




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (1): 80-95.DOI: 10.13304/j.nykjdb.2023.0156
李贤国1,2( ), 戴麒1, 王泽鹏1,2, 陈兆龙1,2, 闫会转2, 李宁1(
), 戴麒1, 王泽鹏1,2, 陈兆龙1,2, 闫会转2, 李宁1( )
)
收稿日期:2023-03-06
接受日期:2023-06-14
出版日期:2025-01-15
发布日期:2025-01-21
通讯作者:
李宁
作者简介:李贤国 E-mail:15099198840@163.com;
基金资助:
Xianguo LI1,2( ), Qi DAI1, Zepeng WANG1,2, Zhaolong CHEN1,2, Huizhuan YAN2, Ning LI1(
), Qi DAI1, Zepeng WANG1,2, Zhaolong CHEN1,2, Huizhuan YAN2, Ning LI1( )
)
Received:2023-03-06
Accepted:2023-06-14
Online:2025-01-15
Published:2025-01-21
Contact:
Ning LI
摘要:
CCCH(C3H)型锌指蛋白是进化上保守的RNA结合蛋白质,主要与DNA和RNA结合来调控基因表达,不仅参与植物的生长发育和激素调控过程,同时也响应生物和非生物胁迫。运用生物信息学方法,在番茄(Solanum Lycopersicum)中鉴定出47个SlC3H基因,不均匀地分布在12条染色体上,主要定位于细胞核中。根据系统进化树将其分为3个亚族(Ⅰ~Ⅲ)。共线性分析发现,SlC3H基因家族存在基因复制现象,表明在进化过程中SlC3H基因可能通过复制进行家族成员扩增。番茄SlC3H基因含有1~14个外显子,其启动子区域存在与激素和非生物胁迫响应的元件。Ka/Ks分析表明,SlC3H基因家族成员和C3H基因在进化过程中经历了强烈的进化选择。qRT-PCR分析发现,在盐胁迫下,SlC3H基因家族有一半以上的基因表达上调,其中SlC3H1在8 h时表达量最高,是0 h的30倍;用0.1 mol·L-1 2,4-表油菜素内酯(2,4-epibrassinolide,EBR)处理后所有SlC3H基因在绿熟期都表达上调,表达量是对照组的8~150倍,尤其是SlC3H28在其他时期表达量几乎为0,但在绿熟期其表达量是对照组的150倍,推测EBR对果实绿熟期的发育有促进作用。上述研究结果为进一步揭示番茄C3H基因家族在响应非生物胁迫以及果实发育中的功能提供了参考依据。
中图分类号:
李贤国, 戴麒, 王泽鹏, 陈兆龙, 闫会转, 李宁. 番茄CCCH类锌指蛋白家族的鉴定及其表达分析[J]. 中国农业科技导报, 2025, 27(1): 80-95.
Xianguo LI, Qi DAI, Zepeng WANG, Zhaolong CHEN, Huizhuan YAN, Ning LI. Identification and Phylogenetic Analysis of Tomato CCCH-like Zinc Finger Protein Family[J]. Journal of Agricultural Science and Technology, 2025, 27(1): 80-95.
基因 Gene | 正向引物序列 Forward primer sequence (5’-3’) | 反向引物序列 Reverse primer sequence (5’-3’) |
|---|---|---|
| SlC3H1 | TGGGTTTCAGTCTTCACCAGTT | TCCACTCGGACTCATAACCG |
| SlC3H2 | TGGCAATGGTGGATTCAAG | ACCAAGCATCTGAGTCACAGC |
| SlC3H3 | GCAGTCTCCTTCTGTTCAATCC | CGGATAAGGAGTCCAACCTGGT |
| SlC3H4 | CTTTCGGTAGCAGTGGAGGCAT | CATAGCAGTGTAGCCAGGAACA |
| SlC3H5 | TTCTGCTATTGATGAGGTGGG | TGGACCACAAGCACGGTTA |
| SlC3H6 | TGGTTCGCTCCACAGACTTT | TGGAACTTTCTTTGCCACTGC |
| SlC3H7 | CTGCGATTACTGCGATAAGGA | GCAGACTCCTTTACCGAACGA |
| SlC3H8 | GATTATGAATGGGCGGACTTG | GATGCCAGCGTCTTGTAGGA |
| SlC3H9 | CAGAGACAACGCCATCCAA | CCTTGATGTTGCTGTTGCG |
| SlC3H10 | GCAAGACAGGTGGTGATGACA | GCTGAAACTCTGATGTGAGCAG |
| SlC3H11 | GCAACAAATGGCAGGAGACT | GAAGTGGCAACGGTGACCATA |
| SlC3H12 | ACGATAAAGGTCTGGGCTGC | GGCAAATCATAGACACGAATGG |
| SlC3H13 | GGAAGCAAATCAAAGCCGT | TGGGAATAGGTGATGCTGGA |
| SlC3H14 | GATGATGAGATGGCGATGC | TCCCTTCAACATTTCCAAGC |
| SlC3H15 | GCAGTTATGGTGAAAGGTGCC | CACTTCATTTCCACCAGACACA |
| SlC3H16 | — | — |
| SlC3H17 | GAAGGGAAGAGGGTATTGGGCT | CTGGTCCTAAACTTGTCCCGT |
| SlC3H18 | GGAAGTAAGAAGGAAGAGGAACCA | CGGCATACCTCAGTCTTGTAACG |
| SlC3H19 | AGAATCAATGAGGAGGTTGGG | ACAGCACAGCACCAACAGAG |
| SlC3H20 | AGTTGCCGCTTTGAAGAGC | ACCGCAAATCTCCTTGGCTG |
| SlC3H21 | AGGGAGAGGAATCACCAAGC | ATAAGTGGCTGGCGGTGGAA |
| SlC3H22 | TTTGGTTACCCGTTGCGA | AACAACCGCTGGCAAAGGA |
| SlC3H23 | TCGGAAAGTGATAGCGACTACG | CCTTAGAAACATTCCGCCTGA |
| SlC3H24 | AATGAGAAGGCTGGTGACGC | GGGTAACTCAACCTCCTCCCAT |
| SlC3H25 | TTCTGACCAGGCAGTTTGGG | TGCGAGCATCTGAATCCTGC |
| SlC3H26 | AATGAGGTGGGCTTATGGTATG | TAGCCCTATCCGAACCACAAGC |
| SlC3H27 | CGATTAGCGTTTAGTCCCGAA | CCAACTTGATGGTTCGTAAAGG |
| SlC3H28 | CCACGATTGGACGGATTGT | AAGCCAACACTCAAACACCC |
| SlC3H29 | TTGCGAGGGAGATTATGTGC | GGGCTACTGAACACTGGATTTG |
| SlC3H30 | CTACACCCTGCTCGCTATCGTA | CCATCAAACGAATCAGACGAC |
| SlC3H31 | TGGCTCCACCAGTTGTCTCT | TCCAACTTCACTCTCACAAGGC |
| SlC3H32 | AACCTGCTGGTATGTCTGTGCC | GAAGCAACACTGGACCATAAGC |
| SlC3H33 | CTCAGTCTTCAGCACAACCTCA | TGATTGGCTGGAAGCGTCA |
| SlC3H34 | TCAGAACTTCATAGGACGAGGC | GGTGCTTTGGAGAATCGCT |
| SlC3H35 | GCTCCAGCATCAAGAAATGTG | GGCACAATCGGCTTACCAAT |
| SlC3H36 | CCTATGGTTATCGCCCTCTTGA | TCCAAGAGAAGGCGAAAGTG |
| SlC3H37 | GAACTTGCTGCCAACGATG | CGTAGCAGCAACCATCAAAGG |
| SlC3H38 | CAGCACCGCCTTATTCCAA | TCTTGTGAAGGAATGGTAGGCT |
| SlC3H39 | CGCAGTCATAGTAGAAGCCGA | TGTGTCCACTCCTTCGTCCA |
| SlC3H40 | TTATCCTGTCCGTGAAGGTGA | GGCATTCGGGTTGTCCTATTC |
| SlC3H41 | GGATTTCTTGAGGATGAAGGC | CCCATCATAAATGTCGCAGC |
| SlC3H42 | GCTATGAAGACCGCTCTCACA | CTGCCATCCCTAACTGGACTT |
| SlC3H43 | ATCCTGCCGTTACGCTCAT | AACGCCTCAGTCATCATAACG |
| SlC3H44 | CCTGTTCCCTCGCACTTTATG | TTCGGCGAGTTGCTATCAA |
| SlC3H45 | ACCACCTATGTCTCCGATGACG | CCGAATACAGGTGAACCCATT |
| SlC3H46 | GCAGCAGACAAAGTCCAACC | AGGCTGATTGTCGGTTTGC |
| SlC3H47 | CGGAATAGTAGCCGCCAATAC | AACACCAGGAGGTCCATAACC |
表 1 qRT-PCR的引物序列 (续表Continued)
Table 1 Primer sequences for qRT-PCR
基因 Gene | 正向引物序列 Forward primer sequence (5’-3’) | 反向引物序列 Reverse primer sequence (5’-3’) |
|---|---|---|
| SlC3H1 | TGGGTTTCAGTCTTCACCAGTT | TCCACTCGGACTCATAACCG |
| SlC3H2 | TGGCAATGGTGGATTCAAG | ACCAAGCATCTGAGTCACAGC |
| SlC3H3 | GCAGTCTCCTTCTGTTCAATCC | CGGATAAGGAGTCCAACCTGGT |
| SlC3H4 | CTTTCGGTAGCAGTGGAGGCAT | CATAGCAGTGTAGCCAGGAACA |
| SlC3H5 | TTCTGCTATTGATGAGGTGGG | TGGACCACAAGCACGGTTA |
| SlC3H6 | TGGTTCGCTCCACAGACTTT | TGGAACTTTCTTTGCCACTGC |
| SlC3H7 | CTGCGATTACTGCGATAAGGA | GCAGACTCCTTTACCGAACGA |
| SlC3H8 | GATTATGAATGGGCGGACTTG | GATGCCAGCGTCTTGTAGGA |
| SlC3H9 | CAGAGACAACGCCATCCAA | CCTTGATGTTGCTGTTGCG |
| SlC3H10 | GCAAGACAGGTGGTGATGACA | GCTGAAACTCTGATGTGAGCAG |
| SlC3H11 | GCAACAAATGGCAGGAGACT | GAAGTGGCAACGGTGACCATA |
| SlC3H12 | ACGATAAAGGTCTGGGCTGC | GGCAAATCATAGACACGAATGG |
| SlC3H13 | GGAAGCAAATCAAAGCCGT | TGGGAATAGGTGATGCTGGA |
| SlC3H14 | GATGATGAGATGGCGATGC | TCCCTTCAACATTTCCAAGC |
| SlC3H15 | GCAGTTATGGTGAAAGGTGCC | CACTTCATTTCCACCAGACACA |
| SlC3H16 | — | — |
| SlC3H17 | GAAGGGAAGAGGGTATTGGGCT | CTGGTCCTAAACTTGTCCCGT |
| SlC3H18 | GGAAGTAAGAAGGAAGAGGAACCA | CGGCATACCTCAGTCTTGTAACG |
| SlC3H19 | AGAATCAATGAGGAGGTTGGG | ACAGCACAGCACCAACAGAG |
| SlC3H20 | AGTTGCCGCTTTGAAGAGC | ACCGCAAATCTCCTTGGCTG |
| SlC3H21 | AGGGAGAGGAATCACCAAGC | ATAAGTGGCTGGCGGTGGAA |
| SlC3H22 | TTTGGTTACCCGTTGCGA | AACAACCGCTGGCAAAGGA |
| SlC3H23 | TCGGAAAGTGATAGCGACTACG | CCTTAGAAACATTCCGCCTGA |
| SlC3H24 | AATGAGAAGGCTGGTGACGC | GGGTAACTCAACCTCCTCCCAT |
| SlC3H25 | TTCTGACCAGGCAGTTTGGG | TGCGAGCATCTGAATCCTGC |
| SlC3H26 | AATGAGGTGGGCTTATGGTATG | TAGCCCTATCCGAACCACAAGC |
| SlC3H27 | CGATTAGCGTTTAGTCCCGAA | CCAACTTGATGGTTCGTAAAGG |
| SlC3H28 | CCACGATTGGACGGATTGT | AAGCCAACACTCAAACACCC |
| SlC3H29 | TTGCGAGGGAGATTATGTGC | GGGCTACTGAACACTGGATTTG |
| SlC3H30 | CTACACCCTGCTCGCTATCGTA | CCATCAAACGAATCAGACGAC |
| SlC3H31 | TGGCTCCACCAGTTGTCTCT | TCCAACTTCACTCTCACAAGGC |
| SlC3H32 | AACCTGCTGGTATGTCTGTGCC | GAAGCAACACTGGACCATAAGC |
| SlC3H33 | CTCAGTCTTCAGCACAACCTCA | TGATTGGCTGGAAGCGTCA |
| SlC3H34 | TCAGAACTTCATAGGACGAGGC | GGTGCTTTGGAGAATCGCT |
| SlC3H35 | GCTCCAGCATCAAGAAATGTG | GGCACAATCGGCTTACCAAT |
| SlC3H36 | CCTATGGTTATCGCCCTCTTGA | TCCAAGAGAAGGCGAAAGTG |
| SlC3H37 | GAACTTGCTGCCAACGATG | CGTAGCAGCAACCATCAAAGG |
| SlC3H38 | CAGCACCGCCTTATTCCAA | TCTTGTGAAGGAATGGTAGGCT |
| SlC3H39 | CGCAGTCATAGTAGAAGCCGA | TGTGTCCACTCCTTCGTCCA |
| SlC3H40 | TTATCCTGTCCGTGAAGGTGA | GGCATTCGGGTTGTCCTATTC |
| SlC3H41 | GGATTTCTTGAGGATGAAGGC | CCCATCATAAATGTCGCAGC |
| SlC3H42 | GCTATGAAGACCGCTCTCACA | CTGCCATCCCTAACTGGACTT |
| SlC3H43 | ATCCTGCCGTTACGCTCAT | AACGCCTCAGTCATCATAACG |
| SlC3H44 | CCTGTTCCCTCGCACTTTATG | TTCGGCGAGTTGCTATCAA |
| SlC3H45 | ACCACCTATGTCTCCGATGACG | CCGAATACAGGTGAACCCATT |
| SlC3H46 | GCAGCAGACAAAGTCCAACC | AGGCTGATTGTCGGTTTGC |
| SlC3H47 | CGGAATAGTAGCCGCCAATAC | AACACCAGGAGGTCCATAACC |
基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 氨基酸 Amino acid | 分子量 Molecular weight/Da | 等电点 Isoelectric point | 亲疏水性 Gravy | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| SlC3H1 | Solyc01G000285 | Chr1 | 细胞核Nucleus | ||||
| SlC3H2 | Solyc01G000732 | Chr1 | 细胞质Cytoplasm | ||||
| SlC3H3 | Solyc01G001958 | Chr1 | 细胞核Nucleus | ||||
| SlC3H4 | Solyc01G002357 | Chr1 | 细胞核Nucleus | ||||
| SlC3H5 | Solyc01G002417 | Chr1 | 细胞核Nucleus | ||||
| SlC3H6 | Solyc01G002511 | Chr1 | 细胞核Nucleus | ||||
| SlC3H7 | Solyc01G003423 | Chr1 | 叶绿体Chloroplast | ||||
| SlC3H8 | Solyc01G003523 | Chr1 | 细胞核Nucleus | ||||
| SlC3H9 | Solyc02G000685 | Chr2 | 细胞核Nucleus | ||||
| SlC3H10 | Solyc02G001874 | Chr2 | 细胞核Nucleus | ||||
| SlC3H11 | Solyc02G002224 | Chr2 | 细胞核Nucleus | ||||
| SlC3H12 | Solyc03G000439 | Chr3 | 细胞核Nucleus | ||||
| SlC3H13 | Solyc03G001290 | Chr3 | 细胞质Cytoplasm | ||||
| SlC3H14 | Solyc03G002428 | Chr3 | 细胞核Nucleus | ||||
| SlC3H15 | Solyc03G002917 | Chr3 | 细胞核Nucleus | ||||
| SlC3H16 | Solyc03G003234 | Chr3 | 细胞核Nucleus | ||||
| SlC3H17 | Solyc04G002040 | Chr4 | 细胞核Nucleus | ||||
| SlC3H18 | Solyc05G000368 | Chr5 | 细胞核Nucleus | ||||
基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 氨基酸 Amino acid | 分子量 Molecular weight/Da | 等电点 Isoelectric point | 亲疏水性 Gravy | 亚细胞定位 Subcellular localization |
| SlC3H19 | Solyc06G000257 | Chr6 | 细胞核Nucleus | ||||
| SlC3H20 | Solyc06G000284 | Chr6 | 细胞核Nucleus | ||||
| SlC3H21 | Solyc06G001227 | Chr6 | 细胞核Nucleus | ||||
| SlC3H22 | Solyc06G001228 | Chr6 | 细胞核Nucleus | ||||
| SlC3H23 | Solyc06G001444 | Chr6 | 细胞核Nucleus | ||||
| SlC3H24 | Solyc06G002030 | Chr6 | 细胞核Nucleus | ||||
| SlC3H25 | Solyc06G002122 | Chr6 | 细胞核Nucleus | ||||
| SlC3H26 | Solyc06G002533 | Chr6 | 细胞核Nucleus | ||||
| SlC3H27 | Solyc07G001794 | Chr7 | 细胞核Nucleus | ||||
| SlC3H28 | Solyc07G002095 | Chr7 | 细胞核Nucleus | ||||
| SlC3H29 | Solyc07G002693 | Chr7 | 细胞核Nucleus | ||||
| SlC3H30 | Solyc08G001378 | Chr8 | 细胞核Nucleus | ||||
| SlC3H31 | Solyc09G001678 | Chr9 | 细胞外基质Extracellular | ||||
| SlC3H32 | Solyc09G002209 | Chr9 | — | ||||
| SlC3H33 | Solyc10G000921 | Chr10 | 细胞核Nucleus | ||||
| SlC3H34 | Solyc10G001254 | Chr10 | 细胞核Nucleus | ||||
| SlC3H35 | Solyc10G002269 | Chr10 | 细胞核Nucleus | ||||
| SlC3H36 | Solyc10G002300 | Chr10 | 细胞核Nucleus | ||||
| SlC3H37 | Solyc10G002434 | Chr10 | 细胞核Nucleus | ||||
| SlC3H38 | Solyc10G002434 | Chr10 | 细胞核Nucleus | ||||
| SlC3H39 | Solyc10G002648 | Chr10 | 细胞核Nucleus | ||||
| SlC3H40 | Solyc11G000372 | Chr11 | 细胞核Nucleus | ||||
基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 氨基酸 Amino acid | 分子量 Molecular weight/Da | 等电点 Isoelectric point | 亲疏水性 Gravy | 亚细胞定位 Subcellular localization |
| SlC3H41 | Solyc11G001986 | Chr11 | 细胞核Nucleus | ||||
| SlC3H42 | Solyc11G002140 | Chr11 | 细胞核Nucleus | ||||
| SlC3H43 | Solyc11G002291 | Chr11 | 细胞核Nucleus | ||||
| SlC3H44 | Solyc11G002364 | Chr11 | 细胞核Nucleus | ||||
| SlC3H45 | Solyc12G000277 | Chr12 | 细胞核Nucleus | ||||
| SlC3H46 | Solyc12G000350 | Chr12 | 细胞核Nucleus | ||||
| SlC3H47 | Solyc12G000810 | Chr12 | 细胞核Nucleus |
表2 番茄SlC3H基因家族成员编码氨基酸序列的理化性质续表Continued
Table 2 Physicochemical properties of amino acid sequences encoded by members of tomato SlC3H gene family
基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 氨基酸 Amino acid | 分子量 Molecular weight/Da | 等电点 Isoelectric point | 亲疏水性 Gravy | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| SlC3H1 | Solyc01G000285 | Chr1 | 细胞核Nucleus | ||||
| SlC3H2 | Solyc01G000732 | Chr1 | 细胞质Cytoplasm | ||||
| SlC3H3 | Solyc01G001958 | Chr1 | 细胞核Nucleus | ||||
| SlC3H4 | Solyc01G002357 | Chr1 | 细胞核Nucleus | ||||
| SlC3H5 | Solyc01G002417 | Chr1 | 细胞核Nucleus | ||||
| SlC3H6 | Solyc01G002511 | Chr1 | 细胞核Nucleus | ||||
| SlC3H7 | Solyc01G003423 | Chr1 | 叶绿体Chloroplast | ||||
| SlC3H8 | Solyc01G003523 | Chr1 | 细胞核Nucleus | ||||
| SlC3H9 | Solyc02G000685 | Chr2 | 细胞核Nucleus | ||||
| SlC3H10 | Solyc02G001874 | Chr2 | 细胞核Nucleus | ||||
| SlC3H11 | Solyc02G002224 | Chr2 | 细胞核Nucleus | ||||
| SlC3H12 | Solyc03G000439 | Chr3 | 细胞核Nucleus | ||||
| SlC3H13 | Solyc03G001290 | Chr3 | 细胞质Cytoplasm | ||||
| SlC3H14 | Solyc03G002428 | Chr3 | 细胞核Nucleus | ||||
| SlC3H15 | Solyc03G002917 | Chr3 | 细胞核Nucleus | ||||
| SlC3H16 | Solyc03G003234 | Chr3 | 细胞核Nucleus | ||||
| SlC3H17 | Solyc04G002040 | Chr4 | 细胞核Nucleus | ||||
| SlC3H18 | Solyc05G000368 | Chr5 | 细胞核Nucleus | ||||
基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 氨基酸 Amino acid | 分子量 Molecular weight/Da | 等电点 Isoelectric point | 亲疏水性 Gravy | 亚细胞定位 Subcellular localization |
| SlC3H19 | Solyc06G000257 | Chr6 | 细胞核Nucleus | ||||
| SlC3H20 | Solyc06G000284 | Chr6 | 细胞核Nucleus | ||||
| SlC3H21 | Solyc06G001227 | Chr6 | 细胞核Nucleus | ||||
| SlC3H22 | Solyc06G001228 | Chr6 | 细胞核Nucleus | ||||
| SlC3H23 | Solyc06G001444 | Chr6 | 细胞核Nucleus | ||||
| SlC3H24 | Solyc06G002030 | Chr6 | 细胞核Nucleus | ||||
| SlC3H25 | Solyc06G002122 | Chr6 | 细胞核Nucleus | ||||
| SlC3H26 | Solyc06G002533 | Chr6 | 细胞核Nucleus | ||||
| SlC3H27 | Solyc07G001794 | Chr7 | 细胞核Nucleus | ||||
| SlC3H28 | Solyc07G002095 | Chr7 | 细胞核Nucleus | ||||
| SlC3H29 | Solyc07G002693 | Chr7 | 细胞核Nucleus | ||||
| SlC3H30 | Solyc08G001378 | Chr8 | 细胞核Nucleus | ||||
| SlC3H31 | Solyc09G001678 | Chr9 | 细胞外基质Extracellular | ||||
| SlC3H32 | Solyc09G002209 | Chr9 | — | ||||
| SlC3H33 | Solyc10G000921 | Chr10 | 细胞核Nucleus | ||||
| SlC3H34 | Solyc10G001254 | Chr10 | 细胞核Nucleus | ||||
| SlC3H35 | Solyc10G002269 | Chr10 | 细胞核Nucleus | ||||
| SlC3H36 | Solyc10G002300 | Chr10 | 细胞核Nucleus | ||||
| SlC3H37 | Solyc10G002434 | Chr10 | 细胞核Nucleus | ||||
| SlC3H38 | Solyc10G002434 | Chr10 | 细胞核Nucleus | ||||
| SlC3H39 | Solyc10G002648 | Chr10 | 细胞核Nucleus | ||||
| SlC3H40 | Solyc11G000372 | Chr11 | 细胞核Nucleus | ||||
基因名称 Gene name | 基因ID Gene ID | 染色体定位 Chromosome location | 氨基酸 Amino acid | 分子量 Molecular weight/Da | 等电点 Isoelectric point | 亲疏水性 Gravy | 亚细胞定位 Subcellular localization |
| SlC3H41 | Solyc11G001986 | Chr11 | 细胞核Nucleus | ||||
| SlC3H42 | Solyc11G002140 | Chr11 | 细胞核Nucleus | ||||
| SlC3H43 | Solyc11G002291 | Chr11 | 细胞核Nucleus | ||||
| SlC3H44 | Solyc11G002364 | Chr11 | 细胞核Nucleus | ||||
| SlC3H45 | Solyc12G000277 | Chr12 | 细胞核Nucleus | ||||
| SlC3H46 | Solyc12G000350 | Chr12 | 细胞核Nucleus | ||||
| SlC3H47 | Solyc12G000810 | Chr12 | 细胞核Nucleus |
重复基因对 Duplicated gene pairs | 同义替换率 Ka | 非同义替换率 Ks | Ka/Ks |
|---|---|---|---|
| SlC3H1/SlC3H41 | 0.177 401 | 0.647 337 | 0.274 048 |
| SlC3H35/SlC3H38 | 0.113 408 | 0.685 379 | 0.165 467 |
| SlC3H36/SlC3H37 | 0.152 286 | 0.535 449 | 0.284 408 |
| SlC3H45/SlC3H30 | 0.298 310 | 1.423 712 | 0.209 530 |
| SlC3H14/SlC3H24 | 0.283 640 | 0.744 457 | 0.381 003 |
| SlC3H19/SlC3H32 | 0.204 448 | 0.524 756 | 0.389 606 |
表 3 番茄SlC3H基因Ka/Ks分析
Table 3 Ka/Ks analysis of tomato SlC3H genes
重复基因对 Duplicated gene pairs | 同义替换率 Ka | 非同义替换率 Ks | Ka/Ks |
|---|---|---|---|
| SlC3H1/SlC3H41 | 0.177 401 | 0.647 337 | 0.274 048 |
| SlC3H35/SlC3H38 | 0.113 408 | 0.685 379 | 0.165 467 |
| SlC3H36/SlC3H37 | 0.152 286 | 0.535 449 | 0.284 408 |
| SlC3H45/SlC3H30 | 0.298 310 | 1.423 712 | 0.209 530 |
| SlC3H14/SlC3H24 | 0.283 640 | 0.744 457 | 0.381 003 |
| SlC3H19/SlC3H32 | 0.204 448 | 0.524 756 | 0.389 606 |
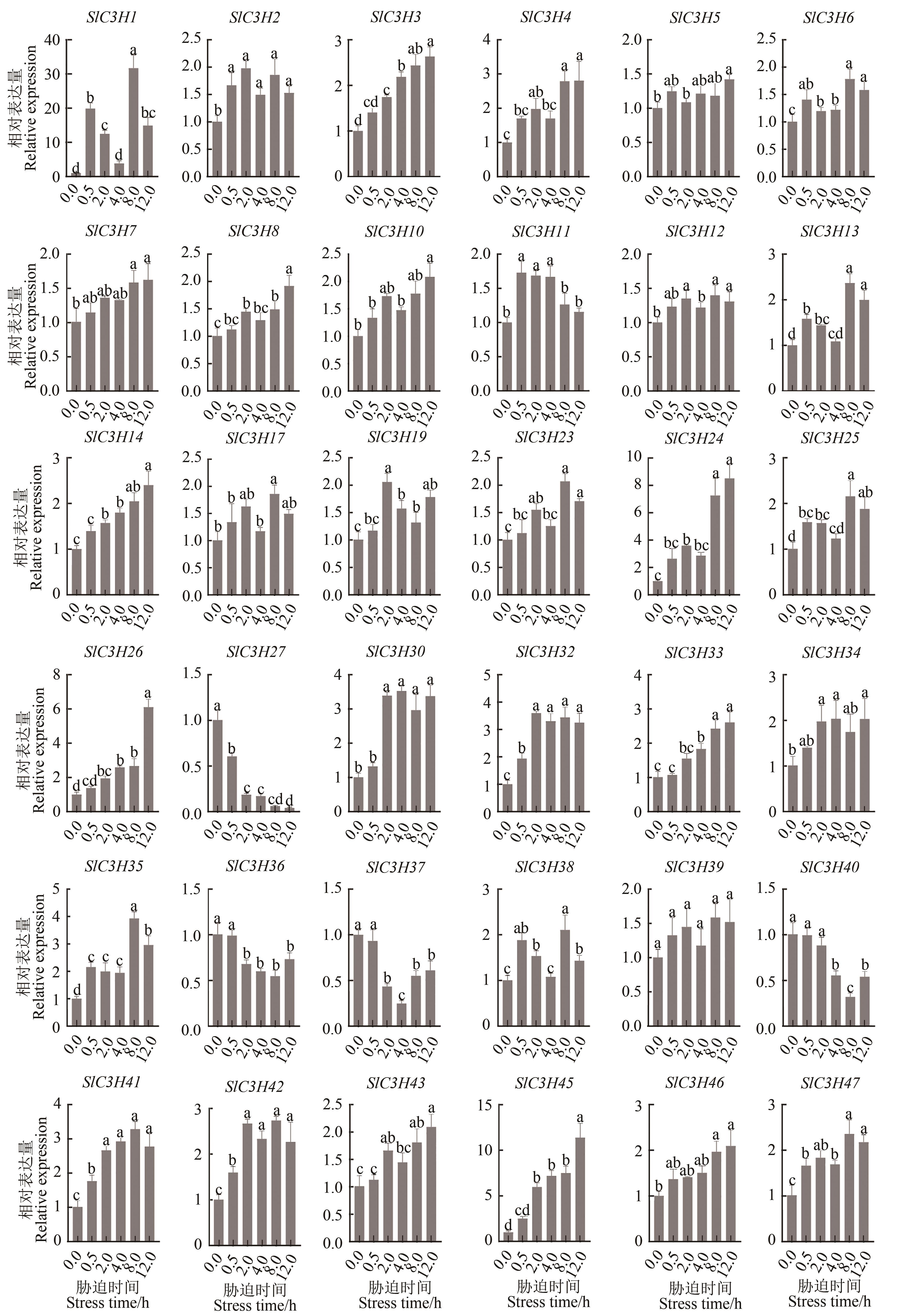
图7 番茄SlC3H家族基因响应盐胁迫的表达注:不同小写字母表示不同胁迫时长间在P<0.05水平差异显著。
Fig. 7 Expression of tomato SlC3H family genes in response to salt stressNote: Different lowercase letters indicate significant differences between different stress times at P<0.05 level.
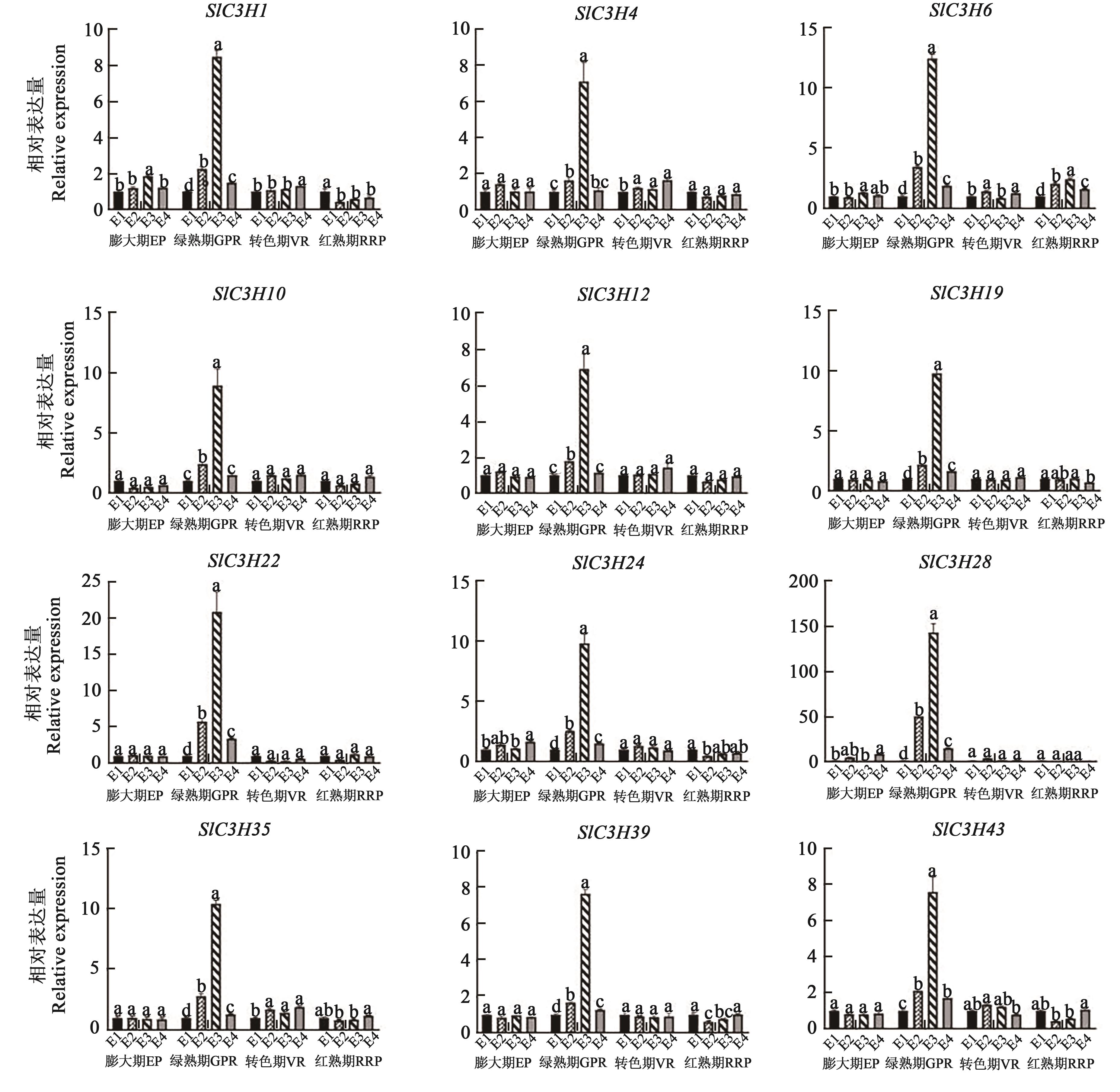
图8 番茄SlC3H家族基因果实发育时期激素胁迫下的表达分析注:不同小写字母表示同一时期不同处理间在P<0.05水平差异显著。
Fig. 8 Expression analysis of tomato SlC3H family genes under hormonal stress during fruit developmentNote: Different lowercase letters indicate significant differences between different treatments of same period at P<0.05 level.
| 1 | GAO G, GUO X, GOFF S P, et al.. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein [J]. Science, 2002, 297(5587):1703-1706. |
| 2 | MAZUMDAR P, LAU S E, WEE W Y, et al.. Genome-wide analysis of the CCCH zinc-finger gene family in banana (Musa acuminata): an insight in to motif and gene structure arrangement, evolution and salt stress responses [J]. Trop. Plant Biol., 2017, 10(11):177-193. |
| 3 | DRÖGE-LASER W, SNOEK B L, SNEL B, et al.. The Arabidopsis bZIP transcription factor family-an update [J]. Curr. Opin. Plant Biol., 2018, 45(PtA):36-49. |
| 4 | SU L Y, XIAO X C, JIANG M Q, et al.. Genome-wide analysis of the CCCH zinc finger family in longan: characteristic identification and expression profiles in Dimocarpus longan Lour [J]. J. Integr. Agric., 2022, 21(1):113-130. |
| 5 | DELANEY K J, XU R, ZHANG J, et al.. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit [J]. Plant Physiol., 2006, 140 (4):1507-1521. |
| 6 | CARRICK D M, LAI W S, BLACKSHEAR P J. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis [J]. Arthritis Res. Ther., 2004, 6 (6):248-264. |
| 7 | DE J, LAI W S, THORN J M, et al.. Identification of four CCCH zinc finger proteins in Xenopus, including a novel vertebrate protein with four zinc fingers and severely restricted expression [J]. Gene, 1999, 228(1-2):133-145. |
| 8 | BOGAMUWA S P, JANG J C. Tandem CCCH zinc finger proteins in plant growth, development and stress response [J]. Plant Cell Physiol., 2014, 55(8):1367‐1375. |
| 9 | ZHANG L Y, BAI M Y, WU J, et al.. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis [J]. Plant Cell, 2009, 21(12):3767-3780. |
| 10 | ZHANG H, GAO X, ZHI Y, et al.. A non‐tandem CCCH‐type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato [J]. New Phytol., 2019, 223(4):1918‐1936. |
| 11 | KIM D H, YAMAGUCHI S, LIM S, et al.. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5 [J]. Plant Cell, 2008, 20(5):1260-1277. |
| 12 | LI J, JIA D, CHEN X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein [J]. Plant Cell, 2001, 13(10):2269-2281. |
| 13 | JIANG M, JIANG J J, MIAO L X, et al.. Over-expression of a C3H-type zinc finger gene contributes to salt stress tolerance in transgenic broccoli plants [J]. Plant Cell, 2017, 130(2):239-254. |
| 14 | PRADHAN S, KANT C, VERMA S, et al.. Genome-wide analysis of the CCCH zinc finger family identifies tissue specific and stress responsive candidates in chickpea (Cicer arietinum L.) [J/OL]. PloS One, 2017, 12(7):e0180469 [2023-02-05]. . |
| 15 | GUO Y H, YU YP, WANG D, et al.. GhZFP1, a novel CCCH-type zinc fifinger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5 [J]. New Phytol., 2009,183(1):62-75. |
| 16 | 刘雨轩.番茄WRKY基因家族成员鉴定及表达分析[D].沈阳:沈阳农业大学,2020. |
| LIU Y X. Identification and expression analysis of tomato WRKY gene family members [D]. Shenyang: Shenyang Agricultural University, 2020. | |
| 17 | WANG D, GUO Y, WU C, et al.. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice [J]. BMC Genomics, 2008, 9(1):44-64. |
| 18 | PENG X, ZHAO Y, CAO J, et al.. CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments [J/OL]. PloS One, 2012, 7(7): e40120 [2023-02-05]. . |
| 19 | LI C, FANG Q, ZHANG W, et al.. Genome-wide identification of the CCCH gene family in rose (Rosa chinensis Jacq.) reveals its potential functions [J]. Biotechnol. Equip., 2021, 35(1):517-526. |
| 20 | 唐春闺,邓兆龙,刘琼,等.普通烟草CCCH类锌指蛋白家族的全基因组鉴定和表达分析[J]. 河南农业科学, 2022, 51(4): 48-58. |
| TANG C G, DENG Z L, LIU Q, et al.. Genome wide identification and expression analysis of CCCH zinc finger like protein family in tobacco [J]. J. Henan Agric. Sci., 2022, 51(4):48-58. | |
| 21 | XU R. Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Solanum lycopersicum [J]. Mol. Genet. Genomics, 2014, 289(5):965-979. |
| 22 | PI B, HE X, RUAN Y, et al.. Genome-wide analysis and stress-responsive expression of CCCH zinc finger family genes in Brassica rapa [J/OL]. BMC Plant Biol., 2018, 18(1):7 [2023-02-05]. . |
| 23 | PI B, PAN J, XIAO M, et al.. Systematic analysis of CCCH zinc finger family in Brassica napus showed that BnRR-TZFs are involved in stress resistance [J/OL]. BMC Plant Biol., 2021, 21(1):8 [2023-02-05]. . |
| 24 | LIU S, KHAN M R G, LI Y, et al.. Comprehensive analysis of CCCH-type zinc finger gene family in citrus (Clementine mandarin) by genome-wide characterization [J]. Mol. Genet. Genomics, 2014, 289(5): 855-872. |
| 25 | WANG X L, ZHONG Y, CHENG Z M. Evolution and expression analysis of the CCCH zinc finger gene family in Vitis vinifera [J]. Plant Genome, 2014, 7(3):16-25. |
| 26 | PRAKASH A, JEFFRYES M, BATEMAN A, et al.. The HMMER web server for protein sequence similarity search [J]. Curr. Prot. Bioinf., 2017, 60:15-23. |
| 27 | GASTEIGER E, GATTIKER A, HOOGLAND C, et al.. ExPASy: the proteomics server for in-depth protein knowledge and analysis [J]. Nucl. Acids Res., 2003, 31(13):3784-3788. |
| 28 | HORTON P, PARK K J, OBAYASHI T, et al.. WoLF PSORT: protein localization predictor [J]. Nucl. Acids Res., 2007, 35:585-587. |
| 29 | KUMAR S, STECHER G, LI M, et al.. MEGA X: molecular evolutionary genetics analysis across computing platforms [J]. Mol. Biol. Evol., 2018, 35(6):1547-1579. |
| 30 | BAILEY T L, BODEN M, BUSKE F A, et al.. MEME SUITE: tools for motif discovery and searching [J]. Nucl. Acids Res., 2009, 37:202-208. |
| 31 | CHEN C, CHEN H, ZHANG Y, et al.. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol. Plant, 2020, 13(8):1191-1202. |
| 32 | WANG Y P, TANG H B, DEBARRY J D, et al.. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J/OL]. Nucl. Acids Res., 2012, 40(7):e49 [2023-02-05]. . |
| 33 | KRZYWINSKI M, SCHEIN J, BIROL I, et al.. Circos: an information aesthetic for comparative genomics [J]. Genome Res., 2009, 19(9):1639-1645. |
| 34 | WANG D, ZHANG Y, ZHANG Z, et al.. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies [J]. Genom Proteom. Bioinf., 2010, 8(1):77-80. |
| 35 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method [J]. Methods, 2001, 25(4):402-408. |
| 36 | YAO Q, BAI Y, KUMAR S, et al.. Minimal residual disease detection by next-generation sequencing in multiple myeloma: A omparison with real-time quantitative PCR [J/OL]. Front. Oncol., 2020, 10: 611021 [2023-02-05]. . |
| 37 | CHAI G, HU R, ZHANG D, et al.. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa ) [J/OL]. BMC Genomics, 2012, 13(1):253 [2023-02-05]. . |
| 38 | JOUFFREY V, LEONARD A S, AHNERT S E. Gene duplication and subsequent diversification strongly affect phenotypic evolvability and robustness [J]. Roy Soc. Open Sci., 2021, 8(6):1636-1642. |
| 39 | WANG W, JIANG W, LIU J G, et al.. Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response [J/OL]. BMC Genomics, 2017, 18(1):518 [2023-02-05]. . |
| 40 | 魏梦园,刘爱丽,黎冬华,等.芝麻CCCH锌指蛋白基因SiC3H1的克隆及表达分析[J].分子植物育种,2020,18(24):7982-7988. |
| WEI M Y, LIU A L, LI D H, et al.. Cloning and expression analysis of CCCH zinc finger protein gene SiC3H1 from sesame [J]. Mol. Plant Breed., 2020, 18(24):7982-7988. | |
| 41 | 刘小艳,孙艳侠,王亚男,等.水稻CCCH型锌指蛋白亚家族Ⅰ基因的表达分析[J]. 山东农业科学,2015,47(2):7-11. |
| LIU X Y, SUN Y X, WANG Y N, et al.. Expression analysis of CCCH zinc finger protein subfamily Ⅰ genes in rice [J]. Shandong Agric. Sci., 2015, 47(2):7-11. | |
| 42 | VERMA V, RAVINDRAN P, KUMAR P P. Plant hormone-mediated regulation of stress responses [J]. BMC Plant Biol., 2016, 16:1-10. |
| 43 | LIN P C, POMERANZ M C, JIKUMARU Y, et al.. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA‐and GA‐mediated growth, stress and gene expression responses [J]. Plant J., 2011, 65(2):253-268. |
| [1] | 米春娇, 孙洪仁, 张吉萍, 吕玉才, 张砚迪. 我国番茄土壤有效磷丰缺指标和推荐施磷量初步研究[J]. 中国农业科技导报, 2025, 27(1): 222-232. |
| [2] | 郭肖蓉, 刘颖, 樊佳珍, 黄涛, 周荣. 猪CREBRF基因生物信息学和表达规律分析[J]. 中国农业科技导报, 2024, 26(9): 44-53. |
| [3] | 张俊蕾, 盖晓彤, 赵正婷, 刘弟, 王金凤, 姜宁, 刘雅婷. 烟草番茄斑萎病毒RT-LAMP检测体系的建立及优化[J]. 中国农业科技导报, 2024, 26(8): 140-150. |
| [4] | 张福林, 奚瑞, 刘宇翔, 陈兆龙, 余庆辉, 李宁. 番茄BURP结构域基因家族全基因组鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(8): 51-62. |
| [5] | 王子凡, 李燕, 张庆银, 王丹丹, 师建华, 耿晓彬, 田东良, 钟增明, 赵晓明, 齐连芬. 微生物菌剂对设施番茄主要病害及土壤微生物群落的影响[J]. 中国农业科技导报, 2024, 26(6): 102-112. |
| [6] | 鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21. |
| [7] | 陈明迪, 胡桂花, 张海文, 王旺田. 水稻RR基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(5): 20-29. |
| [8] | 李名博, 刘玉乐, 穆志民, 郭俊旺, 卫勇, 任东悦, 贾济深, 卫泽中, 栗宇红. 基于YOLOX-L-TN模型的番茄果实识别[J]. 中国农业科技导报, 2024, 26(4): 97-105. |
| [9] | 吴占清, 陈威, 赵展, 许海良, 李豪远, 彭星星, 陈东旭, 张明月. 玉米GRAS基因家族的全基因组鉴定及生物信息学分析[J]. 中国农业科技导报, 2024, 26(3): 15-25. |
| [10] | 刘博, 王旺田, 马骊, 武军艳, 蒲媛媛, 刘丽君, 方彦, 孙万仓, 张岩, 刘睿敏, 曾秀存. 白菜型油菜IPT基因家族鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(2): 56-66. |
| [11] | 黄大野, 余志斌, 万中义, 杨丹, 李金萍, 曹春霞. 产褐黄色链霉菌HEBRC45958菌株防控番茄棒孢叶斑病研究[J]. 中国农业科技导报, 2024, 26(11): 136-142. |
| [12] | 尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤, 黄耿青. 薰衣草 JAZ和MYC2-like基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(11): 79-90. |
| [13] | 张振伟, 董相书, 杨婧, 李学俊, 杞美军, 蒋快乐, 杨永林, 王步天, 施学东, 邱俊超, 陈治华, 葛宇. 小粒咖啡叶绿素合成基因CaPOR的全基因组鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(10): 83-97. |
| [14] | 邓玉荣, 韩联, 王金龙, 韦兴翰, 王旭东, 赵颖, 魏小红, 李朝周. 藜麦SOD家族基因的鉴定及其对混合盐碱胁迫的响应[J]. 中国农业科技导报, 2024, 26(1): 28-39. |
| [15] | 陈春林, 王琳洋, 单梦伟, 裴甜甜, 王吉庆, 肖怀娟, 李娟起, 李猛, 杜清洁. 发酵花生壳和牛粪替代草炭基质的番茄育苗效果分析[J]. 中国农业科技导报, 2023, 25(4): 205-214. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号