




















中国农业科技导报 ›› 2024, Vol. 26 ›› Issue (11): 79-90.DOI: 10.13304/j.nykjdb.2024.0423
尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤( ), 黄耿青(
), 黄耿青( )
)
收稿日期:2024-05-23
接受日期:2024-06-21
出版日期:2024-11-15
发布日期:2024-11-19
通讯作者:
陈永坤,黄耿青
作者简介:第一联系人:尼格拉•阿布都外力 E-mail: 1755877085@qq.com
基金资助:
Nigela Abuduwaili, Xiuzhen WEI, Jingjie MEN, Lingna CHEN, Yongkun CHEN( ), Gengqing HUANG(
), Gengqing HUANG( )
)
Received:2024-05-23
Accepted:2024-06-21
Online:2024-11-15
Published:2024-11-19
Contact:
Yongkun CHEN,Gengqing HUANG
摘要:
JAZ蛋白和MYC2-like转录因子是茉莉酸(jasmonate,JA)类物质信号转导途径的关键调节因子,在植物的生长发育和次级代谢中发挥重要作用。以拟南芥13个JAZ蛋白和11个MYC2-like蛋白的氨基酸序列为参考序列,检索薰衣草基因组中的LaJAZ和LaMYC2-like基因家族成员,并对其进行生物信息学分析。结果表明,在薰衣草基因组中共鉴定到26个LaJAZ和26个LaMYC2-like基因,这些基因不均匀的分布在22条染色体上。LaJAZ和LaMYC2-like基因启动子的顺式作用元件涉及植物对光、茉莉酸、脱落酸、防御和胁迫反应的响应。qRT-PCR显示,选择的6个LaJAZ基因在深紫色萼片中表达较高,而LaMYC2-like基因在浅紫色萼片中表达较高。此外,有15个LaJAZ和LaMYC2-like基因受茉莉酸甲酯(methyl jasmonate,MeJA)诱导。以上研究结果为薰衣草LaJAZ和LaMYC2-like基因的功能分析及其对JA信号的响应机理研究奠定了基础。
中图分类号:
尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤, 黄耿青. 薰衣草 JAZ和MYC2-like基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(11): 79-90.
Nigela Abuduwaili, Xiuzhen WEI, Jingjie MEN, Lingna CHEN, Yongkun CHEN, Gengqing HUANG. Bioinformatics and Expression Pattern Analysis of JAZ and MYC2-like Gene Family in Lavandula angustifolia[J]. Journal of Agricultural Science and Technology, 2024, 26(11): 79-90.
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数量 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAZ1 | La01G01624 | Chr1 | 14 191 132 | 14 192 638 | 334 | 35.531 14 | 9.73 |
| LaJAZ2 | La01G02122 | Chr1 | 22 315 757 | 22 318 426 | 279 | 29.241 02 | 7.64 |
| LaJAZ3 | La01G02748 | Chr1 | 29 933 237 | 29 935 361 | 233 | 25.500 76 | 9.58 |
| LaJAZ4 | La01G02775 | Chr1 | 30 304 246 | 30 305 952 | 270 | 29.719 64 | 8.74 |
| LaJAZ5 | La02G01205 | Chr2 | 27 444 500 | 27 446 515 | 252 | 26.659 79 | 9.34 |
| LaJAZ6 | La04G00443 | Chr4 | 23 907 703 | 2 908 702 | 207 | 21.913 95 | 8.46 |
| LaJAZ7 | La10G01957 | Chr10 | 37 391 837 | 37 394 501 | 255 | 27.540 92 | 9.12 |
| LaJAZ8 | La11G01369 | Chr11 | 30 835 601 | 30 838 263 | 297 | 31.243 32 | 8.31 |
| LaJAZ9 | La12G01015 | Chr12 | 20 257 088 | 20 259 717 | 275 | 29.305 02 | 9.43 |
| LaJAZ10 | La14G00862 | Chr14 | 9 169 268 | 9 170 127 | 211 | 22.548 44 | 9.44 |
| LaJAZ11 | La15G00121 | Chr15 | 866 936 | 870 632 | 345 | 37.302 14 | 8.46 |
| LaJAZ12 | La15G00153 | Chr15 | 1 075 151 | 1 079 126 | 212 | 22.390 18 | 9.80 |
| LaJAZ13 | La15G00189 | Chr15 | 1 330 626 | 1 333 013 | 283 | 30.562 52 | 9.45 |
| LaJAZ14 | La15G01076 | Chr15 | 25 198 964 | 25 199 826 | 213 | 22.829 80 | 9.93 |
| LaJAZ15 | La16G00168 | Chr16 | 1 435 513 | 1 440 135 | 156 | 17.603 23 | 10.38 |
| LaJAZ16 | La17G00791 | Chr17 | 17 083 687 | 17 085 577 | 226 | 24.465 89 | 9.93 |
| LaJAZ17 | La18G00414 | Chr18 | 4 272 331 | 4 275 830 | 330 | 34.990 72 | 9.87 |
| LaJAZ18 | La19G01387 | Chr19 | 22 164 844 | 22 167 383 | 136 | 15.788 10 | 9.52 |
| LaJAZ19 | La20G01484 | Chr20 | 16 672 832 | 16 673 901 | 214 | 22.483 34 | 9.08 |
| LaJAZ20 | La20G02795 | Chr20 | 29 446 294 | 29 450 179 | 190 | 20.995 00 | 10.19 |
| LaJAZ21 | La21G00568 | Chr21 | 6 010 126 | 6 014 598 | 413 | 45.401 64 | 8.20 |
| LaJAZ22 | La21G00790 | Chr21 | 16 075 757 | 16 078 401 | 278 | 29.511 56 | 9.81 |
| LaJAZ23 | La24G00748 | Chr24 | 6 890 348 | 6 890 710 | 121 | 13.522 38 | 9.31 |
| LaJAZ24 | La00G00334 | scaffold171 | 144 787 | 145 649 | 213 | 22.829 80 | 9.93 |
| LaJAZ25 | La00G00563 | scaffold214 | 58 478 | 62 950 | 413 | 45.391 54 | 8.27 |
| LaJAZ26 | La00G03185 | scaffold362 | 51 131 | 53 153 | 244 | 25.608 65 | 10.10 |
表1 薰衣草JAZ 基因的基本信息
Table 1 Basic information of JAZ genes in Lavandula angustifolia
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数量 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAZ1 | La01G01624 | Chr1 | 14 191 132 | 14 192 638 | 334 | 35.531 14 | 9.73 |
| LaJAZ2 | La01G02122 | Chr1 | 22 315 757 | 22 318 426 | 279 | 29.241 02 | 7.64 |
| LaJAZ3 | La01G02748 | Chr1 | 29 933 237 | 29 935 361 | 233 | 25.500 76 | 9.58 |
| LaJAZ4 | La01G02775 | Chr1 | 30 304 246 | 30 305 952 | 270 | 29.719 64 | 8.74 |
| LaJAZ5 | La02G01205 | Chr2 | 27 444 500 | 27 446 515 | 252 | 26.659 79 | 9.34 |
| LaJAZ6 | La04G00443 | Chr4 | 23 907 703 | 2 908 702 | 207 | 21.913 95 | 8.46 |
| LaJAZ7 | La10G01957 | Chr10 | 37 391 837 | 37 394 501 | 255 | 27.540 92 | 9.12 |
| LaJAZ8 | La11G01369 | Chr11 | 30 835 601 | 30 838 263 | 297 | 31.243 32 | 8.31 |
| LaJAZ9 | La12G01015 | Chr12 | 20 257 088 | 20 259 717 | 275 | 29.305 02 | 9.43 |
| LaJAZ10 | La14G00862 | Chr14 | 9 169 268 | 9 170 127 | 211 | 22.548 44 | 9.44 |
| LaJAZ11 | La15G00121 | Chr15 | 866 936 | 870 632 | 345 | 37.302 14 | 8.46 |
| LaJAZ12 | La15G00153 | Chr15 | 1 075 151 | 1 079 126 | 212 | 22.390 18 | 9.80 |
| LaJAZ13 | La15G00189 | Chr15 | 1 330 626 | 1 333 013 | 283 | 30.562 52 | 9.45 |
| LaJAZ14 | La15G01076 | Chr15 | 25 198 964 | 25 199 826 | 213 | 22.829 80 | 9.93 |
| LaJAZ15 | La16G00168 | Chr16 | 1 435 513 | 1 440 135 | 156 | 17.603 23 | 10.38 |
| LaJAZ16 | La17G00791 | Chr17 | 17 083 687 | 17 085 577 | 226 | 24.465 89 | 9.93 |
| LaJAZ17 | La18G00414 | Chr18 | 4 272 331 | 4 275 830 | 330 | 34.990 72 | 9.87 |
| LaJAZ18 | La19G01387 | Chr19 | 22 164 844 | 22 167 383 | 136 | 15.788 10 | 9.52 |
| LaJAZ19 | La20G01484 | Chr20 | 16 672 832 | 16 673 901 | 214 | 22.483 34 | 9.08 |
| LaJAZ20 | La20G02795 | Chr20 | 29 446 294 | 29 450 179 | 190 | 20.995 00 | 10.19 |
| LaJAZ21 | La21G00568 | Chr21 | 6 010 126 | 6 014 598 | 413 | 45.401 64 | 8.20 |
| LaJAZ22 | La21G00790 | Chr21 | 16 075 757 | 16 078 401 | 278 | 29.511 56 | 9.81 |
| LaJAZ23 | La24G00748 | Chr24 | 6 890 348 | 6 890 710 | 121 | 13.522 38 | 9.31 |
| LaJAZ24 | La00G00334 | scaffold171 | 144 787 | 145 649 | 213 | 22.829 80 | 9.93 |
| LaJAZ25 | La00G00563 | scaffold214 | 58 478 | 62 950 | 413 | 45.391 54 | 8.27 |
| LaJAZ26 | La00G03185 | scaffold362 | 51 131 | 53 153 | 244 | 25.608 65 | 10.10 |
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaGL1 | La03G00379 | Chr3 | 2 443 649 | 2 447 074 | 537 | 59.435 64 | 4.74 |
| LaGL2 | La13G01461 | Chr13 | 28 879 003 | 28 882 769 | 564 | 62.550 64 | 5.89 |
| LaGL3 | La22G02008 | Chr22 | 19 300 412 | 19 303 804 | 615 | 68.438 72 | 6.34 |
| LaGL4 | La23G02048 | Chr23 | 15 406 101 | 15 409 527 | 615 | 68.475 87 | 7.08 |
| LaGL5 | La00G04274 | scaffold463 | 39 967 | 46 925 | 1 260 | 139.787 86 | 7.35 |
| LaJAM1 | La04G02776 | Chr4 | 19 290 825 | 19 295 838 | 488 | 54.222 49 | 6.37 |
| LaJAM2 | La04G01048 | Chr4 | 6 403 237 | 6 405 850 | 607 | 67.699 30 | 7.29 |
| LaJAM3 | La09G00555 | Chr9 | 10 822 064 | 10 824 378 | 586 | 65.382 98 | 8.34 |
| LaJAM4 | La09G02346 | Chr9 | 26 062 346 | 26 064 746 | 500 | 56.370 18 | 7.70 |
表2 薰衣草MYC2-like基因的基本信息
Table 2 Basic information of MYC2-like genes in Lavandula angustifolia
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaGL1 | La03G00379 | Chr3 | 2 443 649 | 2 447 074 | 537 | 59.435 64 | 4.74 |
| LaGL2 | La13G01461 | Chr13 | 28 879 003 | 28 882 769 | 564 | 62.550 64 | 5.89 |
| LaGL3 | La22G02008 | Chr22 | 19 300 412 | 19 303 804 | 615 | 68.438 72 | 6.34 |
| LaGL4 | La23G02048 | Chr23 | 15 406 101 | 15 409 527 | 615 | 68.475 87 | 7.08 |
| LaGL5 | La00G04274 | scaffold463 | 39 967 | 46 925 | 1 260 | 139.787 86 | 7.35 |
| LaJAM1 | La04G02776 | Chr4 | 19 290 825 | 19 295 838 | 488 | 54.222 49 | 6.37 |
| LaJAM2 | La04G01048 | Chr4 | 6 403 237 | 6 405 850 | 607 | 67.699 30 | 7.29 |
| LaJAM3 | La09G00555 | Chr9 | 10 822 064 | 10 824 378 | 586 | 65.382 98 | 8.34 |
| LaJAM4 | La09G02346 | Chr9 | 26 062 346 | 26 064 746 | 500 | 56.370 18 | 7.70 |
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAM5 | La11G00149 | Chr11 | 952 209 | 952 997 | 214 | 23 742.54 | 6.64 |
| LaJAM6 | La14G01835 | Chr14 | 35 018 055 | 35 020 479 | 487 | 53 926.27 | 5.73 |
| LaJAM7 | La15G00077 | Chr15 | 455 788 | 457 275 | 496 | 54 676.16 | 6.17 |
| LaJAM8 | La20G01069 | Chr20 | 13 368 557 | 13 371 145 | 488 | 54 015.28 | 6.39 |
| LaJAM9 | La23G01124 | Chr23 | 8 221 465 | 8 223 969 | 604 | 67 064.71 | 7.03 |
| LaJAM10 | La26G00764 | Chr26 | 4 697 504 | 4 699 730 | 509 | 57 161.84 | 7.48 |
| LaJAM11 | La27G00171 | Chr27 | 2 153 629 | 2 156 648 | 571 | 63 635.30 | 8.47 |
| LaJAM12 | La00G05647 | scaffold88 | 71 229 | 74 222 | 604 | 67 036.69 | 7.03 |
| LaJAM13 | La00G04428 | scaffold479 | 84 467 | 87 484 | 571 | 63 579.26 | 8.52 |
| LaMYC1 | La01G01976 | Chr1 | 19 882 771 | 19 885 227 | 659 | 71 878.19 | 5.25 |
| LaMYC2 | La11G01192 | Chr11 | 26 776 504 | 26 778 951 | 655 | 71 246.38 | 5.11 |
| LaMYC3 | La13G00765 | Chr13 | 12 863 123 | 12 864 978 | 476 | 52 637.39 | 6.80 |
| LaMYC4 | La13G00767 | Chr13 | 13 043 198 | 13 044 704 | 476 | 52 623.36 | 6.80 |
| LaMYC5 | La14G01082 | Chr14 | 19 151 335 | 19 158 157 | 678 | 75 041.97 | 6.39 |
| LaMYC6 | La14G01770 | Chr14 | 34 640 706 | 34 642 877 | 617 | 66 918.74 | 5.00 |
| LaMYC7 | La15G00833 | Chr15 | 16 057 347 | 16 059 330 | 598 | 66 439.99 | 5.93 |
| LaMYC8 | La19G01463 | Chr19 | 25 336 517 | 25 338 519 | 474 | 52 241.72 | 6.06 |
表2 薰衣草MYC2-like基因的基本信息 ( 续表Continued)
Table 2 Basic information of MYC2-like genes in Lavandula angustifolia
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAM5 | La11G00149 | Chr11 | 952 209 | 952 997 | 214 | 23 742.54 | 6.64 |
| LaJAM6 | La14G01835 | Chr14 | 35 018 055 | 35 020 479 | 487 | 53 926.27 | 5.73 |
| LaJAM7 | La15G00077 | Chr15 | 455 788 | 457 275 | 496 | 54 676.16 | 6.17 |
| LaJAM8 | La20G01069 | Chr20 | 13 368 557 | 13 371 145 | 488 | 54 015.28 | 6.39 |
| LaJAM9 | La23G01124 | Chr23 | 8 221 465 | 8 223 969 | 604 | 67 064.71 | 7.03 |
| LaJAM10 | La26G00764 | Chr26 | 4 697 504 | 4 699 730 | 509 | 57 161.84 | 7.48 |
| LaJAM11 | La27G00171 | Chr27 | 2 153 629 | 2 156 648 | 571 | 63 635.30 | 8.47 |
| LaJAM12 | La00G05647 | scaffold88 | 71 229 | 74 222 | 604 | 67 036.69 | 7.03 |
| LaJAM13 | La00G04428 | scaffold479 | 84 467 | 87 484 | 571 | 63 579.26 | 8.52 |
| LaMYC1 | La01G01976 | Chr1 | 19 882 771 | 19 885 227 | 659 | 71 878.19 | 5.25 |
| LaMYC2 | La11G01192 | Chr11 | 26 776 504 | 26 778 951 | 655 | 71 246.38 | 5.11 |
| LaMYC3 | La13G00765 | Chr13 | 12 863 123 | 12 864 978 | 476 | 52 637.39 | 6.80 |
| LaMYC4 | La13G00767 | Chr13 | 13 043 198 | 13 044 704 | 476 | 52 623.36 | 6.80 |
| LaMYC5 | La14G01082 | Chr14 | 19 151 335 | 19 158 157 | 678 | 75 041.97 | 6.39 |
| LaMYC6 | La14G01770 | Chr14 | 34 640 706 | 34 642 877 | 617 | 66 918.74 | 5.00 |
| LaMYC7 | La15G00833 | Chr15 | 16 057 347 | 16 059 330 | 598 | 66 439.99 | 5.93 |
| LaMYC8 | La19G01463 | Chr19 | 25 336 517 | 25 338 519 | 474 | 52 241.72 | 6.06 |
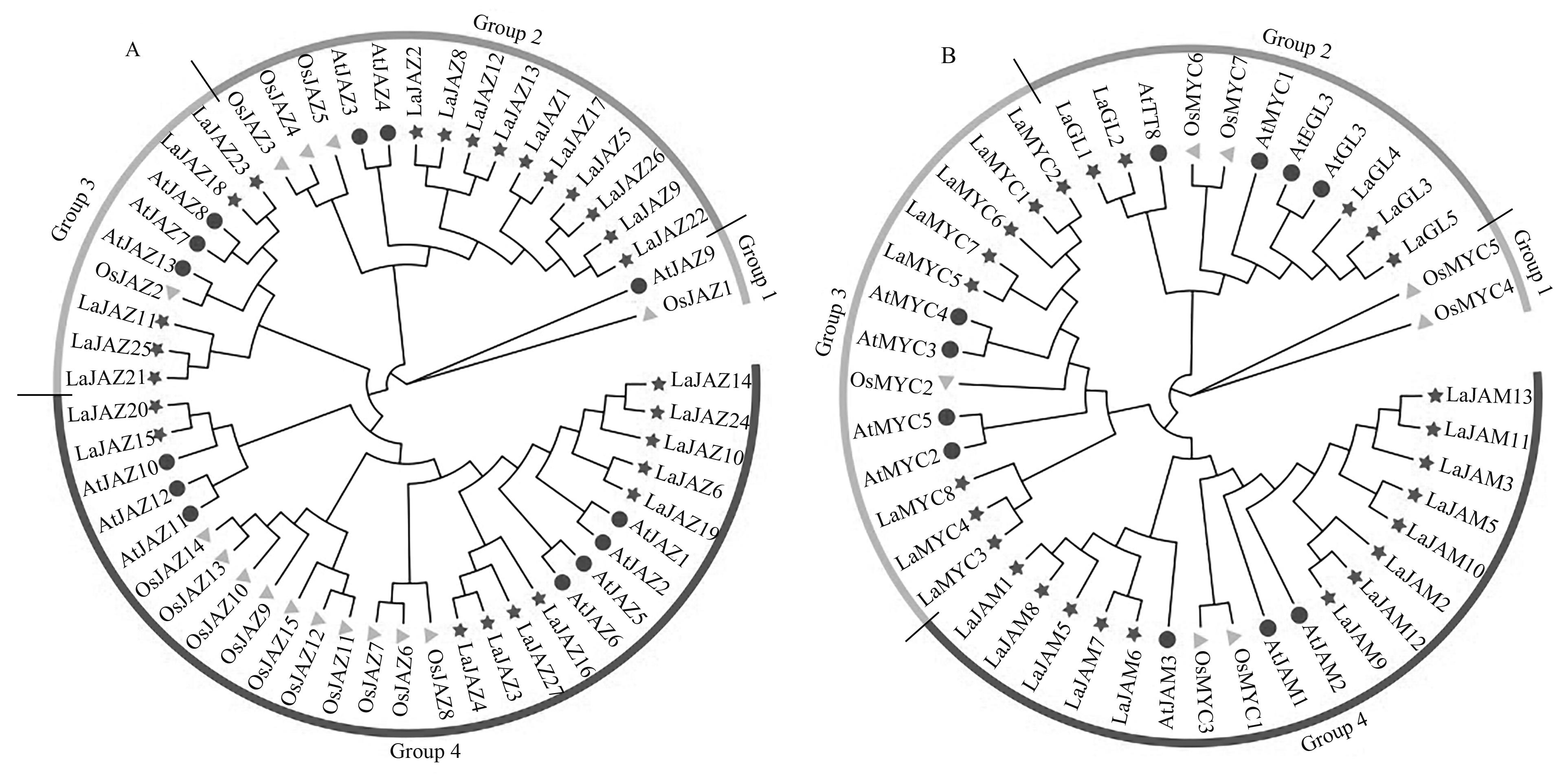
图 2 JAZ 和MYC2-like蛋白的进化树分析A:JAZ蛋白的系统发育树;B:MYC2-like蛋白的系统发育树; —薰衣草;—拟南芥;—水稻
Fig. 2 Phylogenetic tree analysis of JAZ and MYC2-like proteinA: Phylogenetic tree of JAZ proteins; B: Phylogenetic tree of MYC2-like proteins; —Lavandula angustifolia; —Arabidopsis thaliana; —Oryza sativa
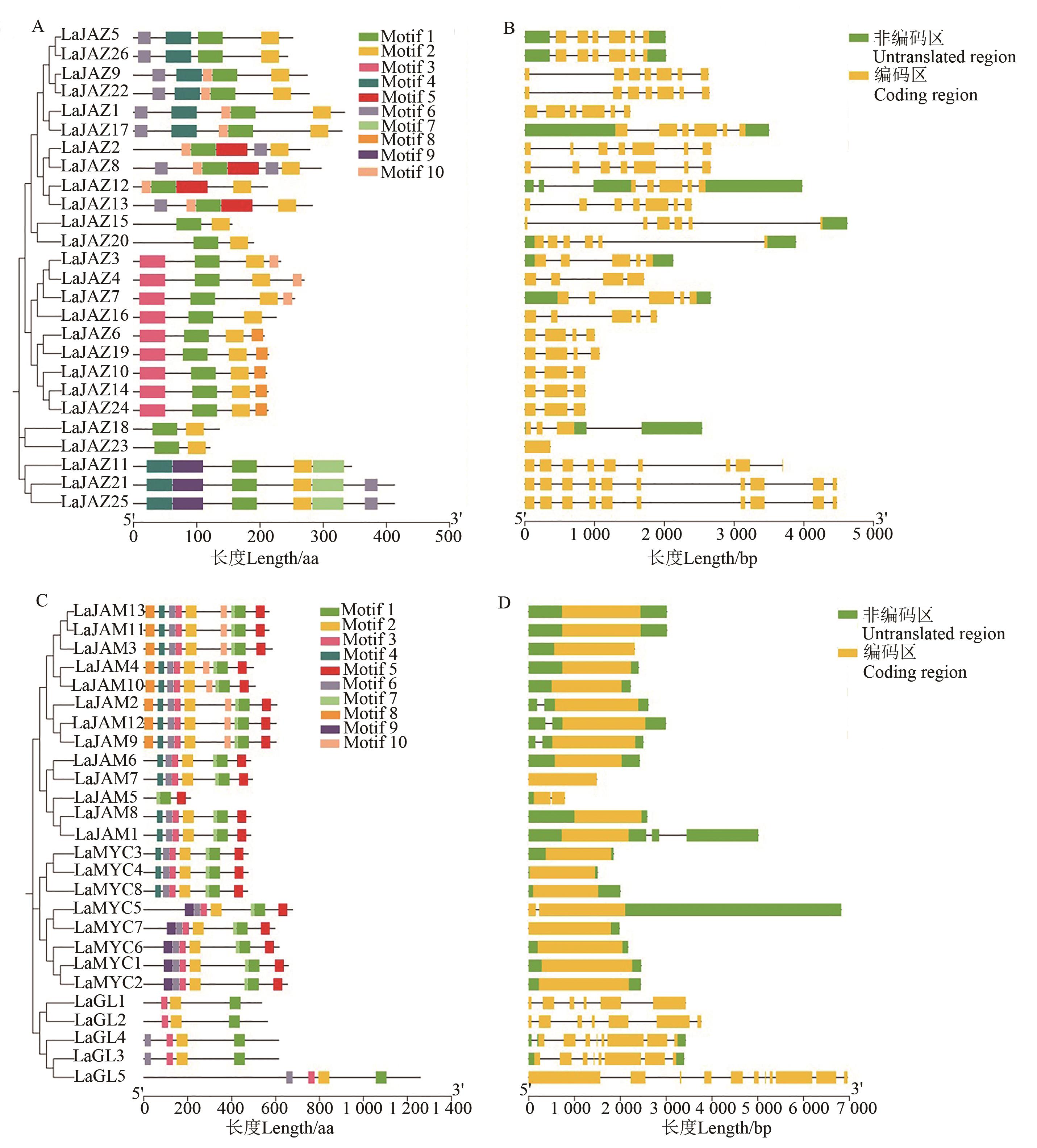
图3 LaJAZ 和 LaMYC2-like 基因编码蛋白的保守基序和基因结构分析A: LaJAZ 蛋白的保守基序组成;B: LaJAZ基因的结构;C: LaMYC2-like 蛋白的保守基序组成;D: LaMYC2-like 基因的结构
Fig. 3 Conserved motif and gene structure analysis of proteins encoded by LaJAZ and LaMYC2-like genesA: Conserved motif composition of LaJAZ proteins; B: Structure of LaJAZ genes; C: Conserved motif composition of LaMYC2-like proteins; D: Structure of LaMYC2-like genes

图4 LaJAZ和LaMYC2-like基因 2 000 bp上游启动子中顺式作用元件的分析A: LaJAZ启动子中顺式作用元件的分布;B: LaMYC2-like 启动子中顺式作用元件的分布
Fig. 4 Analysis of cis-elements in the 2 000 bp upstream promoter of LaJAZ and LaMYC2-like genesA: Distribution of cis-acting elements in the LaJAZ promoters; B: Distribution of cis-acting elements in the LaMYC2-like promoters

图5 LaJAZ和 LaMYC2-like基因在不同组织中的表达分析注:Petal—开花期深紫色花瓣;Sepal1—花蕾期深紫色萼片;Sepal2—开花期深紫色萼片;Sepal3—凋萎期深紫色萼片;Stem—2月龄的茎;Leaf—2 月龄的叶片。
Fig. 5 Expression analysis of LaJAZ and LaMYC2-like genes in different tissuesNote:Petal—Deep purple petals at the flowering stage; Sepal1—Deep purple sepals at the flower bud stage; Sepal2—Deep purple sepals at the blooming stage; Sepal3—Deep purple sepals at fade stage; Stem—Stems at 2-month-old stage;Leaf—Leaves at 2-month-old stage.

图 6 LaJAZ和LaMYC2-like基因在薰衣草不同组织中的表达分析A:用于基因表达分析的不同薰衣草组织;B:不同薰衣草组织中 LaJAZ 和 LaMYC2-like 基因的qRT-PCR表达分析。1—深紫色花瓣;2—花蕾期深紫色萼片;3—盛花期深紫色萼片;4—凋落期深紫色萼片;5—浅紫色花瓣;6—花蕾期浅紫色萼片;7—盛花期浅紫色萼片;8—凋落期浅紫色萼片;9—2月龄的根;10—2月龄的茎;11—2月龄的叶;不同小写字母表示在P<0.05水平差异显著。
Fig. 6 Expression analysis of LaJAZ and LaMYC2-like genes in different tissues of lavenderA: Different lavender tissues used for gene expression analysis; B: qRT-PCR analysis of expression of the LaJAZ and LaMYC2-like genes in different lavender tissues. 1—Deep purple petals; 2—Deep purple sepals in bud stage; 3—Deep purple sepals at blooming stage; 4—Deep purple sepals at fade stage; 5—Light purple petals; 6—Light purple sepals at bud stage; 7—Light purple sepals at blooming stage; 8—Light purple sepals at fade stage; 9—2-month-old roots; 10—2-month-old stems; 11—2-month-old leaves; different small letters indicate significant differences at P<0.05 level.
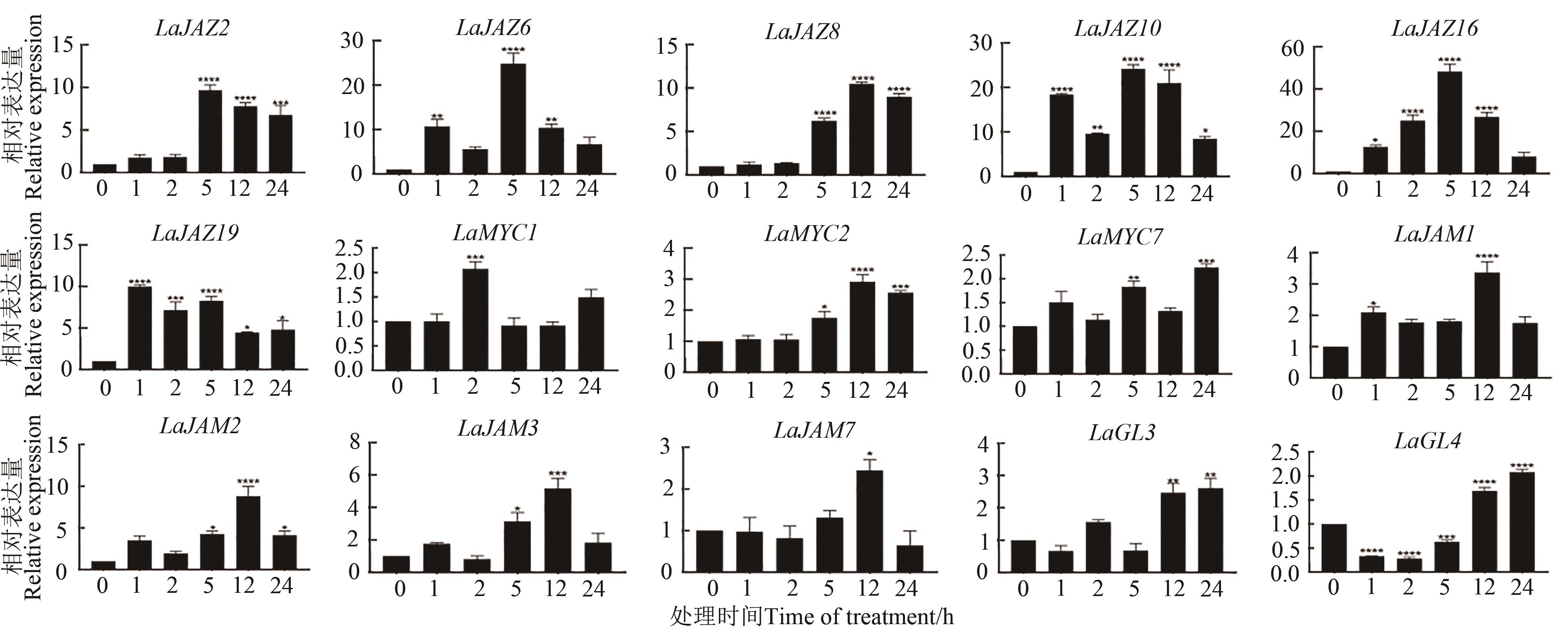
图 7 MeJA 处理 2 月龄薰衣草叶片后LaJAZ 和 LaMYC2-like基因的相对表达量注:*、**、***和****分别表示与0 h间在P<0.05、P<0.01、P<0.001、P<0.000 1水平差异显著。
Fig. 7 Relative expression of LaJAZ and LaMYC2-like genes was observed after MeJA treatment of 2-month-old lavender leavesNote: *, **, *** and **** indicate significant differences at P<0.05, P<0.01, P<0.001 and P<0.000 1 levels, repectively.
| 1 | ZHANG W Y, LI J R, DONG Y M,et al..Genome-wide identification and expression of BAHD acyltransferase gene family shed novel insights into the regulation of linalyl acetate and lavandulyl acetate in lavender [J/OL]. J.Plant Physiol.,2024,292:154143 [2024-04-20]. . |
| 2 | SOHN S I, PANDIAN S, RAKKAMMAL K, et al.. Jasmonates in plant growth and development and elicitation of secondary metabolites:an updated overview [J/OL].Plant Sci., 2022,13:942789 [2024-04-20]. . |
| 3 | CHEN X Y, WANG D D, FANG X,et al..Plant specialized metabolism regulated by jasmonate signaling [J].Plant Cell Physiol., 2019,60(12):2638-2647. |
| 4 | CHINI A, FONSECA S, FERNÁNDEZ G, et al.. The JAZ family of repressors is the missing link in jasmonate signaling [J].Nature, 2007, 448:666-671. |
| 5 | KATSIR L, CHUNG H S, KOO A J, et al.. Jasmonate signaling:a conserved mechanism of hormone sensing [J].Curr. Opin. Plant Biol., 2008, 11(4):428-435. |
| 6 | BAI Y H, MENG Y J, HUANG D L,et al..Origin and evolutionary analysis of the plant-specific TIFY transcription factor family [J]. Genomics, 2011, 98(2):128-136. |
| 7 | YAN Y X, STOLZ S, CHÉTELAT A, et al.. A downstream mediator in the growth repression limb of the jasmonate pathway [J].Plant Cell, 2007, 19(8):2470-2483. |
| 8 | YE H Y, DU H, TANG N,et al..Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice [J]. Plant Mol. Biol., 2009, 71(3):291-305. |
| 9 | ZHANG Z B, LI X L, YU R,et al..Isolation,structural analysis,and expression characteristics of the maize TIFY gene family [J].Mol. Genet. Genomics,2015,290(5):1849-1858. |
| 10 | WANG Y, QIAO L, BAI J, et al.. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.) [J/OL].BMC Genomics,2017,18(1):152 [2024-04-20]. . |
| 11 | SASAKI-SEKIMOTO Y, JIKUMARU Y, OBAYASHI T,et al..Basic Helix-loop-Helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1),JAM2,and JAM3 are negative regulators of jasmonate responses in Arabidopsis [J].Plant Physiol., 2013, 163(1):291-304. |
| 12 | GARRIDO-BIGOTES A, FIGUEROA N E, FIGUEROA P M, et al..Jasmonate signalling pathway in strawberry:genome-wide identification,molecular characterization and expression of JAZs and MYCs during fruit development and ripening [J/OL].PLoS One,2018,13(5):e0197118 [2024-04-20].. |
| 13 | WANG S, WANG Y, YANG R, et al.. Genome-wide identification and analysis uncovers the potential role of JAZ and MYC families in potato under abiotic stress [J/OL]. Int. J. Mol. Sci., 2023,24(7):6706 [2024-04-20]. . |
| 14 | LI J R, WANG Y M, DONG Y M, et al.. The chromosome-based lavender genome provides new insights into Lamiaceae evolution and terpenoid biosynthesis [J/OL]. Hortic. Res., 2021, 8:53 [2024-04-20]. . |
| 15 | CHEN C J, CHEN H, ZHANG Y,et al..TBtools:an integrative toolkit developed for interactive analyses of big biological data [J].Mol. Plant, 2020, 13(8):1194-1202. |
| 16 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002, 30(1):325-327. |
| 17 | DONG Y M, LI J R, ZHANG W Y, et al.. Exogenous application of methyl jasmonate affects the emissions of volatile compounds in lavender (Lavandula angustifolia) [J].Plant Physiol.Biochem.,2022,185:25-34. |
| 18 | HUANG G Q, GONG S Y, XU W L,et al..A fasciclin-like arabinogalactan protein,GhFLA1,is involved in fiber initiation and elongation of cotton [J]. Plant Physiol.,2013,161(3):1278-1290. |
| 19 | HONG G J, XUE X Y, MAO Y B,et al.. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression [J]. Plant Cell, 2012, 24(6):2635-2648. |
| 20 | RADHIKA V, KOST C, MITHÖFER A,et al..Regulation of extrafloral nectar secretion by jasmonates in Lima bean is light dependent [J]. Proc. Natl. Acad. Sci. USA, 2010, 107(40):17228-17233. |
| 21 | GANGAPPA S N, PRASAD V B R, CHATTOPADHYAY S.Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis [J].Plant Physiol., 2010, 154(3):1210-1219. |
| 22 | XU J S, VAN HERWIJNEN Z O, DRÄGER D B, et al.. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells [J].Plant Cell,2019,30(12):2988-3005. |
| 23 | SHEN Q, LU X, YAN T,et al..The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua [J]. New Phytol., 2016, 210(4):1269-1281. |
| 24 | DONG Y M, ZHANG W Y, LI J R,et al.. The transcription factor LaMYC4 from lavender regulates volatile terpenoid biosynthesis [J/OL].BMC Plant Biol., 2022, 22(1):289 [2024-04-20]. . |
| 25 | MA Y N, XU D B, LI L, et al.. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua [J/OL]. Sci. Adv., 2018, 4(11):eaas9357 [2024-04-20]. . |
| 26 | SHI M, ZHOU W, ZHANG J L,et al..Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins [J/OL]. Sci. Rep., 2016, 6:20919 [2024-04-20]. . |
| 27 | GUO D L, KANG K C, WANG P,et al..Transcriptome profiling of spike provides expression features of genes related to terpene biosynthesis in lavender [J/OL]. Sci.Rep., 2020, 10:6933 [2024-04-20]. . |
| 28 | KAZAN K, MANNERS J M.MYC2: the master in action [J]. Mol. Plant, 2013, 6(3):686-703. |
| [1] | 郭肖蓉, 刘颖, 樊佳珍, 黄涛, 周荣. 猪CREBRF基因生物信息学和表达规律分析[J]. 中国农业科技导报, 2024, 26(9): 44-53. |
| [2] | 鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21. |
| [3] | 陈明迪, 胡桂花, 张海文, 王旺田. 水稻RR基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(5): 20-29. |
| [4] | 吴占清, 陈威, 赵展, 许海良, 李豪远, 彭星星, 陈东旭, 张明月. 玉米GRAS基因家族的全基因组鉴定及生物信息学分析[J]. 中国农业科技导报, 2024, 26(3): 15-25. |
| [5] | 刘博, 王旺田, 马骊, 武军艳, 蒲媛媛, 刘丽君, 方彦, 孙万仓, 张岩, 刘睿敏, 曾秀存. 白菜型油菜IPT基因家族鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(2): 56-66. |
| [6] | 张振伟, 董相书, 杨婧, 李学俊, 杞美军, 蒋快乐, 杨永林, 王步天, 施学东, 邱俊超, 陈治华, 葛宇. 小粒咖啡叶绿素合成基因CaPOR的全基因组鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(10): 83-97. |
| [7] | 邓玉荣, 韩联, 王金龙, 韦兴翰, 王旭东, 赵颖, 魏小红, 李朝周. 藜麦SOD家族基因的鉴定及其对混合盐碱胁迫的响应[J]. 中国农业科技导报, 2024, 26(1): 28-39. |
| [8] | 岳俊齐, 约日耶提·萨力, 克拉热木·克里木江, 陈永坤. 薰衣草LaGGPPS5基因在大肠杆菌中催化萜类物质合成[J]. 中国农业科技导报, 2023, 25(12): 85-92. |
| [9] | 武博琼, 崔东遥, 焦仁和, 宋坚, 湛垚垚, 常亚青. 中间球海胆己糖激酶基因克隆及高温-酸化胁迫对其表达影响的初步研究[J]. 中国农业科技导报, 2022, 24(7): 205-217. |
| [10] | 王琴琴, 陈修贵, 陆许可, 王帅, 张悦新, 范亚朋, 陈全家, 叶武威. 陆地棉GhPKE1的生物信息学分析及功能验证[J]. 中国农业科技导报, 2022, 24(1): 38-45. |
| [11] | 关思静, 王楠, 徐蓉蓉, 葛甜甜, 高静, 颜永刚, 张岗, 陈莹, 张明英. 甘草PRR基因家族鉴定及表达分析[J]. 中国农业科技导报, 2021, 23(12): 66-75. |
| [12] | 李霞1,2,范鑫2,梁成真2,王远2,张锐2,孟志刚2*,刘晓东1*,孙国清2*. BS2基因在烟草中的功能解析[J]. 中国农业科技导报, 2019, 21(11): 43-50. |
| [13] | 杨笑敏,芮存,张悦新,王德龙,王俊娟,陆许可,陈修贵,郭丽雪,王帅,陈超,叶武威*. 棉花DNA甲基转移酶GhDMT9抗逆性分析[J]. 中国农业科技导报, 2019, 21(10): 12-19. |
| [14] | 商艳鹏,田燚*,李晓雨,蒋亚男,常亚青. 仿刺参7个盐度相关基因在低盐胁迫下的表达模式[J]. 中国农业科技导报, 2018, 20(11): 145-153. |
| [15] | 刘赟1,2,卢海强2,石鹏君2,谢响明1*. Achaetomium sp.Xz8来源的甘露聚糖酶Man5Xz8 热稳定性的研究[J]. , 2015, 17(3): 49-55. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号