




















中国农业科技导报 ›› 2023, Vol. 25 ›› Issue (10): 22-34.DOI: 10.13304/j.nykjdb.2023.0108
张会1,2( ), 王越越2,3, 赵波2,3, 张丽玲3, 郄倩茹3, 韩渊怀2,3, 李旭凯1,2(
), 王越越2,3, 赵波2,3, 张丽玲3, 郄倩茹3, 韩渊怀2,3, 李旭凯1,2( )
)
收稿日期:2023-02-17
接受日期:2023-06-14
出版日期:2023-10-15
发布日期:2023-10-27
通讯作者:
李旭凯
作者简介:张会 E-mail:huizhang@sxau.edu.cn;
基金资助:
Hui ZHANG1,2( ), Yueyue WANG2,3, Bo ZHAO2,3, Liling ZHANG3, Qianru QIE3, Yuanhuai HAN2,3, Xukai LI1,2(
), Yueyue WANG2,3, Bo ZHAO2,3, Liling ZHANG3, Qianru QIE3, Yuanhuai HAN2,3, Xukai LI1,2( )
)
Received:2023-02-17
Accepted:2023-06-14
Online:2023-10-15
Published:2023-10-27
Contact:
Xukai LI
摘要:
研究植物响应低温胁迫的分子基础及选育与种植耐冷作物品种对于保障粮食安全具有重要的现实意义,但目前谷子(Setaria italica)耐冷相关研究比较匮乏。为深入研究谷子苗期响应低温胁迫的分子机制,利用谷子33份不同发育阶段和不同冷处理时间的RNA-Seq数据,筛选获得32 017个高表达基因并构建基因表达矩阵。利用R软件的WGCNA包构建共表达网络,进一步将网络划分为44个模块,发现谷子冷胁迫应答基因分布于多个模块中。选取包含冷胁迫基因数目排名前3的模块进一步分析,对预测到的靶基因进行GO富集分析,并对此3个模块进行基因调控网络的构建及核心基因注释,发现大多候选基因与非生物胁迫响应相关。以上结果为谷子抗逆研究提供了新思路,发掘的冷胁迫关键基因为谷子耐冷新品种选育提供了基因资源。
中图分类号:
张会, 王越越, 赵波, 张丽玲, 郄倩茹, 韩渊怀, 李旭凯. 基于WGCNA的谷子苗期冷胁迫应答基因网络构建与核心因子发掘[J]. 中国农业科技导报, 2023, 25(10): 22-34.
Hui ZHANG, Yueyue WANG, Bo ZHAO, Liling ZHANG, Qianru QIE, Yuanhuai HAN, Xukai LI. Identification of Co-expression Genes Related to Cold Stress in Foxtail Millet by WGCNA[J]. Journal of Agricultural Science and Technology, 2023, 25(10): 22-34.
基因 Gene | 上游引物 Forward primer (5’-3’) | 下游引物 Reverse primer (5’-3’) |
|---|---|---|
| Actin | TGCTCAGTGGAGGCTCAACA | CCAGACACTGTACTTGCGCTC |
| Si2g38970 | GCCGGATGTTCTGGTGGTAT | CCTTGAAACTGCGTTCACCG |
| Si3g02940 | TATACCGCACCAGCGATGAC | TCCTTTGGGACCTGATTGCC |
| Si3g08050 | CTCCGACACCGACTCATTCT | AAGAAGATGACCGACACCCG |
| Si5g36480 | CAGCATCCCCAGAGAGCAAA | GCACCTTGGGAACTGTGTCT |
| Si6g03220 | GAGCATCGTGCTGTCAGAGG | CGTTGGGTCCAGGAACAGAT |
| Si9g50340 | CCTCACAGACATGCCCATCG | TCGACATCGACTCAGTTGCC |
表1 RT-qPCR引物序列
Table 1 RT-qPCR primer sequence
基因 Gene | 上游引物 Forward primer (5’-3’) | 下游引物 Reverse primer (5’-3’) |
|---|---|---|
| Actin | TGCTCAGTGGAGGCTCAACA | CCAGACACTGTACTTGCGCTC |
| Si2g38970 | GCCGGATGTTCTGGTGGTAT | CCTTGAAACTGCGTTCACCG |
| Si3g02940 | TATACCGCACCAGCGATGAC | TCCTTTGGGACCTGATTGCC |
| Si3g08050 | CTCCGACACCGACTCATTCT | AAGAAGATGACCGACACCCG |
| Si5g36480 | CAGCATCCCCAGAGAGCAAA | GCACCTTGGGAACTGTGTCT |
| Si6g03220 | GAGCATCGTGCTGTCAGAGG | CGTTGGGTCCAGGAACAGAT |
| Si9g50340 | CCTCACAGACATGCCCATCG | TCGACATCGACTCAGTTGCC |
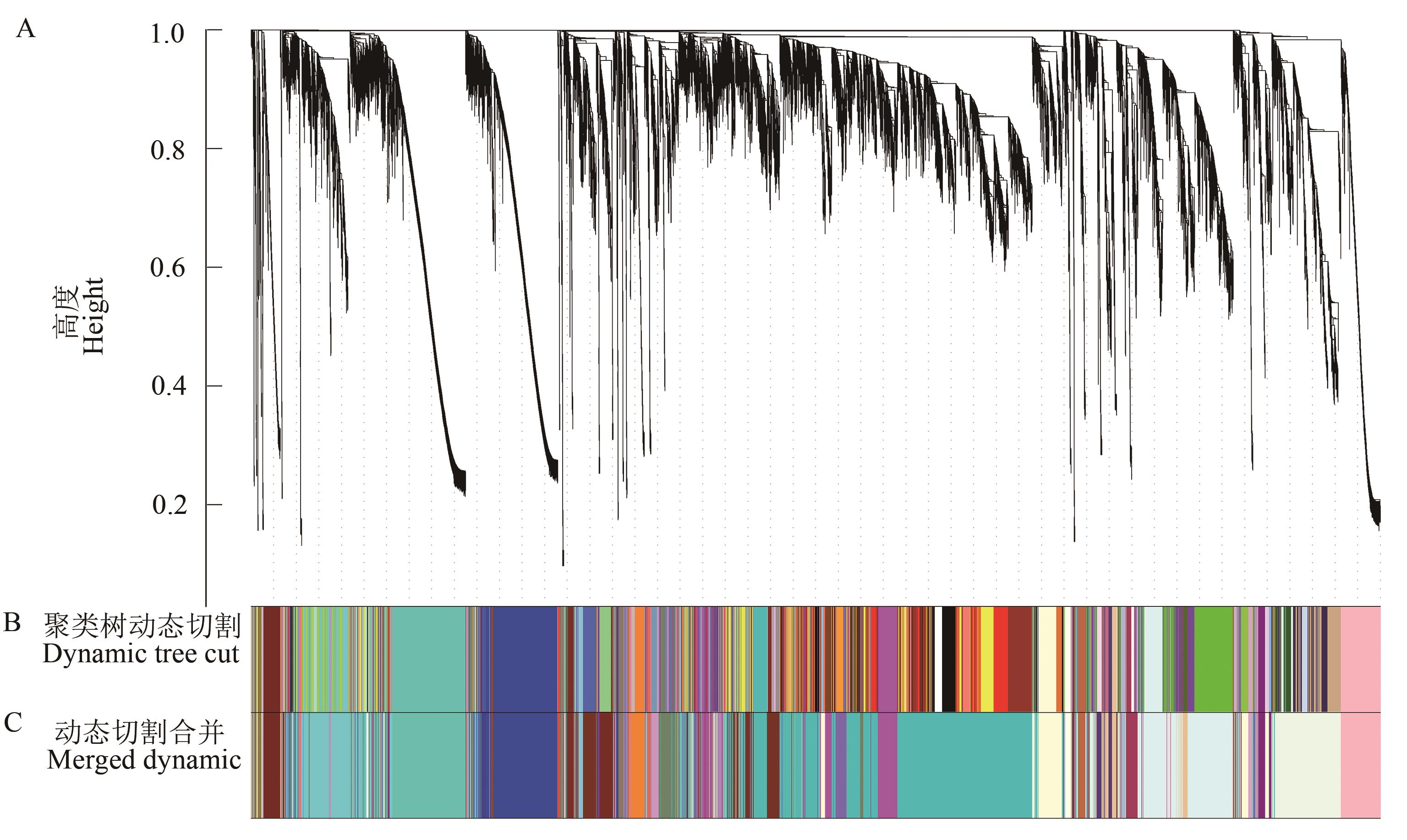
图1 基因聚类树和样品分割A: 基于拓扑重叠构建的基因聚类树;B: 基于各基因的表达量计算划分的模块;C: 在B模块的基础上合并相似模块后的结果
Fig. 1 Gene cluster dendrogram and module detectingA: Clustering of genes based on the topological overlap; B: Dynamic tree cut represents the module divided according to the expression of each gene, colors represent modules; C: Merged dynamic is the result of merging similar modules according to the dynamic tree cut
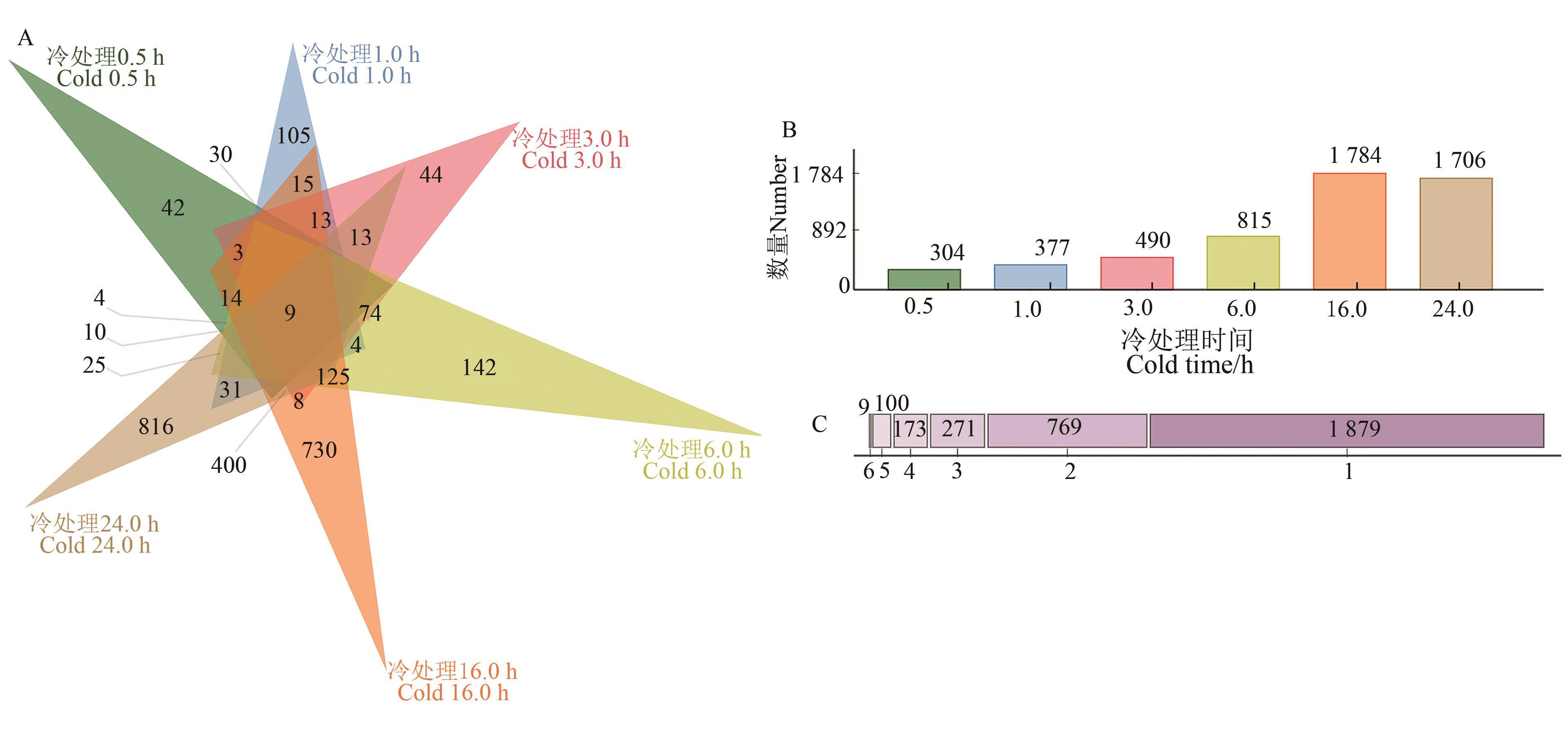
图2 谷子6个冷胁迫时间差异表达基因分析A:6个冷胁迫处理条件下差异表达基因的韦恩图;B:6个冷胁迫处理条件下的差异基因数量;C:在6个冷胁迫处理中特异或同时出现的差异基因数目,1~6表示差异基因在6个冷胁迫处理中出现的频次
Fig. 2 Analysis of differentially expressed genes in foxtail millet under cold stressA: Venn diagram of number of the differentially expressed genes among 6 different cold stress conditions; B: Number of the differentially expressed genes in 6 different cold stress conditions; C:Number of the differentially expressed genes that are unique or shared under 6 different cold stress conditions,1~6 in the X-axis mean the frequency of the differentially expressed genes among 6 different cold stress conditions
基因 Gene | 水稻同源基因 Homologous gene in rice | 功能注释 Function annotation | 拟南芥同源基因 Homologous gene in Arabidopsis thaliana | 功能注释 Function annotation |
|---|---|---|---|---|
| Si2g38970 | LOC_Os07g42650 | 保守假蛋白 Conserved hypothetical protein | At4g33980 | 冷调控基因28 Cold-regulate gene 28 |
| Si3g01950 | LOC_Os04g58710 | 4-香豆酸-Co-A连接酶类蛋白 4-coumarate-Co-A ligase (4CL) like protein | At1g77230 | 四肽重复类超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein |
| Si3g02940 | LOC_Os04g57550 | 多胺氧化酶,种子萌发 Polyamine oxidase (PAO2), seed germination | At2g43020 | 多肽氧化酶2 Polyamine oxidase 2, ATPAO2 |
| Si3g08050 | LOC_Os05g11810 | 类赤霉素2-β-双加氧酶 Similar to gibberellin 2-beta-dioxygenase | At1g30040 | 赤霉素2-氧化酶 Gibberellin 2-oxidase |
| Si5g36480 | LOC_Os01g60600 | WRKY 转录因子 WRKY transcription factor | At5g22570 | WRKY DNA 结合蛋白38 WRKY DNA-binding protein 38 |
| Si5g39150 | LOC_Os05g37060 | R-R型MYB类转录因子 R-R-type MYB-like transcription factor | At5g67420 | 含LOB结构域的蛋白37 LOB-domain-containing protein 37 |
| Si6g03220 | LOC_Os04g52090 | APETALA 2 类转录因子 APETALA-2-like transcription factor | At5g44210 | ERF 结构域蛋白9 ERF domain protein- 9 |
| Si7g06300 | LOC_Os04g23550 | 螺旋-环-螺旋转录激活因子 Basic helix-loop-helix (bHLH) transcription activator | At4g16430 | bHLH DNA 结合超家族蛋白 bHLH DNA-binding superfamily protein |
| Si9g50340 | LOC_Os07g42370 | 含TIFY结构域蛋白 TIFY domain containing protein | At1g74950 | TIFY结构域/发散CCT基序家族蛋白 TIFY domain/divergent CCT motif family protein |
表2 谷子6个冷胁迫处理条件下同时差异表达基因
Table 2 Differentially expressed genes under 6 cold stress conditions in foxtail millet
基因 Gene | 水稻同源基因 Homologous gene in rice | 功能注释 Function annotation | 拟南芥同源基因 Homologous gene in Arabidopsis thaliana | 功能注释 Function annotation |
|---|---|---|---|---|
| Si2g38970 | LOC_Os07g42650 | 保守假蛋白 Conserved hypothetical protein | At4g33980 | 冷调控基因28 Cold-regulate gene 28 |
| Si3g01950 | LOC_Os04g58710 | 4-香豆酸-Co-A连接酶类蛋白 4-coumarate-Co-A ligase (4CL) like protein | At1g77230 | 四肽重复类超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein |
| Si3g02940 | LOC_Os04g57550 | 多胺氧化酶,种子萌发 Polyamine oxidase (PAO2), seed germination | At2g43020 | 多肽氧化酶2 Polyamine oxidase 2, ATPAO2 |
| Si3g08050 | LOC_Os05g11810 | 类赤霉素2-β-双加氧酶 Similar to gibberellin 2-beta-dioxygenase | At1g30040 | 赤霉素2-氧化酶 Gibberellin 2-oxidase |
| Si5g36480 | LOC_Os01g60600 | WRKY 转录因子 WRKY transcription factor | At5g22570 | WRKY DNA 结合蛋白38 WRKY DNA-binding protein 38 |
| Si5g39150 | LOC_Os05g37060 | R-R型MYB类转录因子 R-R-type MYB-like transcription factor | At5g67420 | 含LOB结构域的蛋白37 LOB-domain-containing protein 37 |
| Si6g03220 | LOC_Os04g52090 | APETALA 2 类转录因子 APETALA-2-like transcription factor | At5g44210 | ERF 结构域蛋白9 ERF domain protein- 9 |
| Si7g06300 | LOC_Os04g23550 | 螺旋-环-螺旋转录激活因子 Basic helix-loop-helix (bHLH) transcription activator | At4g16430 | bHLH DNA 结合超家族蛋白 bHLH DNA-binding superfamily protein |
| Si9g50340 | LOC_Os07g42370 | 含TIFY结构域蛋白 TIFY domain containing protein | At1g74950 | TIFY结构域/发散CCT基序家族蛋白 TIFY domain/divergent CCT motif family protein |
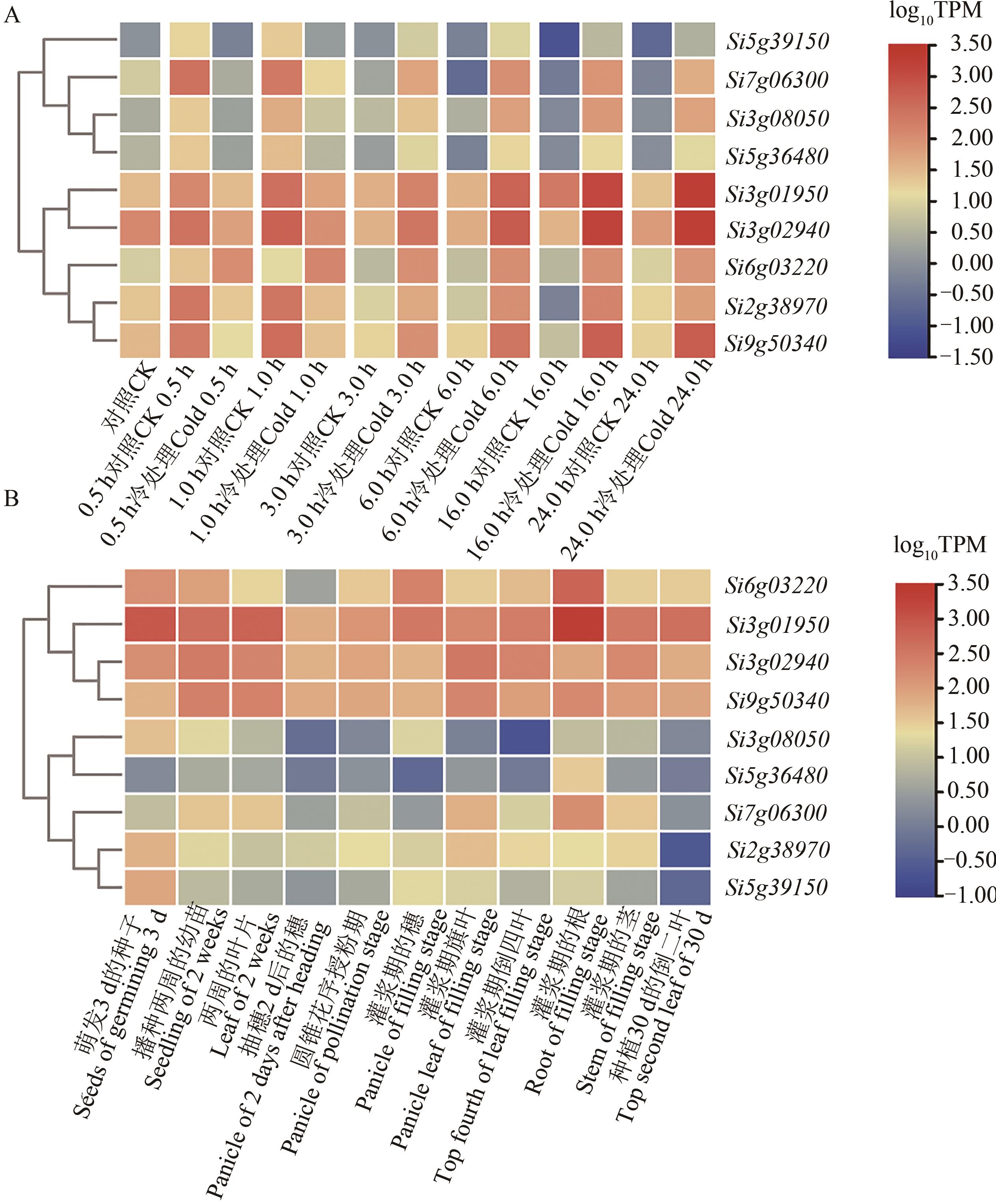
图3 差异表达基因的表达模式A:9个差异表达基因在‘豫谷1号’6个冷胁迫处理条件下的表达模式; B:9个差异表达基因在突变体xiaomi多个生育期内的表达模式
Fig. 3 Expression patterns of differentially expressed genesA: Expression patterns of the 9 differentially expressed genes in ‘Yugu 1’ among 6 cold stress conditions; B: Expression patterns of the 9 differentially expressed genes in mutant xiaomi during 11 developmental stages

图4 模块特有的基因GO富集结果A:Darkturquoise模块; B:Turquoise模块;C:Blue模块
Fig. 4 GO enrichment results of module specific geneA: Darkturquoise module; B: Turquoise module; C: Blue module
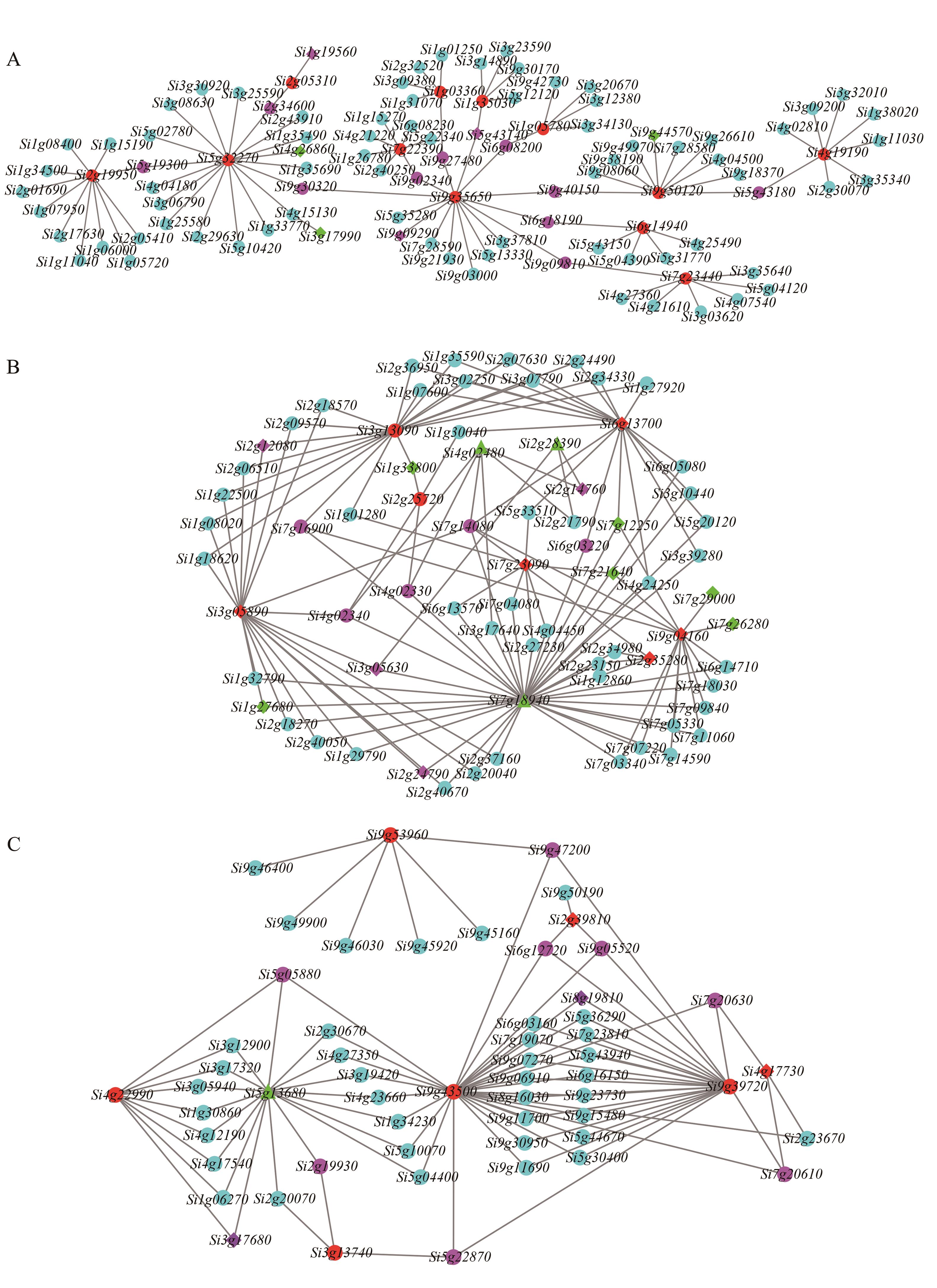
图5 3个模块中与冷胁迫相关基因的共表达网络A:Darkturquoise模块; B:Turquoise模块;C:Blue模块。图中红色菱形表示既注释到转录因子又属于网络核心基因,红色圆形表示注释到的核心基因,紫色圆形表示注释的候选基因,紫色菱形表示既是注释的候选基因又是转录因子,绿色圆形表示结合基序分析的转录因子,绿色菱形表示既是结合基序分析的转录因子又是候选基因,绿色三角形表示既是结合基序分析的转录因子又是核心基因
Fig. 5 Co-expression network of genes relative to cold stress in 3 modulesA: Darkturquoise module; B: Turquoise module; C: Blue module. The red diamond indicates the core gene that is both annotated to the transcription factor and belongs to the network and red circle indicates the annotated core genes, the purple circle indicates the annotated candidate genes and the purple diamond represents annotated candidate gene transcription factors, the green circle indicates transcription factors associated with motif analysis, the green diamond represents annotated candidate gene transcription factors and binding motif analysis transcription factors, the green triangle represents both transcription factors that bind to motif analysis and core genes
模块 Module | 基因名称 Gene name | 水稻Rice | 拟南芥Arabidopsis thaliana | |||
|---|---|---|---|---|---|---|
同源基因 Homologous gene | 功能注释 Function annotation | 同源基因 Homologous gene | 功能注释 Function annotation | |||
| Darkturquoise | Si3g17990 (TCP) | LOC_Os07g05720 | TCP 家族转录因子 TCP family transcription factor | At1g30210 | TCP 家族24 TCP family 24 | |
| Si7g22390 | LOC_Os04g49510 | CAMK-CAMK27钙/钙调素依赖性蛋白激酶 CAMK_CAMK_like.27-CAMK includes calcium/calmodulin depedent protein kinases | At2g17290 | 钙依赖性蛋白激酶家族蛋白 Calcium-dependent protein kinase family protein | ||
| Si9g50120 | LOC_Os03g08570 | 胺氧化酶,含黄素,含结构域蛋白 Amine oxidase, flavin-containing, domain containing protein | At4g14210 | 八氢番茄红素脱氢酶3 Phytoene desaturase 3 | ||
| Si1g03360 | LOC_Os01g10840 | CGMC_GSK.2-CGMC 激酶 CGMC_GSK.2-CGMC includes CDA, MAPK, GSK3, and CLKC kinases | At1g06390 | GSK3/SHAGGY 类蛋白激酶1 GSK3/SHAGGY-like protein kinase 1 | ||
| Turquoise | Si4g02480 (ERF) | LOC_Os09g35010 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | At5g51990 | 核碱、核苷、核苷酸和核酸代谢过程 Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | |
| Si2g28390 (ERF) | LOC_Os09g35030 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | — | — | ||
| Si7g18940 (ERF) | LOC_Os09g20350 | 乙烯应答转录因子 Ethylene-responsive transcription factor | — | — | ||
| Si3g13090 | LOC_Os05g50710 | 晚期胚胎蛋白 Late embryogenesis abundant protein | At2g46140 | 晚期胚胎蛋白 Late embryogenesis abundant protein | ||
| Blue | Si4g17730 (bZIP) | LOC_Os06g45140 | 含bZIP转录因子结构域的 蛋白 bZIP transcription factor domain containing protein | — | — | |
| Si2g39810 (bHLH) | LOC_Os07g43530 | 螺旋-环-螺旋DNA结合蛋白 Helix-loop-helix DNA-binding domain containing protein | — | — | ||
| Si5g13680 (WRKY) | LOC_Os02g08440 | WRKY71转录因子 WRKY71 transcription factor | At1g80840 | 病原体诱导转录因子,体外结合 W-box 序列 Pathogen-induced transcription factor. Binds W-box sequences in vitro | ||
| Si9g43500 | LOC_Os03g17690 | 抗坏血酸过氧化物酶1 Ascorbate peroxidase 1 | At1g07890 | 抗坏血酸过氧化物酶1 Ascorbate peroxidase 1 | ||
表3 3个模块中与冷胁迫相关的核心基因的功能注释
Table 3 Functional annotation of hub genes relative to cold stress in 3 modules
模块 Module | 基因名称 Gene name | 水稻Rice | 拟南芥Arabidopsis thaliana | |||
|---|---|---|---|---|---|---|
同源基因 Homologous gene | 功能注释 Function annotation | 同源基因 Homologous gene | 功能注释 Function annotation | |||
| Darkturquoise | Si3g17990 (TCP) | LOC_Os07g05720 | TCP 家族转录因子 TCP family transcription factor | At1g30210 | TCP 家族24 TCP family 24 | |
| Si7g22390 | LOC_Os04g49510 | CAMK-CAMK27钙/钙调素依赖性蛋白激酶 CAMK_CAMK_like.27-CAMK includes calcium/calmodulin depedent protein kinases | At2g17290 | 钙依赖性蛋白激酶家族蛋白 Calcium-dependent protein kinase family protein | ||
| Si9g50120 | LOC_Os03g08570 | 胺氧化酶,含黄素,含结构域蛋白 Amine oxidase, flavin-containing, domain containing protein | At4g14210 | 八氢番茄红素脱氢酶3 Phytoene desaturase 3 | ||
| Si1g03360 | LOC_Os01g10840 | CGMC_GSK.2-CGMC 激酶 CGMC_GSK.2-CGMC includes CDA, MAPK, GSK3, and CLKC kinases | At1g06390 | GSK3/SHAGGY 类蛋白激酶1 GSK3/SHAGGY-like protein kinase 1 | ||
| Turquoise | Si4g02480 (ERF) | LOC_Os09g35010 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | At5g51990 | 核碱、核苷、核苷酸和核酸代谢过程 Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | |
| Si2g28390 (ERF) | LOC_Os09g35030 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | — | — | ||
| Si7g18940 (ERF) | LOC_Os09g20350 | 乙烯应答转录因子 Ethylene-responsive transcription factor | — | — | ||
| Si3g13090 | LOC_Os05g50710 | 晚期胚胎蛋白 Late embryogenesis abundant protein | At2g46140 | 晚期胚胎蛋白 Late embryogenesis abundant protein | ||
| Blue | Si4g17730 (bZIP) | LOC_Os06g45140 | 含bZIP转录因子结构域的 蛋白 bZIP transcription factor domain containing protein | — | — | |
| Si2g39810 (bHLH) | LOC_Os07g43530 | 螺旋-环-螺旋DNA结合蛋白 Helix-loop-helix DNA-binding domain containing protein | — | — | ||
| Si5g13680 (WRKY) | LOC_Os02g08440 | WRKY71转录因子 WRKY71 transcription factor | At1g80840 | 病原体诱导转录因子,体外结合 W-box 序列 Pathogen-induced transcription factor. Binds W-box sequences in vitro | ||
| Si9g43500 | LOC_Os03g17690 | 抗坏血酸过氧化物酶1 Ascorbate peroxidase 1 | At1g07890 | 抗坏血酸过氧化物酶1 Ascorbate peroxidase 1 | ||
| 1 | WANG C F, CHEN J F, ZHI H, et al.. Population genetics of foxtail millet and its wild ancestor [J/OL]. BMC Genetics, 2010, 11:90 [2023-01-10]. . |
| 2 | BENNETZEN J L, SCHMUTZ J, WANG H, et al.. Reference genome sequence of the model plant Setaria [J]. Nat. Biotechnol., 2012, 30(6):555-561. |
| 3 | ZHANG G Y, LIU X, QUAN Z W, et al.. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential [J]. Nat. Biotechnol., 2012, 30(6):549-554. |
| 4 | YANG Z R, ZHANG H S, LI X K, et al.. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system [J]. Nat. Plants, 2020, 6(9):1167-1178. |
| 5 | DIAO X M, SCHNABLE J, BENNETZEN J L, et al.. Initiation of Setaria as a model plant [J]. Front. Agric. Sci. Eng., 2014, 1(1):16-20. |
| 6 | 曹爽.玉米抗冷基因ZmGDH2的功能分析及遗传转化的研究[D].长春:吉林大学,2017. |
| CAO S. Cloning, characterization, and transformation of cold resistant gene ZmGDH 2 in maize (Zea mays L.) [D]. Changchun: Jilin University, 2017. | |
| 7 | 秦东玲,李钊,尉菊萍,等.作物抗冷性及其化学控制机理研究进展[J].作物杂志,2016,32(4):26-35. |
| QIN D L, LI Z, YU J P, et al.. Progress on cold resistance and chemical control mechanism of crops [J]. Crops, 2016, 32(4):26-35. | |
| 8 | LI J L, ZENG Y W, PAN Y H, et al.. Stepwise selection of natural variations at CTB2 and CTB4a improves cold adaptation during domestication of japonica rice [J]. New phytol., 2021, 231(3):1056-1072. |
| 9 | MA Y, DAI X Y, XU Y Y, et al.. COLD1 confers chilling tolerance in rice [J]. Cell, 2015, 160(6):1209-1221. |
| 10 | LUO W, HUAN Q, XU Y Y, et al.. Integrated global analysis reveals a vitamin E-vitamin K1 sub-network, downstream of COLD 1, underlying rice chilling tolerance divergence [J/OL]. Cell Rep., 2021, 36(3):109397 [2023-01-10]. . |
| 11 | WANG J C, REN Y L, LIU X, et al.. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice [J]. Mol. Plant., 2021, 14(2):315-329. |
| 12 | LIU J, SHI Y, YANG S H. Insights into the regulation of C-repeat binding factors in plant cold signaling [J]. J. Integr. Plant. Biol., 2018, 60(9):780-795. |
| 13 | LI J H, ZHANG Z Y, CHONG K, et al.. Chilling tolerance in rice: past and present [J/OL]. J. Plant. Physiol., 2022, 268(8):153576 [2023-01-10]. . |
| 14 | 郑加兴,覃嘉明,王兵伟,等.玉米芽期耐冷性QTL鉴定与分子定位[J/OL].分子植物育种,2022:1-12.. |
| ZHENG J X, QIN J M, WANG B W, et al.. Identification and mapping of quantitative trait loci for cold tolerance at the budding stage in maize [J/OL]. Mol. Plant Breeding, 2022: 1-12 [2023-01-]. . | |
| 15 | ZHANG B, HORVATH S. A general framework for weighted gene co-expression network analysis [J/OL]. Stat. Appl. Genet. Mol. Biol., 2005, 4(1):17 [2023-01-10]. . |
| 16 | MENG X X, LIANG Z K, DAI X R, et al.. Predicting transcriptional responses to cold stress across plant species [J/OL]. Proc. Natl. Acad. Sci. USA, 2021, 118(10):e2026330118 [2023-01-10]. . |
| 17 | GOLDBERG D H, VICTOR J D, GARDNER E P, et al.. Spike train analysis toolkit: enabling wider application of information-theoretic techniques to neurophysiology [J]. Neuroinformatics, 2009, 7(3):165-178. |
| 18 | KROLL K W, MOKARAM N E, PELLETIER A R, et al.. Quality control for RNA-Seq (QuaCRS): an integrated quality control pipeline [J]. Cancer Inform., 2014, 13(S3):7-14. |
| 19 | BOLGER A M, LOHSE M, USADEL B. Trimmomatic: a flexible trimmer for illumina sequence data [J]. Bioinformatics, 2014, 30(15):2114-2120. |
| 20 | KIM D, LANGMEAD B, SALZBERG S L. HISAT: a fast spliced aligner with low memory requirements [J]. Nat. Methods, 2015, 12(4):357-360. |
| 21 | LIAO Y, SMYTH G K, SHI W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features [J]. Bioinformatics, 2014, 30(7):923-930. |
| 22 | 李旭凯,李任建,张宝俊.利用WGCNA鉴定非生物胁迫相关基因共表达网络[J].作物学报,2019,45(9):1349-1364. |
| LI X K, LI R J, ZHANG B J. Identification of rice stress-related gene co-expression modules by WGCNA [J]. Acta Agron. Sin., 2019, 45(9):1349-1364. | |
| 23 | LANGFELSER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis [J/OL]. BMC Bioinform., 2008, 9:559 [2023-01-10]. . |
| 24 | ALTSCHUL S F, MADDEN T L, SCHAFFER A A, et al.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs [J]. Nucl. Acids Res., 1997, 25(17):3389-3402. |
| 25 | BARDOU P, MARIETTR J, ESCUDIE F, et al.. jvenn: an interactive venn diagram viewer [J/OL]. BMC Bioinform., 2014, 15(1):293 [2023-01-10]. . |
| 26 | CHEN C J, XIA R, CHEN H, et al.. TBtools, a Toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface [J/OL]. BioRxiv, 2018, 14:48 [2023-01-10]. . |
| 27 | SU G, MORRIS J H, DEMCHAK B, et al.. Biological network exploration with cytoscape 3 [J/OL]. Curr. Prot. Bioinform., 2014, 47(1):47 [2023-01-10]. . |
| 28 | THOMAS D S, KENNETH J L. Analyzing real-time PCR data by the comparative CT method [J]. Nat. Protoc., 2008, 3:1101-1108. |
| 29 | MOZOI J, SHINOZAKI K, YAMAGUCHI K. AP2/ERF family transcription factors in plant abiotic stress responses [J]. Biochim. Biophys. Acta, 2012, 1819(2):86-96. |
| 30 | GONG Z Z, XIONG L M, SHI H Z, et al.. Plant abiotic stress response and nutrient use efficiency [J]. Sci China Life Sci., 2020, 63(5):635-674. |
| 31 | ZHANG T T, XU Y, DING Y D, et al.. Identification and expression analysis of WRKY gene family in response to abiotic stress in Dendrobium catenatum [J/OL]. Front. Genet., 2022, 13:800019 [2023-01-10]. . |
| 32 | 洪林,杨蕾,杨海健,等.AP2/ERF转录因子调控植物非生物胁迫响应研究进展[J].植物学报,2020,55(4):481-496. |
| HONG L, YANG L, YANG H J, et al.. Research advances in AP2/ERF transcription factors in regulating plant responses to abiotic stress [J]. Bull Bot., 2020, 55(4):481-496. | |
| 33 | 宫照龙,卓勤,吴景欢,等.大豆脱水应答元件结合蛋白(GmDREB1)的非融合原核表达及纯化[J].卫生研究,2013,42(3):392-398. |
| GONG Z L, ZHUO Q, WU J H, et al.. Study on no-fusion expression and purification of glycine max dehydration-responsive element-binding protein (GmDREB1) expressed in E. coli [J]. J. Hygiene Res., 2013, 42(3):392-398. | |
| 34 | 张计育,王庆菊,郭忠.植物AP2/ERF类转录因子研究进展[J].遗传,2012,34(7):835-847. |
| ZHANG J Y, WANG Q J, GUO Z. Progresses on plant AP2/ERF transcription factors [J]. Hereditas, 2012, 34(7):835-847. | |
| 35 | ZHANG H, NI L, LIU Y P, et al.. The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice [J]. J. Integr. Plant. Biol., 2012, 54(7):500-510. |
| 36 | 刘文文,李文学.植物bHLH转录因子研究进展[J].生物技术进展,2013,3(1):7-11. |
| LIU W W, LI W X. Progress of plant bHLH transcription factor [J]. Curr. Biotechnol., 2013, 3(1):7-11. | |
| 37 | 应炎标,朱友银,郭卫东,等.樱桃bHLH转录因子家族基因鉴定及表达分析[J].分子植物育种,2018,16(14):4559-4568. |
| YING Y B, ZHU Y Y, GUO W D, et al.. Identification and expression analysis of bHLH transcription factor gene family in Prunus pseudocerasus Lindl [J]. Mol. Plant Breeding, 2018, 16(14):4559-4568. | |
| 38 | BAI B, WU J, SHENG W T, et al.. Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress [J/OL]. Int. J. Mol. Sci., 2015, 16(5):11398 [2023-01-10]. . |
| 39 | 王凤珠,陈琴芳.过表达水稻MYB转录因子OsARM1对转基因拟南芥冷冻耐受性的提高[J].中国科技论文,2017,12(18):2103-2108. |
| WANG F Z, CHEN Q F. Overexpression of rice MYB transcription factor OsARM1 enhances freezing tolerance in Arabidopsis [J]. China. Sci. Paper, 2017, 12(18):2103-2108. | |
| 40 | 崔荣秀,张议文,陈晓倩,等.植物bZIP参与胁迫应答调控的最新研究进展[J].生物技术通报,2019,35(2):143-155. |
| CUI R X, ZHANG Y W, CHEN X Q, et al.. The latest research progress on the stress responses of bZIP involved in plants [J]. Biotechnol. Bull., 2019, 35(2):143-155. | |
| 41 | TRIPATHI P, RABARA R C, RUSHTON P J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants [J]. Planta, 2014, 239(2):255-266. |
| 42 | LIANG X, ABEL S, KELLER J A, et al.. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana [J]. Proc. Natl. Acad. Sci. USA, 1992, 89(22):11046-11050. |
| 43 | ARTECA J M, ARTECA R N. A multi-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase (ACS6) in mature Arabidopsis leaves [J]. Plant. Mol. Biol., 1999, 39(2):209-219. |
| 44 | SHI Y T, TIAN S W, HOU L Y, et al.. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis [J]. Plant Cell, 2012, 24(6):2578-2595. |
| [1] | 李瑞珍, 姚建民, 王忠祥, 高凤翔, 窦贵新, 杨瑞平, 刘钊, 张继, 张振宇. 全生物降解渗水地膜覆盖冬播谷子产量结构关系分析[J]. 中国农业科技导报, 2023, 25(5): 185-191. |
| [2] | 焦雄飞, 于晋, 冯乐勇, 郭耀东, 樊丽生. 不同播期对谷子DUS测试性状的影响[J]. 中国农业科技导报, 2022, 24(8): 55-64. |
| [3] | 赵晋锋, 余爱丽, 李颜方, 杜艳伟, 王高鸿, 王振华. 谷子SiCBL3对非生物胁迫响应特征分析[J]. 中国农业科技导报, 2022, 24(11): 68-75. |
| [4] | 郭瑞锋, 任月梅, 杨忠, 刘贵山, 任广兵, 张绶, 朱文娟. 草甘膦铵盐诱导谷子雄性不育的转录组分析[J]. 中国农业科技导报, 2022, 24(10): 35-43. |
| [5] | 鱼冰星, 王宏富, 王振华, 张鹏, 成锴, 余爱丽, 闫海丽, 鱼冰洁. 多效唑对谷子茎秆特征及抗倒性的影响[J]. 中国农业科技导报, 2021, 23(8): 37-44. |
| [6] | 李冉, 刘宇航, 梁杉, 张敏, . 硒肥对谷子产量因子及其籽粒富硒效果的影响[J]. 中国农业科技导报, 2021, 23(6): 140-146. |
| [7] | 张宇杰, 郭平毅, 郭美俊, 周浩, 原向阳, 董淑琦, 王玉国. 外源硒矿粉对谷子保护酶活性、产量和籽粒中硒含量的影响[J]. 中国农业科技导报, 2021, 23(5): 153-159. |
| [8] | 岳琳祺,郭佳晖,白雄辉,施卫萍,郭平毅*,郭杰*. 叶面喷施硒肥对不同基因型谷子农艺性状及籽粒硒含量的影响[J]. 中国农业科技导报, 2021, 23(4): 154-163. |
| [9] | 田岗, 刘鑫, 王玉文, 刘永忠, 李会霞, 成锴, 王振华, 刘红. 遮光处理对谷子农艺性状、小米品质及蒸煮特性的影响[J]. 中国农业科技导报, 2021, 23(11): 47-54. |
| [10] | 相吉山1,张恒儒2,刘涵1,索良喜2,贾姝婧1,张颖1,史景奇1,胡利喆1,蔡一宁1. 不同生态区谷子种质资源表型比较分析[J]. 中国农业科技导报, 2020, 22(9): 31-41. |
| [11] | 李会霞1§,郑植尹2§,田岗1,刘鑫1,王玉文1,刘红1,史关燕3*. 7个谷子杂交种及其亲本的抗旱性分析[J]. 中国农业科技导报, 2020, 22(7): 20-28. |
| [12] | 李智,王宏富*,王钰云,杨净,鱼冰星,黄珊珊. 谷子大豆间作对作物光合特性及产量的影响[J]. 中国农业科技导报, 2020, 22(6): 168-175. |
| [13] | 王钰云,王宏富*,李智,段宏凯,黄珊珊. 谷子花生间作对谷子光合特性及产量的影响[J]. 中国农业科技导报, 2020, 22(5): 153-165. |
| [14] | 王振华,刘鑫*,余爱丽,成锴,李会霞,田岗,王玉文,陈新霞,张鹏,刘红. 不同谷子品种萌发期对干旱胁迫生理响应的变化及抗旱指标筛选[J]. 中国农业科技导报, 2020, 22(12): 39-49. |
| [15] | 秦岭,陈二影,杨延兵,张艳亭,孔清华,张华文,王海莲,王润丰,管延安*. 干旱和复水对不同耐旱型谷子品种苗期生理指标的影响[J]. 中国农业科技导报, 2019, 21(3): 146-151. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号