lowercase letters indicate significant differences at P<0.05 level.
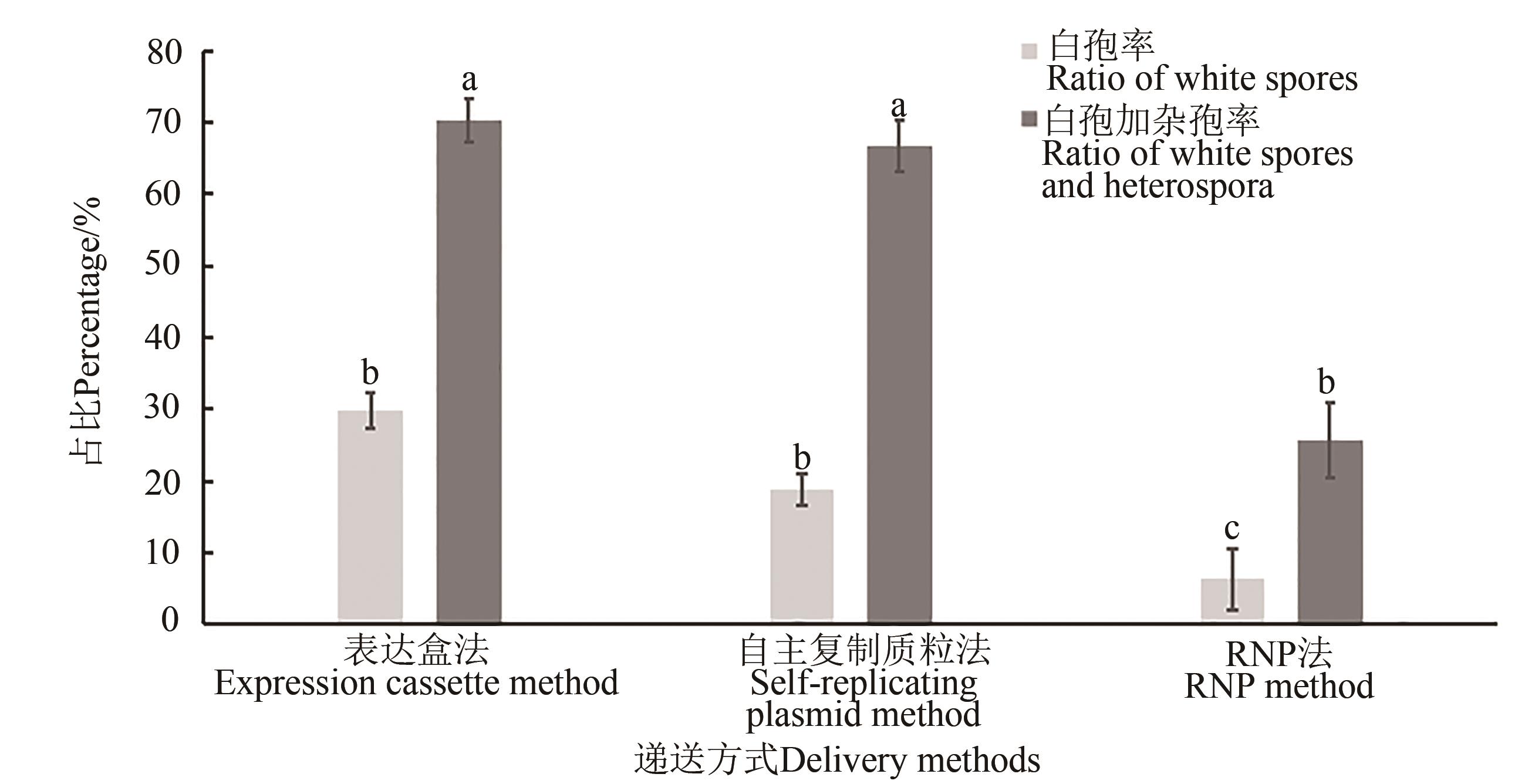
图6 不同递送方式的同源重组效率注:不同小写字母表示在P<0.05水平差异显著。
Fig. 6 HR efficiency of different delivery methodsNote:Different lowercase letters indicate significant difference at P<0.05 level.
| 1 | KHAN S, NADIR S, SHAH Z U, et al.. Biodegradation of polyester polyurethane by Aspergillus tubingensis [J]. Environ. Pollut., 2017, 225(1):469-480. |
| 2 | HUANG D M, SONG Y Y, LIU Y L,et al..A new strain of Aspergillus tubingensis for high-activity pectinase production [J]. Braz. J. Microbiol., 2019,50(1):53-65. |
| 3 | OKADO N, SUGI M, KASAMOTO S, et al.. Safety evaluation of arabinose (arabinan endo-1,5-α-L-arabinanase) from Aspergillus tubingensis [J]. Food Sci. Nutr., 2019, 8(1):456-478. |
| 4 | KRISNAWATI R, CAHYANTO MN, SARDJONO S, et al.. RNA-seq data of Aspergillus tubingensis NBRC 31125 in carbon catabolite repressor related to xylanase production [J/OL]. Data Brief., 2022, 45:108700 [2023-11-20]. . |
| 5 | 郭雷,王聪,郭家才,等.海洋来源塔宾曲霉LWG-42菌株的鉴定及其抗氧化活性化合物的分离[J].微生物学杂志,2017,37(6):30-35. |
| GUO L, WANG C, GUO J C,et al..Identification of marine-derived Aspergillus tubingensis LWG-42 and isolation of its antioxidant active compounds [J]. J. Microbiol., 2017,37(6):30-35. | |
| 6 | WU Y M, YANG X Q, ZHAO T D, et al.. Antifeedant and antifungal activities of metabolites isolated from the coculture of endophytic fungus Aspergillus tubingensis S1120 with red ginseng [J/OL]. Chem Biodivers., 2022, 19(1):e202100608 [2023-11-20]. . |
| 7 | 冯宇航,侯凯丽,姜洋,等.塔宾曲霉L-27菌株代谢产物2,5-呋喃二酮衍生物的分离与除草潜力[J].农药,2023,62(5):387-390. |
| FENG Y H, HOU K L, JIANG Y, et al.. Isolation and herbicidal potential of 2,5-furandione derivatives from Aspergillus tubingensis L-27 metabolites [J]. Agrochemicals, 2023,62(5):387-390. | |
| 8 | SHAN B, HAO R X, XU X Y, et al.. Efficient immobilization behavior and mechanism investigation of Pb(II) by Aspergillus tubingensis [J]. Biotechnol. Lett., 2022,44(5):741-753. |
| 9 | KHAN S, NADIR S, SHAH Z U,et al..Biodegradation of polyester polyurethane by Aspergillus tubingensis [J]. Environ. Pollut., 2017,225:469-480. |
| 10 | MIRHENDI H, ZAREI F, MOTAMEDI M,et al.. Aspergillus tubingensis and Aspergillus niger as the dominant black Aspergillus,use of simple PCR-RFLP for preliminary differentiation [J]. J. De Mycol. Médicale, 2016,26(1):9-16. |
| 11 | 徐圆程.我国南方仓储稻谷中黑曲霉群的分布、产毒与生长特性研究[D].南京:南京财经大学,2021. |
| XU Y C. Distribution, ota-producing and growth characteristics of Aspergillus niger aggregates in stored paddy rice in southern china [D]. Nanjing: Nanjing University of Finance & Economics, 2021. | |
| 12 | SAMSON R A, VISAGIE C M, HOUBRAKEN J, et al.. Phylogeny,identification and nomenclature of the genus Aspergillus [J]. Studies Mycol., 2014,78:141-173. |
| 13 | JIN F J, WANG B T, WANG Z D, et al.. CRISPR/Cas9-Based genome editing and its application in aspergillus species [J]. J Fungi (Basel)., 2022, 8(5):467-478. |
| 14 | SHEN C C, HSU M N, CHANG C W, et al.. Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation [J/OL].Nucl. Acids Res.,2019,47(3):e13 [2023-11-20]. . |
| 15 | KWON M J, SCHÜTZE T, SPOHNER S, et al.. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi [J/OL]. Fungal Biol. Biotechnol., 2019,6:15 [2023-11-20]. . |
| 16 | 石庆楠,曾海英,文安燕,等.贵州薏米致霉菌的分离鉴定[J].食品工业科技,2017,38(16):101-105. |
| SHI Q N, ZENG H Y, WEN A Y,et al..Isolation and identification of moldy fungus from coix seed in Guizhou [J]. Sci. Technol. Food Ind., 2017,38(16):101-105. | |
| 17 | FRISVAD J C, SAMSON R A. Polyphasic taxonomy of Penicillium subgenus Penicillium-a guide to identification of food and airborne terverticillate Penicillia and their mycotoxins [J]. Studies Mycol., 2004, 49:174-186. |
| 18 | 高伟欣,黄火清,赵晶,等.应用于基因编辑的核糖核蛋白复合体的构建与活性验证[J].生物技术通报,2022,38(8):60-68. |
| GAO W X, HUANG H Q, ZHAO J,et al..Construction and activity verification of ribonucleoprotein complex for gene editing [J]. Biotechnol. Bull., 2022,38(8):60-68. | |
| 19 | 孔华忠,齐祖同.曲霉属和散囊菌属的新记录[J].真菌学报, 1995,14 (1):75-78. |
| KONG H Z, QI Z T. New records of Aspergillus and Eurotium in China [J]. Acta Mycol. Sin., 1995, 14(1):75-78. | |
| 20 | SAMSON R, HOUBRAKEN J, KUIJPERS A, et al.. New ochratoxin A or sclerotium producing species in Aspergillus section Nigri [J]. Stud. Mycol., 2004, 50(1):45-61. |
| 21 | FLIPPHI M, MÁRTON A, BÍRÓ V, et al.. Mutations in the second alternative oxidase gene:a new approach to group Aspergillus niger strains [J]. J. Fungi (Basel),2023,9(5):570-582. |
| 22 | BAKER O, TSURKAN S, FU J,et al..The contribution of homology arms to nuclease-assisted genome engineering [J]. Nucl. Acids Res., 2017,45(13):8105-8115. |
| [1] | 杨大兵, 胡亮, 杜雪树, 万丙良, 夏明元, 戚华雄, 李进波. CRISPR/Cas9基因编辑技术创制水稻雄性不育系的研究进展[J]. 中国农业科技导报, 2025, 27(3): 24-34. |
| [2] | 孙志康, 李力群, 郝捷, 吴晗, 吴娜, 郑超, 季嫱, 李选文, 陈晨. CRISPRCas系统在枯草芽孢杆菌基因组编辑中的研究进展[J]. 中国农业科技导报, 2025, 27(2): 24-32. |
| [3] | 翟利敏, 李文通, 冯政, 李华, 裴杨莉. 基因编辑猪的研究现状[J]. 中国农业科技导报, 2022, 24(8): 25-34. |
| [4] | 汪海, 赖锦盛, 王海洋, 李新海. 作物智能设计育种——自然变异的智能组合和人工变异的智能创制[J]. 中国农业科技导报, 2022, 24(6): 1-8. |
| [5] | 陈浩, 周菲, 林拥军. 我国作物新种质创制研究进展[J]. 中国农业科技导报, 2022, 24(12): 112-119. |
| [6] | 闫磊, 张金山, 朱健康, 夏兰琴. 基因编辑技术及其在农作物中的应用进展[J]. 中国农业科技导报, 2022, 24(12): 78-89. |
| [7] | 庄重, 赵龙, 白皓, 毕瑜林, 黄应权, 陈国宏, 常国斌. CRISPR/Cas9技术在家禽育种方面的应用[J]. 中国农业科技导报, 2022, 24(1): 14-23. |
| [8] | 马小倩, 杨涛, 张全, 张洪亮. 水稻新型育种技术研究现状与展望[J]. 中国农业科技导报, 2022, 24(1): 24-30. |
| [9] | 廖嘉明, §, 李春梅§, 张石虎, 李布野, 欧阳昆唏, 陈晓阳. CRISPR/Cas9基因编辑技术的发展及其在植物中的应用[J]. 中国农业科技导报, 2021, 23(12): 20-28. |
| [10] | 薛满德,龙艳,裴新梧*. 基因编辑技术及其在作物育种中的应用与安全管理[J]. 中国农业科技导报, 2018, 20(9): 12-22. |
| [11] | 魏景亮,吴添文,阮进学,牟玉莲*. 基因组编辑技术改良家畜的研究进展[J]. , 2014, 16(1): 32-38. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号