




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (2): 56-66.DOI: 10.13304/j.nykjdb.2022.0373
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Bo LIU1,2( ), Wangtian WANG1,2(
), Wangtian WANG1,2( ), Li MA2, Junyan WU2, Yuanyuan PU2, Lijun LIU2, Yan FANG2, Wancang SUN2(
), Li MA2, Junyan WU2, Yuanyuan PU2, Lijun LIU2, Yan FANG2, Wancang SUN2( ), Yan ZHANG1,2, Ruimin LIU1, Xiucun ZENG3
), Yan ZHANG1,2, Ruimin LIU1, Xiucun ZENG3
Received:2022-05-05
Accepted:2022-07-23
Online:2024-02-15
Published:2024-02-04
Contact:
Wangtian WANG,Wancang SUN
刘博1,2( ), 王旺田1,2(
), 王旺田1,2( ), 马骊2, 武军艳2, 蒲媛媛2, 刘丽君2, 方彦2, 孙万仓2(
), 马骊2, 武军艳2, 蒲媛媛2, 刘丽君2, 方彦2, 孙万仓2( ), 张岩1,2, 刘睿敏1, 曾秀存3
), 张岩1,2, 刘睿敏1, 曾秀存3
通讯作者:
王旺田,孙万仓
作者简介:刘博 E-mail:2081826780@qq.com
基金资助:CLC Number:
Bo LIU, Wangtian WANG, Li MA, Junyan WU, Yuanyuan PU, Lijun LIU, Yan FANG, Wancang SUN, Yan ZHANG, Ruimin LIU, Xiucun ZENG. Identification and Characterization of IPT Gene Family in Brassica rapa L.[J]. Journal of Agricultural Science and Technology, 2024, 26(2): 56-66.
刘博, 王旺田, 马骊, 武军艳, 蒲媛媛, 刘丽君, 方彦, 孙万仓, 张岩, 刘睿敏, 曾秀存. 白菜型油菜IPT基因家族鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(2): 56-66.
| 基因名称 | 上游引物序列 | 下游引物序列 |
|---|---|---|
| Gene name | Forward primer sequence (5’-3’) | Reverse primer sequence (5’-3’) |
| BrIPT1 | CAAGATTCTCCCAAGCCTCGTCTG | CGCCTGAGCAAGAAGTGGAAGTC |
| BrIPT2 | TCCTTCATTCATGCCCTCCTTGTTG | CACCCAGAGGAAGCAGCAATCG |
| BrIPT3 | GTGAACGGTTACACGGTGGATTAGG | CCTAACACATATCGGACGGCAACC |
| BrIPT4 | AGGATTGTTCATCGGTGTTGGAGTG | GCAAGTGGTGTTACGTTGTTGTTCC |
| BrIPT5 | CCTCACATTCCCTTCTTCCCTTCAC | GGTTGCTCCTAAGATGACGACGATC |
| BrIPT6 | GCAACCGCCGTGCCTTATAC | GCCGGTGGGACCAGATATCA |
| BrIPT7 | GCAGCTCGTTTCTCCGGAGA | TGCGGAGTAGATGGTGAGCG |
| BrIPT8 | ATGGGTGCTACCGGAACAGG | GCAACTCTCCTCGGTCGTGA |
| BrIPT9 | GCGGAATCGACGAAGCGAAG | CGCCAAGTCTACGGCTAGCT |
| BrIPT10 | TCGCATGATGGAAGCAGGATTACTC | TGTACCACGGTCCACTAGAGATTCG |
| BrIPT11 | GGAGAAGGAGGGAGAGTGGGATTC | CTTCACTACCTGTCTCTTCGCCAAC |
| BrIPT12 | TCCGACAAGATCCAAGTTCACCAAG | GAGGCGGCAAAACACTGAGGAG |
| BrIPT13 | ATCGTCACCAACAAAGTCACTCCAG | GTAATCCTCCGCCGTGAAATCCTC |
| Actin | TGTGCCAATCTACGAGGGTTT | TTTCCCGCTCGGCTGTTGT |
Table 1 Primer sequences used in the study
| 基因名称 | 上游引物序列 | 下游引物序列 |
|---|---|---|
| Gene name | Forward primer sequence (5’-3’) | Reverse primer sequence (5’-3’) |
| BrIPT1 | CAAGATTCTCCCAAGCCTCGTCTG | CGCCTGAGCAAGAAGTGGAAGTC |
| BrIPT2 | TCCTTCATTCATGCCCTCCTTGTTG | CACCCAGAGGAAGCAGCAATCG |
| BrIPT3 | GTGAACGGTTACACGGTGGATTAGG | CCTAACACATATCGGACGGCAACC |
| BrIPT4 | AGGATTGTTCATCGGTGTTGGAGTG | GCAAGTGGTGTTACGTTGTTGTTCC |
| BrIPT5 | CCTCACATTCCCTTCTTCCCTTCAC | GGTTGCTCCTAAGATGACGACGATC |
| BrIPT6 | GCAACCGCCGTGCCTTATAC | GCCGGTGGGACCAGATATCA |
| BrIPT7 | GCAGCTCGTTTCTCCGGAGA | TGCGGAGTAGATGGTGAGCG |
| BrIPT8 | ATGGGTGCTACCGGAACAGG | GCAACTCTCCTCGGTCGTGA |
| BrIPT9 | GCGGAATCGACGAAGCGAAG | CGCCAAGTCTACGGCTAGCT |
| BrIPT10 | TCGCATGATGGAAGCAGGATTACTC | TGTACCACGGTCCACTAGAGATTCG |
| BrIPT11 | GGAGAAGGAGGGAGAGTGGGATTC | CTTCACTACCTGTCTCTTCGCCAAC |
| BrIPT12 | TCCGACAAGATCCAAGTTCACCAAG | GAGGCGGCAAAACACTGAGGAG |
| BrIPT13 | ATCGTCACCAACAAAGTCACTCCAG | GTAATCCTCCGCCGTGAAATCCTC |
| Actin | TGTGCCAATCTACGAGGGTTT | TTTCCCGCTCGGCTGTTGT |
基因ID Gene ID | 基因名 Gene name | 氨基酸 Amino acids/aa | 分子量 Molecular weight/Da | 等电点PI | 带负电荷残基数 Total number of negatively charged residues | 带正电荷残基数 Total number of positively charged residue | 不稳定 指数Instability index | 脂肪指数Aliphatic index |
|---|---|---|---|---|---|---|---|---|
| Brapa01T003069.1 | BrIPT1 | 332 | 37 126.59 | 8.77 | 37 | 42 | 39.85 | 91.87 |
| Brapa01T003426.1 | BrIPT2 | 324 | 36 886.28 | 8.50 | 41 | 44 | 44.02 | 81.45 |
| Brapa02T000813.1 | BrIPT3 | 334 | 37 920.51 | 5.75 | 50 | 44 | 34.85 | 94.55 |
| Brapa02T000886.1 | BrIPT4 | 542 | 61 015.95 | 7.09 | 66 | 66 | 46.35 | 80.76 |
| Brapa02T001911.1 | BrIPT5 | 348 | 39 700.26 | 8.72 | 49 | 52 | 54.61 | 81.24 |
| Brapa03T000921.1 | BrIPT6 | 463 | 52 426.39 | 7.50 | 59 | 60 | 50.28 | 78.98 |
| Brapa03T003913.1 | BrIPT7 | 326 | 37 116.50 | 9.34 | 37 | 45 | 48.52 | 82.48 |
| Brapa04T000031.1 | BrIPT8 | 334 | 37 337.14 | 8.11 | 42 | 44 | 44.57 | 90.42 |
| Brapa04T002174.1 | BrIPT9 | 496 | 56 182.83 | 6.15 | 75 | 68 | 41.78 | 89.78 |
| Brapa07T001138.1 | BrIPT10 | 335 | 37 477.17 | 8.48 | 40 | 44 | 35.54 | 91.34 |
| Brapa07T003284.1 | BrIPT11 | 333 | 37 800.18 | 8.40 | 46 | 48 | 44.07 | 88.68 |
| Brapa09T001382.1 | BrIPT12 | 332 | 37 568.44 | 8.40 | 44 | 47 | 37.94 | 91.27 |
| Brapa10T001241.1 | BrIPT13 | 334 | 37 876.59 | 5.96 | 48 | 44 | 35.22 | 92.22 |
Table 2 Protein physical and chemical properties of IPT in Brassica rapa L.
基因ID Gene ID | 基因名 Gene name | 氨基酸 Amino acids/aa | 分子量 Molecular weight/Da | 等电点PI | 带负电荷残基数 Total number of negatively charged residues | 带正电荷残基数 Total number of positively charged residue | 不稳定 指数Instability index | 脂肪指数Aliphatic index |
|---|---|---|---|---|---|---|---|---|
| Brapa01T003069.1 | BrIPT1 | 332 | 37 126.59 | 8.77 | 37 | 42 | 39.85 | 91.87 |
| Brapa01T003426.1 | BrIPT2 | 324 | 36 886.28 | 8.50 | 41 | 44 | 44.02 | 81.45 |
| Brapa02T000813.1 | BrIPT3 | 334 | 37 920.51 | 5.75 | 50 | 44 | 34.85 | 94.55 |
| Brapa02T000886.1 | BrIPT4 | 542 | 61 015.95 | 7.09 | 66 | 66 | 46.35 | 80.76 |
| Brapa02T001911.1 | BrIPT5 | 348 | 39 700.26 | 8.72 | 49 | 52 | 54.61 | 81.24 |
| Brapa03T000921.1 | BrIPT6 | 463 | 52 426.39 | 7.50 | 59 | 60 | 50.28 | 78.98 |
| Brapa03T003913.1 | BrIPT7 | 326 | 37 116.50 | 9.34 | 37 | 45 | 48.52 | 82.48 |
| Brapa04T000031.1 | BrIPT8 | 334 | 37 337.14 | 8.11 | 42 | 44 | 44.57 | 90.42 |
| Brapa04T002174.1 | BrIPT9 | 496 | 56 182.83 | 6.15 | 75 | 68 | 41.78 | 89.78 |
| Brapa07T001138.1 | BrIPT10 | 335 | 37 477.17 | 8.48 | 40 | 44 | 35.54 | 91.34 |
| Brapa07T003284.1 | BrIPT11 | 333 | 37 800.18 | 8.40 | 46 | 48 | 44.07 | 88.68 |
| Brapa09T001382.1 | BrIPT12 | 332 | 37 568.44 | 8.40 | 44 | 47 | 37.94 | 91.27 |
| Brapa10T001241.1 | BrIPT13 | 334 | 37 876.59 | 5.96 | 48 | 44 | 35.22 | 92.22 |
基因ID Gene ID | 基因名Gene name | 最大亲水值Maximum hydrophilic value | 位置Position/aa | 最大疏水值Maximum hydrophobic value | 位置Position/aa | 亲水性平均系数 Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| Brapa01T003069.1 | BrIPT1 | 1.956 | 40 | -2.922 | 276 | -0.262 |
| Brapa01T003426.1 | BrIPT2 | 1.856 | 47 | -2.311 | 214 | -0.338 |
| Brapa02T000813.1 | BrIPT3 | 2.122 | 318 | -2.489 | 263 | -0.224 |
| Brapa02T000886.1 | BrIPT4 | 3.300 | 63,64 | -3.100 | 124 | -0.390 |
| Brapa02T001911.1 | BrIPT5 | 2.389 | 66 | -3.411 | 57 | -0.412 |
| Brapa03T000921.1 | BrIPT6 | 1.700 | 274 | -3.489 | 50 | -0.433 |
| Brapa03T003913.1 | BrIPT7 | 2.144 | 48 | -2.356 | 214 | -0.331 |
| Brapa04T000031.1 | BrIPT8 | 2.144 | 47 | -2.522 | 153 | -0.137 |
| Brapa04T002174.1 | BrIPT9 | 2.822 | 17 | -3.744 | 482 | -0.458 |
| Brapa07T001138.1 | BrIPT10 | 1.956 | 40 | -2.756 | 276 | -0.210 |
| Brapa07T003284.1 | BrIPT11 | 2.389 | 52 | -3.078 | 308 | -0.343 |
| Brapa09T001382.1 | BrIPT12 | 2.144 | 47 | -2.456 | 153 | -0.187 |
| Brapa10T001241.1 | BrIPT13 | 1.956 | 43 | -2.489 | 263 | -0.225 |
Table 3 Amino acid hydrophilicity and hydrophobicity
基因ID Gene ID | 基因名Gene name | 最大亲水值Maximum hydrophilic value | 位置Position/aa | 最大疏水值Maximum hydrophobic value | 位置Position/aa | 亲水性平均系数 Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| Brapa01T003069.1 | BrIPT1 | 1.956 | 40 | -2.922 | 276 | -0.262 |
| Brapa01T003426.1 | BrIPT2 | 1.856 | 47 | -2.311 | 214 | -0.338 |
| Brapa02T000813.1 | BrIPT3 | 2.122 | 318 | -2.489 | 263 | -0.224 |
| Brapa02T000886.1 | BrIPT4 | 3.300 | 63,64 | -3.100 | 124 | -0.390 |
| Brapa02T001911.1 | BrIPT5 | 2.389 | 66 | -3.411 | 57 | -0.412 |
| Brapa03T000921.1 | BrIPT6 | 1.700 | 274 | -3.489 | 50 | -0.433 |
| Brapa03T003913.1 | BrIPT7 | 2.144 | 48 | -2.356 | 214 | -0.331 |
| Brapa04T000031.1 | BrIPT8 | 2.144 | 47 | -2.522 | 153 | -0.137 |
| Brapa04T002174.1 | BrIPT9 | 2.822 | 17 | -3.744 | 482 | -0.458 |
| Brapa07T001138.1 | BrIPT10 | 1.956 | 40 | -2.756 | 276 | -0.210 |
| Brapa07T003284.1 | BrIPT11 | 2.389 | 52 | -3.078 | 308 | -0.343 |
| Brapa09T001382.1 | BrIPT12 | 2.144 | 47 | -2.456 | 153 | -0.187 |
| Brapa10T001241.1 | BrIPT13 | 1.956 | 43 | -2.489 | 263 | -0.225 |

Fig. 6 Site map of IPT gene element in Brassica rapa L.Note:GT1-motif—Light responsiveness; LTR—Low-temperature responsiveness; ABRE—Abscisic acid responsiveness; O2-site—Zein metabolism regulation; TGACG-motif—MeJA-responsiveness; TC-rich repeats—Defense and stress responsiveness; TATC-box—Gibberellin-responsiveness; ARE—Anaerobic induction; CGTCA-motif—MeJA-responsiveness; MRE—Light responsiveness; G-box—Light responsiveness; TCA-element—Salicylic acid responsiveness; RY-element—Seed-specific regulation; G-Box—Light responsiveness; AuxRR-core—Auxin responsiveness; MBS—Drought-induced response element; ACE—Light responsiveness; circadian—Circadian control; 3-AF1 binding site—Light responsive element; CAT-box—Meristem expression; MBSI—Flavonoid biosynthetic genes regulation; GC-motif—Anoxic specific inducibility; Sp1—Light responsive element; GCN4-motif—Endosperm expression.
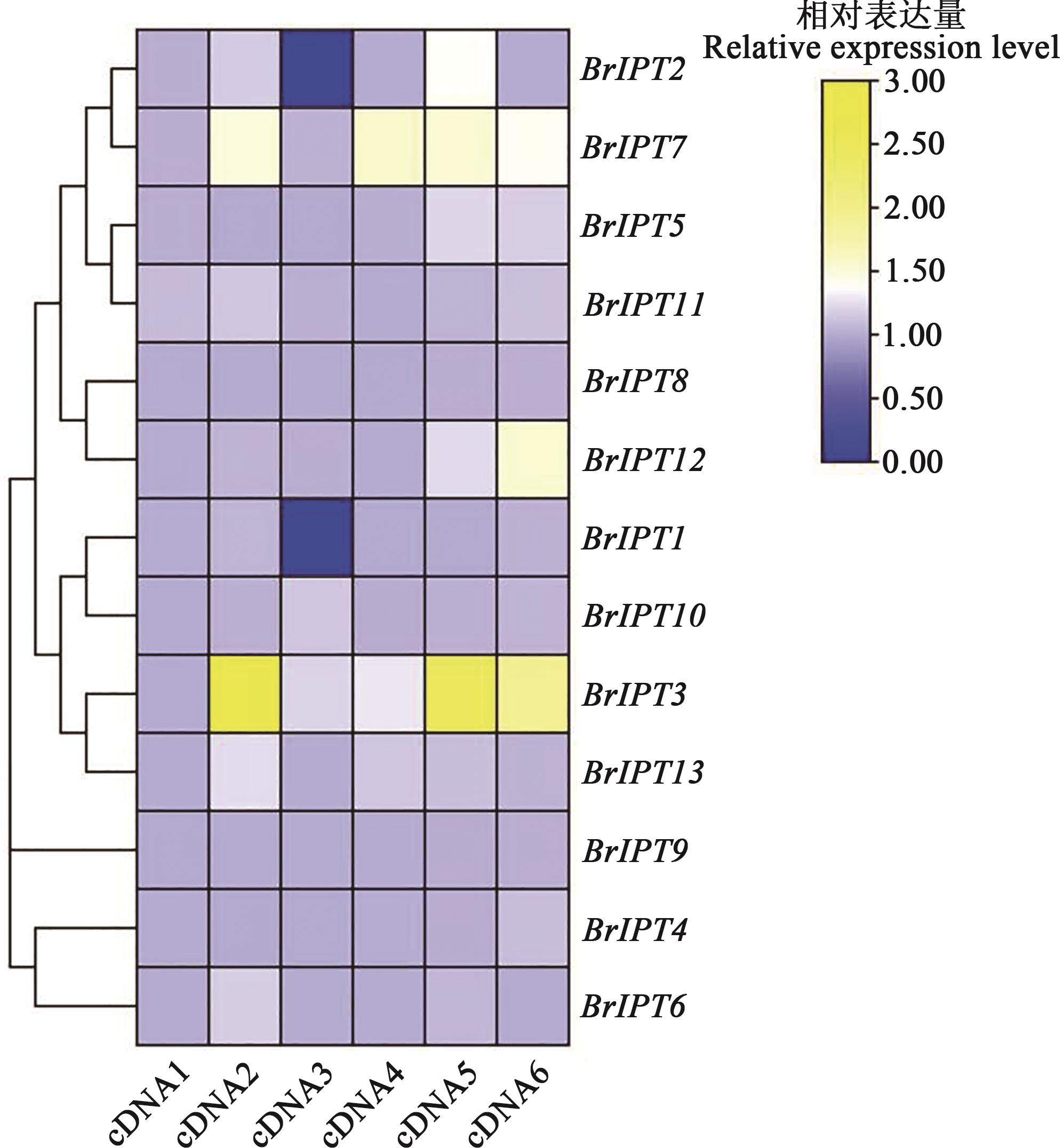
Fig. 7 Expression of IPT gene family in different tissuesNote:cDNA1—Seedling root;cDNA2—Seedling hypocotyl;cDNA3—Seedling cotyledons;cDNA4—Blooming root;cDNA5—Blooming hypocotyl;cDNA6—Blooming leaf.
| 1 | 刘成,冯中朝,肖唐华,等.我国油菜产业发展现状、潜力及对策[J].中国油料作物学报,2019,41(4):485-489. |
| LIU C, FENG Z C, XIAO T H, et al.. Development, potential and adaptation of Chinese rapeseed industry [J]. Chin. J. Oil Crop Sci., 2019, 41(4):485-489. | |
| 2 | 吕艳,黄涌,邹锡玲,等.油菜抗低温的评价指标与分子生理机制研究进展[J].中国油料作物学报,2020,42(4):527-535. |
| LYU Y, HUANG Y, ZOU X L, et al.. Reaserches on evaluation, physiological and molecular mechanism of rapeseed low-tempreature resistance [J]. Chin. J. Oil Crop Sci., 2020, 42(4):527-535. | |
| 3 | 马骊,孙万仓,刘自刚,等.白菜型与甘蓝型冬油菜抗寒机理差异的研究[J].华北农学报,2016,31(1):147-154. |
| MA L, SUN W C, LIU Z G, et al.. Study of difference in mechanism of cold resistance of winter rapeseed of Brassica rape and Brassica napus [J]. Acta Agric. Boreali-Sin., 2016, 31(1):147-154. | |
| 4 | MOK D W, MOK M C. Cytokinin metabolism and action [J]. Annu. Rev. Plant Physiol., 2001, 52:89-118. |
| 5 | HAI N N, LAI N, KISIALA A B, et al.. Isopentenyltransferases as master regulators of crop performance: their function, manipulation, and genetic potential for stress adaptation and yield improvement [J]. Plant Biotechnol. J., 2021, 19:1297-1313. |
| 6 | KAMADA-NOBUSADA T, SAKAKIBARA H. Molecular basis for cytokinin biosynthesis [J]. Phytochemistry, 2009, 70(4):444-449. |
| 7 | BEZNEC A, FACCIO P, MIRALLES D J, et al.. Stress-induced expression of IPT gene in transgenic wheat reduces grain yield penalty under drought [J]. J. Genet. Eng. Biotech., 2021, 19(1):67-84. |
| 8 | 赵佩,高成香,张娜,等.转IPT基因棉花衰老相关基因的差异表达分析[J].分子植物育种,2012,10(3):324-330. |
| ZHAO P, GAO C X, ZHANG N, et al.. Study on differential expression patterns of senescence-associated genes in transgenic cotton with IPT gene [J]. Mol. Plant Breeding, 2012, 10(3):324-330. | |
| 9 | QIAN C L, REN N N, WANG J Y, et al.. Effects of exogenous application of CPPU, NAA and GA4+7 on parthenocarpy and fruit quality in cucumber (Cucumis sativus L.) [J]. Food Chem., 2018, 243(3):410-413. |
| 10 | CORTLEVEN A, LEUENDORF J E, FRANK M, et al.. Cytokinin action in response to abiotic and biotic stress in plants [J]. Plant Cell Environ., 2019, 42(11):998-1018. |
| 11 | 李海林, 赵德刚. 转IPT基因油菜提高抗蚜虫能力[J].分子植物育种,2012,9(3):343-349. |
| LI H L, ZHAO D G. IPT gene expression increased the aphid-resistant of transgenic plant in oilseed [J]. Mol. Plant Breeding, 2012, 9(3):343-349. | |
| 12 | MIYAWAKI K, TARKOWSKI P, MATSUMOTO-KITANO M, et al.. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis [J]. Proc. Natl. Acad. Sci. USA, 2006, 103(44):16598-16603. |
| 13 | 冯杰.水稻RPL1基因相关突变体鉴定及水稻IPT蛋白家族分析[D].南宁:广西大学,2020. |
| FENG J. Identification of rice RPL1 gene related mutants and analysis of rice IPT protein family [D]. Nanning: Guangxi University, 2020. | |
| 14 | 黄文晓,翁飞,査满荣,等.水稻突变体D12W191多分蘖表型产生与细胞分裂素的关系[J].南京农业大学学报,2016,39(5):711-721. |
| HUANG W X, WENG F, ZHA M R, et al.. Relationship of the multi-tiller phenotype of a rice mutant D12W191 with cytokinin [J]. J. Nanjing Agric. Univ., 2016, 39(5):711-721. | |
| 15 | 许明,黄昕,郑华坤,等.高世代转PSAG12-IPT基因水稻生育后期的延衰性[J].福建农林大学学报(自然科学版),2010,39(1):1-5. |
| XU M, HUANG X, ZHENG H K, et al.. Study on senescence delay characteristics of transgenic rice containing anti-senescence chimeric gene PSAG12-IPT in high generation at later developmental stages [J]. J. Fujian Agric. For. Univ. (Nat. Sci.), 2010, 39(1):1-5. | |
| 16 | 玉山江·麦麦提,熊叶辉,麦合木提江·米吉提,等.转IPT基因水稻的抗旱性研究[J].中国农业科技导报,2012,14(6):30-35. |
| Maimaiti Yushanjiang, XIONG Y H, Mijiti Maihemutijiang, et al.. Studies on drought tolerance of IPT transgenic rice [J]. J. Agric. Sci. Technol., 2012, 14(6):30-35. | |
| 17 | 徐春梅,邹娅,刘自刚,等.白菜型冬油菜萌动种子低温春化的生理生化特征[J].中国农业科学,2020,53(5):929-941. |
| XU C M, ZOU Y, LIU Z G, et al.. Physiological and biochemical characteristics of low temperature vernalization of germinating seeds of Brassica rapa [J]. Sci. Agric. Sin., 2020, 53(5):929-941. | |
| 18 | 张停林,李季,崔利,等.黄瓜细胞分裂素合成关键酶IPT基因家族序列特征及其表达分析[J].园艺学报,2013,40(1):58-68. |
| ZHANG T L, LI J, CUI L, et al.. Identification and characterization of CsIPT genes in cucumber [J]. Acta Hortic. Sin., 2013, 40(1):58-68. | |
| 19 | TAN M, LI G F, QI S Y, et al.. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.) [J]. Gene, 2018, 651:106-117. |
| 20 | 蔡兆明,程春红,廖静静,等.茎瘤芥异戊烯基转移酶家族基因全基因组鉴定及表达分析[J].西北植物学报,2022,42(1):38-47. |
| CAI Z M, CHENG C H, LIAO J J, et al.. Genome-wide identification and expression analysis of isopentenyl transferases genes in tuber mustard [J]. Acta Bot. Bor-Occid. Sin., 2022, 42(1):38-47. | |
| 21 | MUN J H, KWON S J, YANG T J, et al.. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication [J/OL]. Genome Biol., 2009, 10(10):R111 [2022-03-15]. . |
| 22 | LIU Z N, LYU Y X, ZHANG M, et al.. Identification, expression and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis) [J]. BMC Genomics, 2013, 14(1):594-614. |
| 23 | LE D T, NISHIYAMA R, WATANABE Y, et al.. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels [J/OL]. PLoS One, 2012, 7(8):e42411 [2022-03-15]. . |
| 24 | SAKAMOTO T, SAKAKIBARA H, KOJIMA M, et al.. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice [J]. Plant Physiol., 2006, 142(1):54-62. |
| 25 | BRUGIERE N, HUMBERT S, RIZZO N, et al.. A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development [J]. Plant Mol. Biol., 2008, 67(3):215-229. |
| 26 | 吕享,叶睿华,田海露,等.生长素介导细胞分裂素(玉米素)调控杜鹃兰侧芽萌发[J].农业生物技术学报,2018,26(11):1872-1879. |
| LYU X, YE R H, TIAN H L, et al.. Auxin regulated lateral buds germination in cremastra appendiculata via the regulation of cytokinin (zeatin) [J]. J. Agric. Biotech., 2018, 26(11):1872-1879. | |
| 27 | HIROSE N, TAKEI K, KUROHA T. Regulation of cytokinin biosynthesis, compartmentalization and translocation [J]. J. Exp. Bot., 2008, 59(1):75-83. |
| 28 | MIYAWAKI K, MATSUMOTO-KITANO M, KAKIMOTO T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate [J]. Plant J., 2004, 37(1):128-138. |
| 29 | YE C J, WU S W, KONG F N, et al.. Identification and characterization of an isopentenyltransferase (IPT) gene in soybean (Glycine max L.) [J]. Plant Sci., 2006, 170(3):542-550. |
| [1] | Yurong DENG, Lian HAN, Jinlong WANG, Xinghan WEI, Xudong WANG, Ying ZHAO, Xiaohong WEI, Chaozhou LI. Identification of SOD Family Genes in Chenopodium quinoa and Their Response to Mixed Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 28-39. |
| [2] | Qinqin WANG, Xiugui CHEN, Xuke LU, Shuai WANG, Yuexin ZHANG, Yapeng FAN, Quanjia CHEN, Wuwei YE. Bioinformatics Analysis and Functional Verification of GhPKE1 inUpland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 38-45. |
| [3] | GUAN Sijing, WANG Nan, XU Rongrong, GE Tiantian, GAO Jing, YAN Yonggang, ZHANG Gang, CHEN Ying, ZHANG Mingying. [J]. Journal of Agricultural Science and Technology, 2021, 23(12): 66-75. |
| [4] | LI Xia1,2, FAN Xin2, LIANG Chengzhen2, WANG Yuan2, ZHANG Rui2, MENG Zhigang2*, LIU Xiaodong1*, SUN Guoqing2*. Functional Analysis of BS2 Gene in Tobacco [J]. Journal of Agricultural Science and Technology, 2019, 21(11): 43-50. |
| [5] | YANG Xiaomin, RUI Cun, ZHANG Yuexin, WANG Delong, WANG Junjuan, LU Xuke, CHEN Xiugui, GUO Lixue, WANG Shuai, CHEN Chao, YE Wuwei*. Cloning and Stress Resistance Analysis of Cotton DNA Methyltransferase GhDMT9 Gene [J]. Journal of Agricultural Science and Technology, 2019, 21(10): 12-19. |
| [6] | ZHAO Yan1, SHENG Yunlong1, SONG Yafei1, ZHANG Jiakuo1, WENG Qiaoyun1, YUAN Jincheng1, ZHAO Zhihai2*, LIU Yinghui1*. Genome-wide Identification and Bio-informatics Analysis of Superoxide Dismutase Gene Family in Setaria italica [J]. Journal of Agricultural Science and Technology, 2018, 20(8): 1-6. |
| [7] | MAIMAITI Yushanjiang1, XIONG Yehui1, MIJITI Maihemutijiang1, SEHROON Khan. Studies on Drought Tolerance of IPT Transgenic Rice [J]. , 2012, 14(6): 30-35. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号