




















中国农业科技导报 ›› 2022, Vol. 24 ›› Issue (11): 76-86.DOI: 10.13304/j.nykjdb.2022.0341
柯会锋1( ), 孙正文1, 王国宁1, 孟成生1, 吴立强2(
), 孙正文1, 王国宁1, 孟成生1, 吴立强2( )
)
收稿日期:2022-04-23
接受日期:2022-05-28
出版日期:2022-11-15
发布日期:2022-11-29
通讯作者:
吴立强
作者简介:柯会锋E-mail: kehuifeng123@126.com;
基金资助:
Huifeng KE1( ), Zhengwen SUN1, Guoning WANG1, Chengsheng MENG1, Liqiang WU2(
), Zhengwen SUN1, Guoning WANG1, Chengsheng MENG1, Liqiang WU2( )
)
Received:2022-04-23
Accepted:2022-05-28
Online:2022-11-15
Published:2022-11-29
Contact:
Liqiang WU
摘要:
棉籽在油脂、饲料加工等领域具有重要用途,且影响纤维产量和品质。为发掘棉籽性状遗传位点与候选基因,利用419份具有丰富遗传变异的棉花品种资源构成的自然群体,在2个年度鉴定其棉籽性状,结合群体重测序SNP基因型进行全基因组关联分析。结果发现,供试群体棉籽长度、宽度、长宽比、面积与周长等性状存在较大遗传变异,变异系数3.91%~9.55%;有7个SNPs在2年同时被检测到与棉籽大小和形状关联,有23个SNPs在1年与3个以上棉籽相关性状关联,其中D11染色体D11∶50561153在2年与粒宽、面积和周长关联,D12染色体D12:56031535在2年与粒长、面积和周长关联;同时,D05染色体在2年检测到4个与籽粒长宽比显著关联SNPs,A07染色体检测到10个与粒长、周长和面积关联SNPs,且单倍型间相关性状差异极显著。进一步分析发现,关联位点附近存在21个候选基因随棉花胚珠发育进程表达量呈现增加趋势,其中Gh_D05G0148编码乙烯调控反应的EBF蛋白(EIN3-binding F-box protein),该基因在棉花开花后1~35 d的胚珠中优势表达,且以开花后10 d表达量最高;同时,候选基因Gh_D05G0144编码胚珠发育相关YABBY转录因子,该基因在棉花开花后20~35 d的胚珠中表达量较高,推测与棉籽后期发育有关。以上结果为棉籽多性状同步改良提供了选择标记与基因,为实现纤维和棉籽性状同步遗传改良奠定了基础。
中图分类号:
柯会锋, 孙正文, 王国宁, 孟成生, 吴立强. 棉籽大小与形状关联标记发掘及候选基因筛选[J]. 中国农业科技导报, 2022, 24(11): 76-86.
Huifeng KE, Zhengwen SUN, Guoning WANG, Chengsheng MENG, Liqiang WU. Mining and Screening of Associated Markers and Candidate Genes Related to Seed Size and Shape in Cotton[J]. Journal of Agricultural Science and Technology, 2022, 24(11): 76-86.
| 性状 Trait | 年度 Year | 均值 Mean | 标准差 SD | 变异系数 CV/% | 最大值 Max. | 最小值 Min. | 偏度系数 Skew | 峰度系数 Kurt |
|---|---|---|---|---|---|---|---|---|
| 粒长 Seed length/mm | 2015 | 8.71 | 0.49 | 5.63 | 10.34 | 7.44 | 0.48 | 0.49 |
| 2017 | 8.20 | 0.45 | 5.49 | 10.54 | 7.09 | 0.59 | 1.52 | |
| 粒宽 Seed width/mm | 2015 | 4.74 | 0.23 | 4.85 | 5.46 | 4.06 | 0.16 | 0.30 |
| 2017 | 4.59 | 0.22 | 4.79 | 5.67 | 3.92 | 0.55 | 1.64 | |
| 籽粒长宽比 Ratio of seed length to width | 2015 | 1.85 | 0.08 | 4.32 | 2.11 | 1.65 | 0.41 | 0.50 |
| 2017 | 1.79 | 0.07 | 3.91 | 2.04 | 1.57 | 0.26 | 1.04 | |
| 籽粒面积 Seed area/mm2 | 2015 | 30.89 | 2.92 | 9.45 | 42.11 | 23.76 | 0.48 | 0.57 |
| 2017 | 28.69 | 2.74 | 9.55 | 45.16 | 22.38 | 0.89 | 3.07 | |
| 籽粒周长 Seed perimeter/mm | 2015 | 22.81 | 1.21 | 5.30 | 28.27 | 20.11 | 0.59 | 1.09 |
| 2017 | 21.43 | 1.10 | 5.13 | 27.32 | 18.96 | 0.65 | 1.71 |
表1 自然群体棉籽相关性状遗传变异
Table 1 Genetic variation of seed related traits in cotton natural population
| 性状 Trait | 年度 Year | 均值 Mean | 标准差 SD | 变异系数 CV/% | 最大值 Max. | 最小值 Min. | 偏度系数 Skew | 峰度系数 Kurt |
|---|---|---|---|---|---|---|---|---|
| 粒长 Seed length/mm | 2015 | 8.71 | 0.49 | 5.63 | 10.34 | 7.44 | 0.48 | 0.49 |
| 2017 | 8.20 | 0.45 | 5.49 | 10.54 | 7.09 | 0.59 | 1.52 | |
| 粒宽 Seed width/mm | 2015 | 4.74 | 0.23 | 4.85 | 5.46 | 4.06 | 0.16 | 0.30 |
| 2017 | 4.59 | 0.22 | 4.79 | 5.67 | 3.92 | 0.55 | 1.64 | |
| 籽粒长宽比 Ratio of seed length to width | 2015 | 1.85 | 0.08 | 4.32 | 2.11 | 1.65 | 0.41 | 0.50 |
| 2017 | 1.79 | 0.07 | 3.91 | 2.04 | 1.57 | 0.26 | 1.04 | |
| 籽粒面积 Seed area/mm2 | 2015 | 30.89 | 2.92 | 9.45 | 42.11 | 23.76 | 0.48 | 0.57 |
| 2017 | 28.69 | 2.74 | 9.55 | 45.16 | 22.38 | 0.89 | 3.07 | |
| 籽粒周长 Seed perimeter/mm | 2015 | 22.81 | 1.21 | 5.30 | 28.27 | 20.11 | 0.59 | 1.09 |
| 2017 | 21.43 | 1.10 | 5.13 | 27.32 | 18.96 | 0.65 | 1.71 |
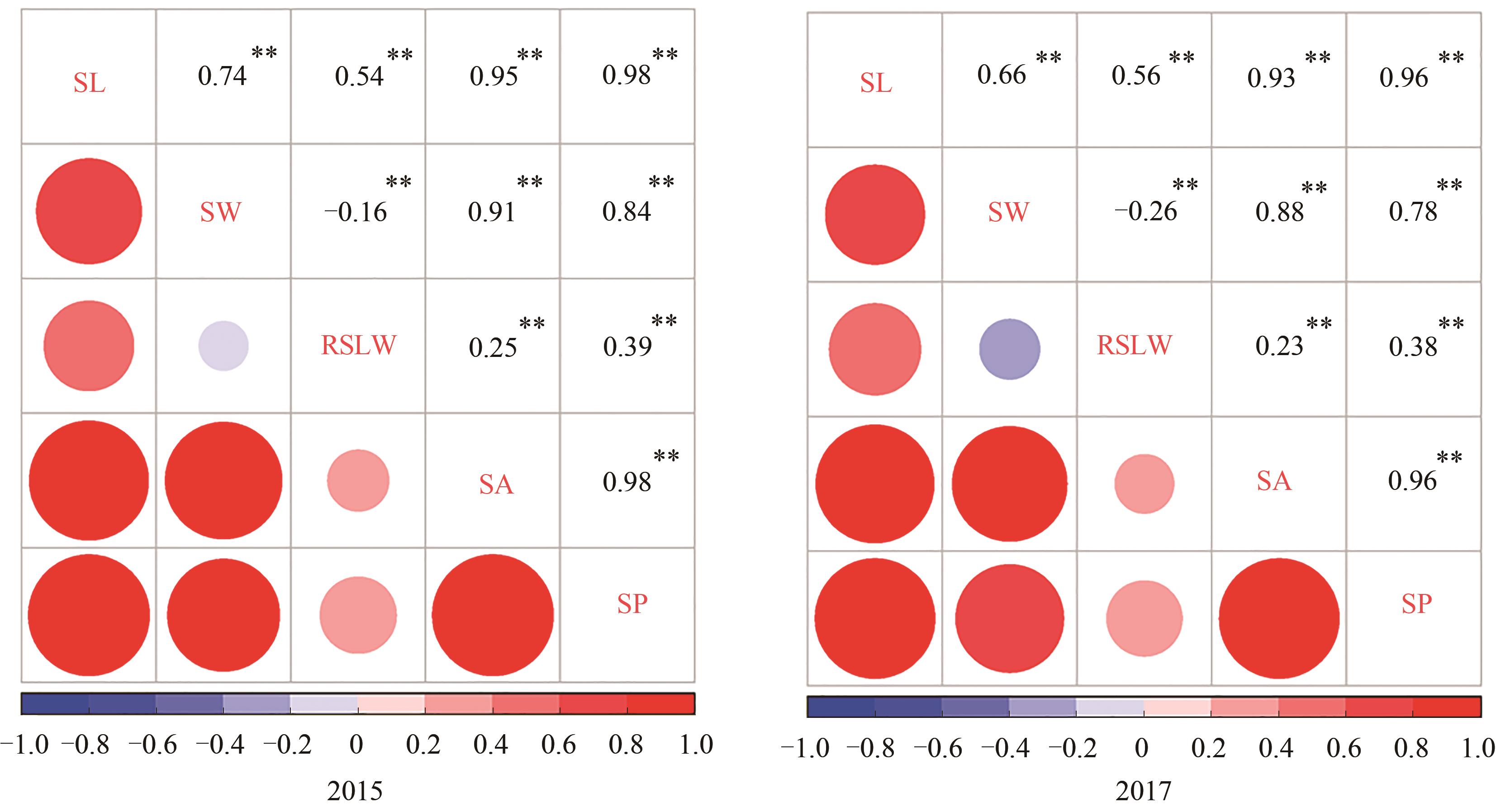
图2 供试棉花自然群体棉籽性状相关性注:SL—粒长;SW—粒宽;RSLW—籽粒长宽比;SA—籽粒面积;SP—籽粒周长;**表示相关性在P<0.01水平显著。
Fig. 2 Correlation of seed related traits in cotton natural populationNote: SL—Seed length; SW—Seed width; RSLW—Ratio of seed length to width; SA—Seed area; SP—Seed perimeter; ** indicates significant correlation at P<0.01 level.
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 5 | D05 | D05∶1496927 | 1 496 927 | T/C | RSLW | 2015, 2017 | 5.21, 5.34 |
| 6 | D05 | D05∶1512276 | 1 512 276 | T/A | RSLW | 2015, 2017 | 5.92, 5.25 |
| 7 | A04 | A04∶54398482 | 54 398 482 | C/A | SP | 2015, 2017 | 5.09, 5.32 |
| SA | 2017 | 5.22 | |||||
| 8# | A07 | A07∶70678488 | 70 678 488 | G/C | SL, SA,SP | 2015 | 6.89, 6.69, 6.72 |
| 9# | A07 | A07∶70678557 | 70 678 557 | A/G | SL,SA,SP | 2015 | 5.72, 5.35, 5.40 |
| 10# | A07 | A07∶70679600 | 70 679 600 | T/C | SL,SA,SP | 2015 | 5.18, 5.49, 5.27 |
| 11# | A07 | A07∶70679917 | 70 679 917 | C/T | SL,SA,SP | 2015 | 6.27, 5.87, 5.96 |
| 12# | A07 | A07∶70680268 | 70 680 268 | A/C | SL,SA,SP | 2015 | 6.14, 5.83, 5.84 |
| 13# | A07 | A07∶70680314 | 70 680 314 | A/G | SL,SA,SP | 2015 | 6.45, 6.38, 6.39 |
| 14# | A07 | A07∶70682150 | 70 682 150 | C/A | SL,SA,SP | 2015 | 5.87, 5.22, 5.51 |
| 15# | A07 | A07∶70682969 | 70 682 969 | T/C | SL,SA,SP | 2015 | 5.25, 5.19, 5.29 |
| 16# | A07 | A07∶70700642 | 70 700 642 | C/T | SL,SA,SP | 2015 | 5.56, 5.02, 5.16 |
| 17 | A07 | A07∶70706541 | 70 706 541 | C/G | SL,SA,SP | 2015 | 6.03, 5.21, 5.66 |
| 18 | A07 | A07∶72096450 | 72 096 450 | A/G | RSLW,SL,SP | 2015 | 5.10, 6.10, 5.12 |
| 19 | A07 | A07∶44855893 | 44 855 893 | G/A | SL,SA,SP | 2015 | 5.98, 6.06, 6.22 |
| 20 | A08 | A08∶28488249 | 28 488 249 | T/C | SL,SA,SP | 2015 | 5.17, 5.68, 5.64 |
| 21 | A08 | A08∶32230490 | 32 230 490 | T/C | SL,SA,SP | 2015 | 5.41, 5.48, 5.28 |
| 22 | A08 | A08∶97095613 | 97 095 613 | G/C | SL,SA,SP | 2015 | 5.23, 5.45, 5.76 |
| 23 | A01 | A01∶1540366 | 1 540 366 | C/T | SL,SA,SP | 2017 | 5.80, 5.25, 5.09 |
| 24 | A01 | A01∶12448791 | 12 448 791 | A/G | SL,SW,SA,SP | 2015 | 5.00, 5.89, 6.43, 5.85 |
| 25 | A02 | A02∶6005362 | 6 005 362 | C/T | SL,SA,SP | 2015 | 6.51, 5.03, 6.07 |
| 26 | A02 | A02∶10265596 | 10 265 596 | G/A | SL,SA,SP | 2017 | 6.20, 5.36, 5.90 |
| 27 | A05 | A05∶20705321 | 20 705 321 | C/G | SW,SA,SP | 2017 | 5.06, 5.04, 5.39 |
| 28 | A05 | A05∶37081549 | 37 081 549 | A/G | SL,SA,SP | 2017 | 6.63, 5.06, 6.17 |
| 29 | A06 | A06∶70904173 | 70 904 173 | A/G | SL,SA,SP | 2015 | 5.07, 5.05, 5.11 |
| 30 | D08 | D08∶34053083 | 34 053 083 | T/C | SL,SA,SP | 2017 | 5.90, 5.73, 5.74 |
表2 供试自然群体棉籽大小和形状关联SNP标记 (续表Continued)
Table 2 Associated SNP markers of seed size and shape in cotton natural population
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 5 | D05 | D05∶1496927 | 1 496 927 | T/C | RSLW | 2015, 2017 | 5.21, 5.34 |
| 6 | D05 | D05∶1512276 | 1 512 276 | T/A | RSLW | 2015, 2017 | 5.92, 5.25 |
| 7 | A04 | A04∶54398482 | 54 398 482 | C/A | SP | 2015, 2017 | 5.09, 5.32 |
| SA | 2017 | 5.22 | |||||
| 8# | A07 | A07∶70678488 | 70 678 488 | G/C | SL, SA,SP | 2015 | 6.89, 6.69, 6.72 |
| 9# | A07 | A07∶70678557 | 70 678 557 | A/G | SL,SA,SP | 2015 | 5.72, 5.35, 5.40 |
| 10# | A07 | A07∶70679600 | 70 679 600 | T/C | SL,SA,SP | 2015 | 5.18, 5.49, 5.27 |
| 11# | A07 | A07∶70679917 | 70 679 917 | C/T | SL,SA,SP | 2015 | 6.27, 5.87, 5.96 |
| 12# | A07 | A07∶70680268 | 70 680 268 | A/C | SL,SA,SP | 2015 | 6.14, 5.83, 5.84 |
| 13# | A07 | A07∶70680314 | 70 680 314 | A/G | SL,SA,SP | 2015 | 6.45, 6.38, 6.39 |
| 14# | A07 | A07∶70682150 | 70 682 150 | C/A | SL,SA,SP | 2015 | 5.87, 5.22, 5.51 |
| 15# | A07 | A07∶70682969 | 70 682 969 | T/C | SL,SA,SP | 2015 | 5.25, 5.19, 5.29 |
| 16# | A07 | A07∶70700642 | 70 700 642 | C/T | SL,SA,SP | 2015 | 5.56, 5.02, 5.16 |
| 17 | A07 | A07∶70706541 | 70 706 541 | C/G | SL,SA,SP | 2015 | 6.03, 5.21, 5.66 |
| 18 | A07 | A07∶72096450 | 72 096 450 | A/G | RSLW,SL,SP | 2015 | 5.10, 6.10, 5.12 |
| 19 | A07 | A07∶44855893 | 44 855 893 | G/A | SL,SA,SP | 2015 | 5.98, 6.06, 6.22 |
| 20 | A08 | A08∶28488249 | 28 488 249 | T/C | SL,SA,SP | 2015 | 5.17, 5.68, 5.64 |
| 21 | A08 | A08∶32230490 | 32 230 490 | T/C | SL,SA,SP | 2015 | 5.41, 5.48, 5.28 |
| 22 | A08 | A08∶97095613 | 97 095 613 | G/C | SL,SA,SP | 2015 | 5.23, 5.45, 5.76 |
| 23 | A01 | A01∶1540366 | 1 540 366 | C/T | SL,SA,SP | 2017 | 5.80, 5.25, 5.09 |
| 24 | A01 | A01∶12448791 | 12 448 791 | A/G | SL,SW,SA,SP | 2015 | 5.00, 5.89, 6.43, 5.85 |
| 25 | A02 | A02∶6005362 | 6 005 362 | C/T | SL,SA,SP | 2015 | 6.51, 5.03, 6.07 |
| 26 | A02 | A02∶10265596 | 10 265 596 | G/A | SL,SA,SP | 2017 | 6.20, 5.36, 5.90 |
| 27 | A05 | A05∶20705321 | 20 705 321 | C/G | SW,SA,SP | 2017 | 5.06, 5.04, 5.39 |
| 28 | A05 | A05∶37081549 | 37 081 549 | A/G | SL,SA,SP | 2017 | 6.63, 5.06, 6.17 |
| 29 | A06 | A06∶70904173 | 70 904 173 | A/G | SL,SA,SP | 2015 | 5.07, 5.05, 5.11 |
| 30 | D08 | D08∶34053083 | 34 053 083 | T/C | SL,SA,SP | 2017 | 5.90, 5.73, 5.74 |
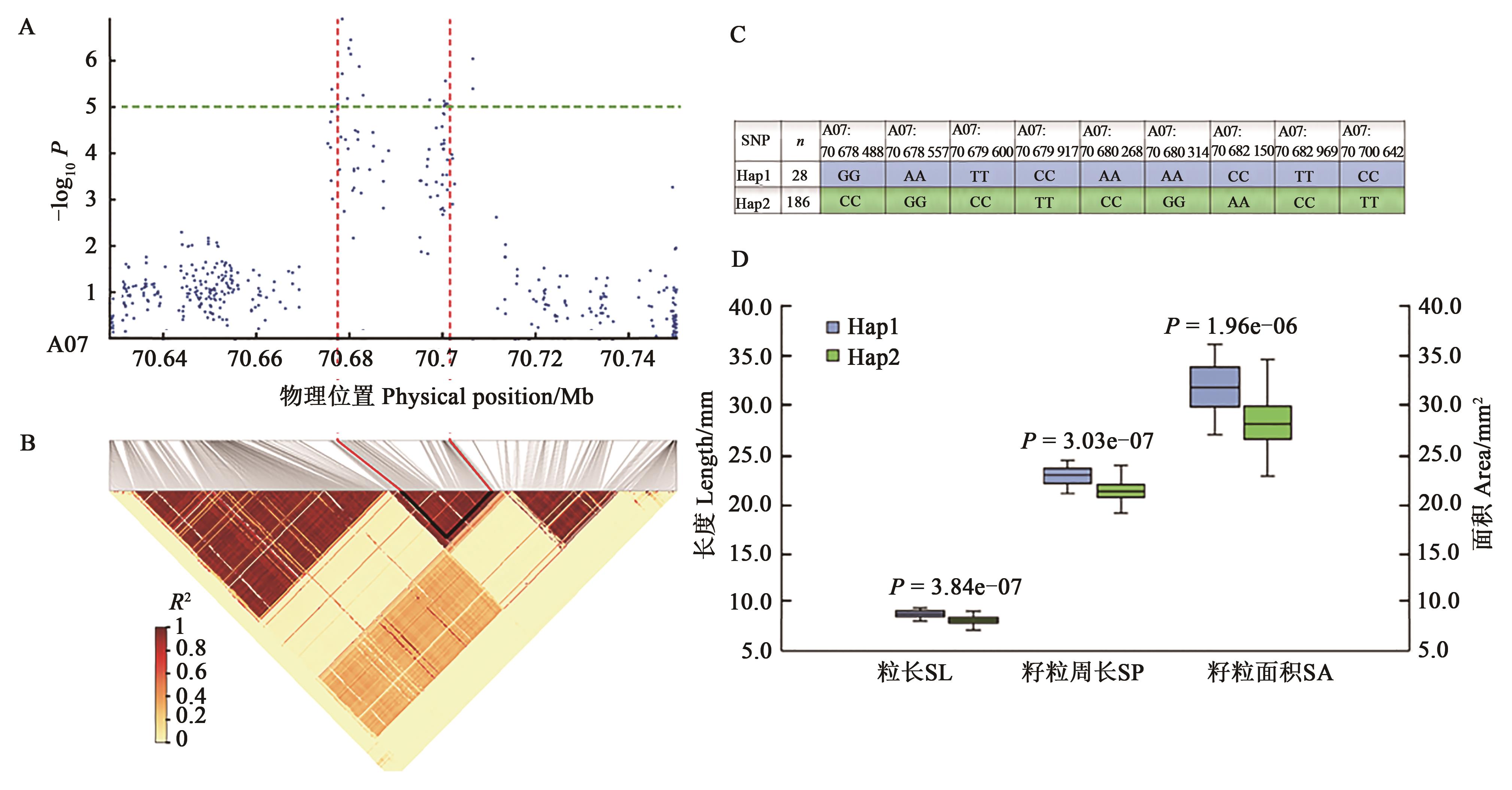
图3 棉花A07染色体棉籽性状关联SNPs单倍型分析A:A07染色体关联SNPs;B:A07染色体LD分析;C:关联SNPs单倍型分析;D:不同单倍型间的性状差异分析
Fig. 3 Associated SNPs and haplotypes on chromosome A07 for cotton seed related traitsA: Associated SNPs on chromosome A07; B: LD analysis of chromosome A07; C: Haplotypes of associated SNPs; D: Different related traits between haplotypes
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 1 | D11 | D11∶50561153 | 50 561 153 | C/T | SW | 2015, 2017 | 5.32, 6.53 |
| SA | 2015, 2017 | 6.06, 6.45 | |||||
| SP | 2015, 2017 | 5.81, 5.76 | |||||
| 2 | D12 | D12∶56031535 | 56 031 535 | A/C | SL | 2015, 2017 | 5.85, 5.35 |
| SP | 2015, 2017 | 5.33, 5.46 | |||||
| SA | 2015 | 5.16 | |||||
| 3 | D05 | D05∶1490099 | 1 490 099 | G/A | RSLW | 2015, 2017 | 8.00, 6.83 |
| 4 | D05 | D05∶1490516 | 1 490 516 | G/T | RSLW | 2015, 2017 | 5.63, 5.36 |
表2 供试自然群体棉籽大小和形状关联SNP标记
Table 2 Associated SNP markers of seed size and shape in cotton natural population
序号 No. | 染色体 Chromosome | SNP | 物理位置 Physical position | 等位变异Allele | 性状 Trait | 年度 Year | -log10P |
|---|---|---|---|---|---|---|---|
| 1 | D11 | D11∶50561153 | 50 561 153 | C/T | SW | 2015, 2017 | 5.32, 6.53 |
| SA | 2015, 2017 | 6.06, 6.45 | |||||
| SP | 2015, 2017 | 5.81, 5.76 | |||||
| 2 | D12 | D12∶56031535 | 56 031 535 | A/C | SL | 2015, 2017 | 5.85, 5.35 |
| SP | 2015, 2017 | 5.33, 5.46 | |||||
| SA | 2015 | 5.16 | |||||
| 3 | D05 | D05∶1490099 | 1 490 099 | G/A | RSLW | 2015, 2017 | 8.00, 6.83 |
| 4 | D05 | D05∶1490516 | 1 490 516 | G/T | RSLW | 2015, 2017 | 5.63, 5.36 |
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation | 表达量增加的时期 Stage with increasing expression |
|---|---|---|---|
| D05 | Gh_D05G0142 | 铜离子结合,电子载体 Copper ion binding, electron carrier | 后期 Later stage |
| Gh_D05G0144 | 植物特异转录因子,YABBY家族蛋白 Plant-specific transcription factor, YABBY family protein | 后期Later stage | |
| Gh_D05G0145 | 核孔蛋白相关 Nucleoporin-related | 后期Later stage | |
| Gh_D05G0146 | TPR超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein | 全期 Whole stage | |
| Gh_D05G0147 | AT1G80133 | 全期Whole stage | |
| Gh_D05G0148 | EBF蛋白 EIN3-binding F box protein | 全期Whole stage | |
| Gh_D05G0149 | AT5G25360 | 前期Earlier stage | |
| Gh_D05G0150 | AT3G15760 | 前期Earlier stage | |
| Gh_D05G0153 | 含SPX结构域基因 SPX domain gene | 全期Whole stage | |
| Gh_D05G0154 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 前期Earlier stage | |
| Gh_D05G0155 | 核糖核酸酶H超家族蛋白 Ribonuclease H-like superfamily protein | 后期Later stage | |
| A07 | Gh_A07G1731 | AT3G13130 | 后期Later stage |
| Gh_A07G1732 | AT3G13130 | 全期Whole stage | |
| A08 | Gh_A08G0768 | 蛋白激酶超家族蛋白 Protein kinase superfamily protein | 全期Whole stage |
| Gh_A08G0782 | 海藻糖磷酸酶/合酶 Trehalose phosphatase/synthase | 前期Earlier stage | |
| A01 | Gh_A01G0158 | 富亮氨酸跨膜蛋白激酶 Leucine-rich repeat transmembrane protein kinase | 全期Whole stage |
| Gh_A01G0160 | 衰老相关基因 Senescence-associated gene | 后期Later stage | |
| Gh_A01G0695 | 含SET结构域蛋白 SET domain group 40 | 全期Whole stage | |
| A02 | Gh_A02G0442 | RING/U-box 超家族蛋白 RING/U-box superfamily protein | 全期Whole stage |
| Gh_A02G0645 | Gigantea蛋白 Gigantea protein | 后期Later stage | |
| A05 | Gh_A05G2586 | 类PsbP蛋白 PsbP-like protein | 后期Later stage |
表3 棉籽大小和形状相关候选基因
Table 3 Candidate genes related to the seed size and shape in cotton
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation | 表达量增加的时期 Stage with increasing expression |
|---|---|---|---|
| D05 | Gh_D05G0142 | 铜离子结合,电子载体 Copper ion binding, electron carrier | 后期 Later stage |
| Gh_D05G0144 | 植物特异转录因子,YABBY家族蛋白 Plant-specific transcription factor, YABBY family protein | 后期Later stage | |
| Gh_D05G0145 | 核孔蛋白相关 Nucleoporin-related | 后期Later stage | |
| Gh_D05G0146 | TPR超家族蛋白 Tetratricopeptide repeat (TPR)-like superfamily protein | 全期 Whole stage | |
| Gh_D05G0147 | AT1G80133 | 全期Whole stage | |
| Gh_D05G0148 | EBF蛋白 EIN3-binding F box protein | 全期Whole stage | |
| Gh_D05G0149 | AT5G25360 | 前期Earlier stage | |
| Gh_D05G0150 | AT3G15760 | 前期Earlier stage | |
| Gh_D05G0153 | 含SPX结构域基因 SPX domain gene | 全期Whole stage | |
| Gh_D05G0154 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 前期Earlier stage | |
| Gh_D05G0155 | 核糖核酸酶H超家族蛋白 Ribonuclease H-like superfamily protein | 后期Later stage | |
| A07 | Gh_A07G1731 | AT3G13130 | 后期Later stage |
| Gh_A07G1732 | AT3G13130 | 全期Whole stage | |
| A08 | Gh_A08G0768 | 蛋白激酶超家族蛋白 Protein kinase superfamily protein | 全期Whole stage |
| Gh_A08G0782 | 海藻糖磷酸酶/合酶 Trehalose phosphatase/synthase | 前期Earlier stage | |
| A01 | Gh_A01G0158 | 富亮氨酸跨膜蛋白激酶 Leucine-rich repeat transmembrane protein kinase | 全期Whole stage |
| Gh_A01G0160 | 衰老相关基因 Senescence-associated gene | 后期Later stage | |
| Gh_A01G0695 | 含SET结构域蛋白 SET domain group 40 | 全期Whole stage | |
| A02 | Gh_A02G0442 | RING/U-box 超家族蛋白 RING/U-box superfamily protein | 全期Whole stage |
| Gh_A02G0645 | Gigantea蛋白 Gigantea protein | 后期Later stage | |
| A05 | Gh_A05G2586 | 类PsbP蛋白 PsbP-like protein | 后期Later stage |
| 1 | NIE X H, TU J L, WANG B, et al.. A BIL population derived from G . hirsutumG.barbadense provides a resource for cotton genetics and breeding [J/OL]. PLoS One, 2015, 10(10): e01410641 [2022-04-19]. . |
| 2 | LIU H Y, QUAMPAH A, CHEN J H, et al.. QTL mapping with different genetic systems for nine nonessential amino acids of cottonseeds [J]. Mol. Genet. Genomics, 2017, 292(10): 671-684. |
| 3 | SHANG L G, ABDUWELI A, WANG Y M, et al.. Genetic analysis and QTL mapping of oil content and seed index using two recombinant inbred lines and two backcross populations in upland cotton [J]. Plant Breeding, 2016, 135(2): 224-231. |
| 4 | 钱玉源,刘祎,张曦,等.棉花种子质量突变体ims-15及其近等基因系种子发育期转录组分析[J].华北农学报, 2021, 36 (3): 50-59. |
| QIAN Y Y, LIU Y, ZHANG X, et al.. Transcriptome analysis of cotton seed weight mutant ims-15 and its near isogenic line during seed development [J]. Acta Agric. Boreali-Sin., 2021, 36 (3): 50-59. | |
| 5 | 赵娟.调控内源生长素和细胞分裂素水平实现棉花种子纤维产量和品质的同步改良[D].重庆:西南大学, 2014. |
| ZHAO J. Manipulation of auxin and cytokinin level in cotton concurrently increases fiber and seed yield [D]. Chongqing: Southwest University, 2014. | |
| 6 | 覃珊.胚中特异性下调细胞分裂素氧化酶基因(GhCKX)促进棉花种子发育[D].重庆:西南大学, 2009. |
| QIN S. Ovule-specific down-regulation of GhCKX gene promoted seed development of cotton (Gossypium hirsutum L.) [D]. Chongqing: Southwest University, 2009. | |
| 7 | 王俊娟,叶武威,樊伟莉,等.不同基因型棉花种子活力的研究[J].种子科技, 2009, 27(9): 19-20. |
| WANG J J, YE, W W, FAN W L, et al.. Study on vitality of different genotypes cotton seed [J]. Seed Sci. Technol., 2009, 27(9): 19-20. | |
| 8 | ZHANG D, LI J P, COMPTON R O, et al.. Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae) [J]. G3: Genes Genom. Genet., 2015, 5(6): 1117-1128. |
| 9 | LI L, LI X H, LI L L, et al.. QTL identification and epistatic effect analysis of seed size and weight-related traits in Zea mays L [J/OL]. Mol. Breeding, 2019, 39(5): 67 [2022-04-20]. . |
| 10 | WILLIAMS K, SORRELLS M E. Three-dimensional seed size and shape QTL in hexaploid wheat (Triticum aestivum L.) populations [J]. Crop Sci., 2014, 54(1): 98-110. |
| 11 | CUI B F, CHEN L, YANG Y Q, et al.. Genetic analysis and map-based delimitation of a major locus qSS3 for seed size in soybean [J]. Plant Breeding, 2020, 139(2): 1145-1157. |
| 12 | HINA A, CAO Y C, SONG S Y, et al.. High-resolution mapping in two RIL populations refines major "QTL hotspot" regions for seed size and shape in soybean (Glycine max L.) [J/OL]. Int. J. Mol. Sci., 2020, 21(3): 1040 [2022-04-20]. . |
| 13 | GERAVANDI M, CHEGHAMIRZA K, FARSHADFAR E, et al.. QTL analysis of seed size and yield-related traits in an inter-gene pool population of common bean (Phaseolus vulgaris L.) [J/OL]. Scientia Hortic., 2020, 274(15): 109678 [2022-05-20]. . |
| 14 | SONG X J, HUANG W, SHI M, et al.. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase [J]. Nat. Genet., 2007, 39(5): 623-630. |
| 15 | 程瑞如.小麦粒形的分子遗传分析及穗长基因HL1的精细定位[D].南京:南京农业大学, 2018. |
| CHENG R R. Molecular genetic analysis of wheat kernel dimensions and fine mapping of spike length QTL HLl [D]. Nanjing: Nanjing Agricultural University, 2018. | |
| 16 | 耿庆河,王兰芬,武晶,等.普通菜豆籽粒大小与形状的QTL定位[J].作物学报,2017, 43(8): 1149-1160. |
| GENG Q H, WANG L F, WU J, et al.. QTL mapping for seed size and shape in common bean [J]. Acta Agron. Sin., 2017, 43(8): 1149-1160. | |
| 17 | 秦伟伟,李永祥,李春辉,等.基于高密度遗传图谱的玉米籽粒性状QTL定位[J].作物学报, 2015, 41(10): 1510-1518. |
| QIN W W, LI Y X, LI C H, et al.. QTL mapping for kernel related traits based on a high-density genetic map [J]. Acta Agron. Sin., 2015, 41(10): 1510-1518. | |
| 18 | 耿青青.大豆籽粒大小与形状性状的全基因组关联分析和DistortedMap软件包的研制[D].南京:南京农业大学, 2014. |
| GENG Q Q. Genome-wide association studies of seed size and shape traits in soybean and development of distortedmap software [D]. Nanjing: Nanjing Agricultural University, 2014. | |
| 19 | 曾雅青.棉花种子活力的基因型差异及生态点影响[D].南京:南京农业大学, 2014. |
| ZENG Y Q. Genotype difference and ecological environmental impact in cotton seed vigor [D]. Nanjing: Nanjing Agricultural University, 2014. | |
| 20 | 陈建华,杨继良,杨伟华,等.棉花种子活力及其影响因素探讨[J].棉花学报, 1995, 7(3): 134-140. |
| CHEN J H, YANG J L, YANG W H, et al.. The study of cottonseed vigor and influence factors [J]. Acta Gossypii Sin., 1995, 7(3): 134-140. | |
| 21 | MA Z Y, HE S P, WANG X F, et al.. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield [J]. Nat. Genet., 2018, 50(6): 803-813. |
| 22 | GU Q S, KE H F, SUN Z W, et al.. Genetic variation associated with the shoot biomass of upland cotton seedlings under contrasting phosphate supplies [J/OL]. Mol. Breeding, 2020, 40(8): 80 [2022-04-20]. . |
| 23 | FANG Y X, ZHANG X Q, ZHANG X, et al.. A high-density genetic linkage map of SLAFs and QTL analysis of grain size and weight in barley (Hordeum vulgare L.) [J/OL]. Front. Plant Sci., 2020, 11: 620922 [2022-04-20]. . |
| 24 | LEI L, WANG L F, WANG S M, et al.. Marker-trait association analysis of seed traits in accessions of common bean (Phaseolus vulgaris L.) in China [J/OL]. Front. Genet., 2020, 11: 698 [2022-04-20]. . |
| 25 | ZHANG T Z, HU Y, JIANG W K, et al.. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement [J]. Nat. Biotechnol., 2015, 33(5): 531-537. |
| 26 | GUO X K, ZHANG Y, TU Y, et al.. Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato [J]. Plant Sci., 2018, 272: 131-141. |
| 27 | 陈秀玲,刘思源,孙婷,等.番茄YABBY基因家族的生物信息学分析[J].东北农业大学学报, 2017, 48(10): 11-19. |
| CHEN X L, LIU S Y, SUN T, et al.. Bioinformatics analysis on YABBY gene family in tomato [J]. J. Northeast Agric. Univ., 2017, 48(10): 11-19. | |
| 28 | ZHANG M, WANG C P, LIN Q F, et al.. A tetratricopeptide repeat domain-containing protein SSR1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis [J]. Plant J., 2015, 83(4): 582-599. |
| 29 | WEI K F, LI Y X. Functional genomics of the protein kinase superfamily from wheat [J/OL]. Mol. Breeding, 2019, 39: 141 [2022-04-20]. . |
| 30 | WANG W W, SUN Y, YANG P, et al.. A high density SLAF-seq SNP genetic map and QTL for seed size, oil and protein content in upland cotton [J/OL]. BMC Genomics, 2019, 20(1): 599 [2022-04-20]. . |
| 31 | FINET C, FLOYD S K, CONWAY S J, et al.. Evolution of the YABBY gene family in seed plants [J]. Evol. Dev., 2016, 18(2): 116-126. |
| 32 | SUN L, WEI Y Q, WU K H, et al.. Restriction of iron loading into developing seeds by a YABBY transcription factor safeguards successful reproduction in Arabidopsis [J]. Mol. Plant, 2021, 14(10): 1624-1639. |
| [1] | 刘艳, 鲍红帅, 尚红燕, 王国宁, 张艳, 王省芬, 马峙英, 吴金华. 棉花枯萎病菌及其培养条件筛选[J]. 中国农业科技导报, 2022, 24(8): 124-132. |
| [2] | 刘海涛, 韩鑫, 兰玉彬, 伊丽丽, 王宝聚, 崔立华. 基于YOLOv4网络的棉花顶芽精准识别方法[J]. 中国农业科技导报, 2022, 24(8): 99-108. |
| [3] | 孙正文, 谷淇深, 张艳, 王省芬, 马峙英. 棉花基因发掘与分子育种研究进展[J]. 中国农业科技导报, 2022, 24(7): 32-38. |
| [4] | 闫成川, 曾庆涛, 陈琴, 付锦程, 王婷伟, 陈全家, 曲延英. 陆地棉花铃期抗旱指标筛选及评价[J]. 中国农业科技导报, 2022, 24(7): 46-57. |
| [5] | 吴楠, 杨君, 张艳, 孙正文, 张冬梅, 李丽花, 吴金华, 马峙英, 王省芬. 过表达棉花葡萄糖醛酸激酶基因GbGlcAK促进拟南芥细胞伸长[J]. 中国农业科技导报, 2022, 24(6): 36-46. |
| [6] | 黄雅婕, 任丹, 李生梅, 崔进鑫, 杨涛, 任姣姣, 高文伟. 陆地棉苗期的耐盐碱性评价及鉴定指标筛选[J]. 中国农业科技导报, 2022, 24(5): 46-55. |
| [7] | 周雨青, 杨永飞, 葛常伟, 沈倩, 张思平, 刘绍东, 马慧娟, 陈静, 刘瑞华, 李士丛, 赵新华, 李存东, 庞朝友. 基于WGCNA的棉花子叶抗冷相关共表达模块鉴定[J]. 中国农业科技导报, 2022, 24(4): 52-62. |
| [8] | 秦宁, 李俊茹, 田蕊, 邵振启, 李喜焕, 张彩英. 大豆籽粒生育酚遗传位点发掘及候选基因筛选[J]. 中国农业科技导报, 2022, 24(3): 48-56. |
| [9] | 石米娟, 张婉婷, 程莹寅, 夏晓勤. 基于全基因组分析技术的鱼类育种技术原理与应用[J]. 中国农业科技导报, 2022, 24(2): 33-41. |
| [10] | 张静, 郭思梦, 韩迎春, 雷亚平, 邢芳芳, 杜文丽, 李亚兵, 冯璐. 基于无人机RGB图像的棉花产量估算[J]. 中国农业科技导报, 2022, 24(11): 112-120. |
| [11] | 李娟, 闫旭宇, 吴香, 李玲. 纤维类作物对农田土壤镉污染修复的研究进展[J]. 中国农业科技导报, 2022, 24(11): 171-178. |
| [12] | 孙逸凡, 黄志磊, 李葆春, 姚立蓉, 汪军成, 司二静, 杨轲, 孟亚雄, 马小乐, 王化俊. 大麦种质资源抗叶斑病评价[J]. 中国农业科技导报, 2022, 24(11): 43-54. |
| [13] | 袁进成, 孟亚轩, 孙颖琦, 赵心月, 王凤霞, 刘颖慧. 基于全基因关联分析的代谢组学在植物中的应用[J]. 中国农业科技导报, 2021, 23(9): 12-18. |
| [14] | 李生梅, 张大伟, 迪丽拜尔·迪力买买提, 魏鑫, 芮存, 杨涛, 耿世伟, 高文伟. 减量灌溉对转ScALDH21基因棉花农艺性状、产量和品质的影响[J]. 中国农业科技导报, 2021, 23(9): 152-159. |
| [15] | 张旭1,何俊峰1,陈佛文1,李继福1*,吴启侠1,谭京红1,邹家龙2. 麦秆还田对直播和移栽棉花产量及氮素吸收的影响[J]. 中国农业科技导报, 2021, 23(3): 122-131. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号