




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (4): 22-35.DOI: 10.13304/j.nykjdb.2024.0620
马蓓1,2( ), 公杰2(
), 公杰2( ), 杜银柯2,3, 甘雨薇1,2, 程蓉2,3, 朱波1, 易丽霞1(
), 杜银柯2,3, 甘雨薇1,2, 程蓉2,3, 朱波1, 易丽霞1( ), 马锦绣2(
), 马锦绣2( ), 高世庆2(
), 高世庆2( )
)
收稿日期:2024-08-02
接受日期:2024-09-18
出版日期:2025-04-15
发布日期:2025-04-15
通讯作者:
易丽霞,马锦绣,高世庆
作者简介:马蓓 E-mail:18034656852@163.com基金资助:
Bei MA1,2( ), Jie GONG2(
), Jie GONG2( ), Yinke DU2,3, Yuwei GAN1,2, Rong CHENG2,3, Bo ZHU1, Lixia YI1(
), Yinke DU2,3, Yuwei GAN1,2, Rong CHENG2,3, Bo ZHU1, Lixia YI1( ), Jinxiu MA2(
), Jinxiu MA2( ), Shiqing GAO2(
), Shiqing GAO2( )
)
Received:2024-08-02
Accepted:2024-09-18
Online:2025-04-15
Published:2025-04-15
Contact:
Lixia YI,Jinxiu MA,Shiqing GAO
摘要:
花粉孔径调控因子在植物萌发孔径形成中发挥着核心作用。为探究小麦花粉表面萌发的孔发育调控机理,深入探索与OsINP1同源的TaINP1基因家族在小麦萌发孔径形成中的调控功能,利用生物信息学策略,在小麦基因组中鉴定出53个小麦TaINP1基因,系统分析了其理化性质、启动子顺式作用元件、基因间的共线性关系。组织表达分析表明,TaINP2.2、TaINP2.5等基因在小麦穗部显著高表达; TaINP2.5、TaINP3.2等基因经低温(10 ℃)处理后在花粉中高表达。结合花粉萌发及纳米磁珠转化技术,将RFP报告基因成功导入小麦花粉中,并利用激光共聚焦显微镜观测不同低温处理花粉的荧光强度,证实TaINP1基因在调控花粉孔开闭中起重要作用。以上研究结果为小麦花粉孔发育的调控机理奠定了基础,也为优化小麦花粉纳米磁珠高效转化体系提供了参考,为小麦分子育种遗传改良开辟了新途径。
中图分类号:
马蓓, 公杰, 杜银柯, 甘雨薇, 程蓉, 朱波, 易丽霞, 马锦绣, 高世庆. 小麦花粉孔发育相关TaINP1基因鉴定及表达分析[J]. 中国农业科技导报, 2025, 27(4): 22-35.
Bei MA, Jie GONG, Yinke DU, Yuwei GAN, Rong CHENG, Bo ZHU, Lixia YI, Jinxiu MA, Shiqing GAO. Identification and Expression Analysis of TaINP1 Gene Related to Pollen Pore Development in Wheat[J]. Journal of Agricultural Science and Technology, 2025, 27(4): 22-35.
基因名称 Gene name | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaINP2.2 | TCGTAGATGGGATAATGGGGC | CAAGCCACAGGAAGCACCTT |
| TaINP2.5 | ATTGGGTTTCAGACGGAGCA | CAAGGGCTTGTCAGACCTGT |
| TaINP2.8 | CCGCTGGAATCTTGTCCGTTA | TTCCTTGCAGCCTCACGATT |
| TaINP3.2 | TCCATGAGTGGAAATGGGGC | ACTACGCAGCTCAGTATCGC |
| TaINP3.7 | TTTTAAAGCTTGAACCCCTAACTGA | ACCCAGCCAATGTTTCTGCC |
| TaINP3.12 | TTTTAAAGCTTGAACCCCTAACTGA | CCGATAGACCCAGCCAACG |
| TaINP3.4 | GCAGTTGATCCTCAGGCACT | AGGGTTTGTAGCCGCAGATT |
| TaINP3.9 | CTCAGCACACCATCCCAACA | TCTTCCACATTCCCGACAGC |
| TaINP3.14 | CAGGCTGATAATCTTCGTCTGC | GTGCAGATTGGCGAGTGGTT |
| TaINP4.1 | CCAAAAGGTGGTATTGCGGA | ACTGCTGCTCTGTTAGAGGC |
| TaINP4.4 | ACTCCAGAAAGCTCGACAGC | CAGAGAGAACTGAGGGCACG |
| 18s | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
表1 荧光定量引物
Table 1 Fluorescence quantification primers
基因名称 Gene name | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaINP2.2 | TCGTAGATGGGATAATGGGGC | CAAGCCACAGGAAGCACCTT |
| TaINP2.5 | ATTGGGTTTCAGACGGAGCA | CAAGGGCTTGTCAGACCTGT |
| TaINP2.8 | CCGCTGGAATCTTGTCCGTTA | TTCCTTGCAGCCTCACGATT |
| TaINP3.2 | TCCATGAGTGGAAATGGGGC | ACTACGCAGCTCAGTATCGC |
| TaINP3.7 | TTTTAAAGCTTGAACCCCTAACTGA | ACCCAGCCAATGTTTCTGCC |
| TaINP3.12 | TTTTAAAGCTTGAACCCCTAACTGA | CCGATAGACCCAGCCAACG |
| TaINP3.4 | GCAGTTGATCCTCAGGCACT | AGGGTTTGTAGCCGCAGATT |
| TaINP3.9 | CTCAGCACACCATCCCAACA | TCTTCCACATTCCCGACAGC |
| TaINP3.14 | CAGGCTGATAATCTTCGTCTGC | GTGCAGATTGGCGAGTGGTT |
| TaINP4.1 | CCAAAAGGTGGTATTGCGGA | ACTGCTGCTCTGTTAGAGGC |
| TaINP4.4 | ACTCCAGAAAGCTCGACAGC | CAGAGAGAACTGAGGGCACG |
| 18s | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP1.4 | TraesCS1B02G127400 | 1B | 333 | 36 564.21 | 6.14 | 54.41 | 83.60 | -0.416 | 细胞核Nucleus |
| TaINP6.2 | TraesCS6A02G248300 | 6A | 232 | 25 301.97 | 9.61 | 67.01 | 93.15 | -0.088 | 细胞核Nucleus |
| TaINP3.6 | TraesCS3B02G120900 | 3B | 260 | 27 937.63 | 5.43 | 54.74 | 86.58 | -0.181 | 细胞核Nucleus |
| TaINP2.4 | TraesCS2A02G526300 | 2A | 419 | 46 270.21 | 6.24 | 59.92 | 71.89 | -0.536 | 细胞核Nucleus |
| TaINP3.3 | TraesCS3A02G306200 | 3A | 296 | 31 824.06 | 5.63 | 49.12 | 88.34 | -0.210 | 细胞核Nucleus |
| TaINP3.11 | TraesCS3D02G105800 | 3D | 260 | 27 976.71 | 5.46 | 55.33 | 85.85 | -0.202 | 细胞核Nucleus |
| TaINP1.9 | TraesCS1D02G400300 | 1D | 226 | 2 449.19 | 5.65 | 46.52 | 98.14 | -0.078 | 线粒体Mitochondria |
| TaINP1.6 | TraesCS1B02G420300 | 1B | 226 | 24 449.82 | 5.65 | 47.19 | 96.86 | -0.098 | 线粒体Mitochondria |
| TaINP5.3 | TraesCS5B02G265300 | 5B | 525 | 58 620.02 | 6.58 | 70.75 | 76.63 | -0.560 | 细胞核Nucleus |
| TaINP4.2 | TraesCS4A02G183400 | 4A | 539 | 58 911.63 | 6.44 | 54.62 | 69.13 | -0.568 | 细胞核Nucleus |
| TaINP5.2 | TraesCS5A02G265600 | 5A | 518 | 57 856.21 | 6.51 | 68.93 | 77.30 | -0.550 | 细胞核Nucleus |
| TaINP1.1 | TraesCS1A02G096300 | 1A | 335 | 36 675.16 | 5.71 | 57.88 | 82.27 | -0.438 | 细胞核Nucleus |
| TaINP1.8 | TraesCS1D02G276100 | 1D | 481 | 53 546.29 | 5.95 | 62.72 | 74.39 | -0.503 | 细胞核Nucleus |
| TaINP4.1 | TraesCS4A02G126300 | 4A | 335 | 37 349.28 | 7.87 | 51.81 | 84.00 | -0.491 | 细胞核Nucleus |
| TaINP7.2 | TraesCS7B02G114300 | 7B | 448 | 48 464.72 | 7.36 | 56.89 | 82.01 | -0.285 | 细胞核Nucleus |
| TaINP1.5 | TraesCS1B02G285800 | 1B | 526 | 58 065.31 | 5.79 | 61.88 | 73.23 | -0.471 | 细胞核Nucleus |
| TaINP6.4 | TraesCS6B02G276400 | 6B | 232 | 25 314.02 | 9.46 | 68.12 | 92.72 | -0.100 | 叶绿体Chloroplast |
| TaINP4.5 | TraesCS4D02G129900 | 4D | 537 | 58 671.34 | 6.41 | 54.16 | 68.47 | -0.567 | 细胞核Nucleus |
| TaINP3.13 | TraesCS3D02G295800 | 3D | 292 | 31 490.63 | 5.37 | 49.44 | 89.55 | -0.191 | 叶绿体Chloroplast |
| TaINP3.9 | TraesCS3B02G365100 | 3B | 477 | 51 918.14 | 6.78 | 50.58 | 75.68 | -0.494 | 细胞核Nucleus |
| TaINP4.3 | TraesCS4B02G135000 | 4B | 566 | 6 217.82 | 6.40 | 56.40 | 66.70 | -0.614 | 细胞核Nucleus |
| TaINP2.9 | TraesCS2D02G529000 | 2D | 425 | 46 843.83 | 5.86 | 57.18 | 73.39 | -0.507 | 细胞核Nucleus |
| TaINP5.1 | TraesCS5A02G174200 | 5A | 467 | 51 251.99 | 7.02 | 53.54 | 77.41 | -0.473 | 细胞核Nucleus |
| TaINP2.6 | TraesCS2B02G512700 | 2B | 240 | 26 178.71 | 9.20 | 44.06 | 86.33 | -0.215 | 细胞质Cytoplasm |
表2 小麦 TaINP1基因家族成员的基本理化性质 (续表Continued)
Table 2 Basic physical and chemical properties of TaINP1 gene family in wheat
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP1.4 | TraesCS1B02G127400 | 1B | 333 | 36 564.21 | 6.14 | 54.41 | 83.60 | -0.416 | 细胞核Nucleus |
| TaINP6.2 | TraesCS6A02G248300 | 6A | 232 | 25 301.97 | 9.61 | 67.01 | 93.15 | -0.088 | 细胞核Nucleus |
| TaINP3.6 | TraesCS3B02G120900 | 3B | 260 | 27 937.63 | 5.43 | 54.74 | 86.58 | -0.181 | 细胞核Nucleus |
| TaINP2.4 | TraesCS2A02G526300 | 2A | 419 | 46 270.21 | 6.24 | 59.92 | 71.89 | -0.536 | 细胞核Nucleus |
| TaINP3.3 | TraesCS3A02G306200 | 3A | 296 | 31 824.06 | 5.63 | 49.12 | 88.34 | -0.210 | 细胞核Nucleus |
| TaINP3.11 | TraesCS3D02G105800 | 3D | 260 | 27 976.71 | 5.46 | 55.33 | 85.85 | -0.202 | 细胞核Nucleus |
| TaINP1.9 | TraesCS1D02G400300 | 1D | 226 | 2 449.19 | 5.65 | 46.52 | 98.14 | -0.078 | 线粒体Mitochondria |
| TaINP1.6 | TraesCS1B02G420300 | 1B | 226 | 24 449.82 | 5.65 | 47.19 | 96.86 | -0.098 | 线粒体Mitochondria |
| TaINP5.3 | TraesCS5B02G265300 | 5B | 525 | 58 620.02 | 6.58 | 70.75 | 76.63 | -0.560 | 细胞核Nucleus |
| TaINP4.2 | TraesCS4A02G183400 | 4A | 539 | 58 911.63 | 6.44 | 54.62 | 69.13 | -0.568 | 细胞核Nucleus |
| TaINP5.2 | TraesCS5A02G265600 | 5A | 518 | 57 856.21 | 6.51 | 68.93 | 77.30 | -0.550 | 细胞核Nucleus |
| TaINP1.1 | TraesCS1A02G096300 | 1A | 335 | 36 675.16 | 5.71 | 57.88 | 82.27 | -0.438 | 细胞核Nucleus |
| TaINP1.8 | TraesCS1D02G276100 | 1D | 481 | 53 546.29 | 5.95 | 62.72 | 74.39 | -0.503 | 细胞核Nucleus |
| TaINP4.1 | TraesCS4A02G126300 | 4A | 335 | 37 349.28 | 7.87 | 51.81 | 84.00 | -0.491 | 细胞核Nucleus |
| TaINP7.2 | TraesCS7B02G114300 | 7B | 448 | 48 464.72 | 7.36 | 56.89 | 82.01 | -0.285 | 细胞核Nucleus |
| TaINP1.5 | TraesCS1B02G285800 | 1B | 526 | 58 065.31 | 5.79 | 61.88 | 73.23 | -0.471 | 细胞核Nucleus |
| TaINP6.4 | TraesCS6B02G276400 | 6B | 232 | 25 314.02 | 9.46 | 68.12 | 92.72 | -0.100 | 叶绿体Chloroplast |
| TaINP4.5 | TraesCS4D02G129900 | 4D | 537 | 58 671.34 | 6.41 | 54.16 | 68.47 | -0.567 | 细胞核Nucleus |
| TaINP3.13 | TraesCS3D02G295800 | 3D | 292 | 31 490.63 | 5.37 | 49.44 | 89.55 | -0.191 | 叶绿体Chloroplast |
| TaINP3.9 | TraesCS3B02G365100 | 3B | 477 | 51 918.14 | 6.78 | 50.58 | 75.68 | -0.494 | 细胞核Nucleus |
| TaINP4.3 | TraesCS4B02G135000 | 4B | 566 | 6 217.82 | 6.40 | 56.40 | 66.70 | -0.614 | 细胞核Nucleus |
| TaINP2.9 | TraesCS2D02G529000 | 2D | 425 | 46 843.83 | 5.86 | 57.18 | 73.39 | -0.507 | 细胞核Nucleus |
| TaINP5.1 | TraesCS5A02G174200 | 5A | 467 | 51 251.99 | 7.02 | 53.54 | 77.41 | -0.473 | 细胞核Nucleus |
| TaINP2.6 | TraesCS2B02G512700 | 2B | 240 | 26 178.71 | 9.20 | 44.06 | 86.33 | -0.215 | 细胞质Cytoplasm |
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP3.4 | TraesCS3A02G334100 | 3A | 475 | 51 766.93 | 6.53 | 52.32 | 74.36 | -0.514 | 细胞核Nucleus |
| TaINP4.4 | TraesCS4B02G178600 | 4B | 334 | 37 268.18 | 7.87 | 50.04 | 82.22 | -0.513 | 细胞核Nucleus |
| TaINP4.5 | TraesCS3B02G220400 | 3B | 327 | 36 381.83 | 7.79 | 59.51 | 76.51 | -0.622 | 细胞核Nucleus |
| TaINP3.5 | TraesCS3A02G372400 | 3A | 557 | 60 348.69 | 6.31 | 59.06 | 70.09 | -0.472 | 细胞核Nucleus |
| TaINP6.6 | TraesCS6D02G230400 | 6D | 232 | 25 274.05 | 9.97 | 65.71 | 96.08 | -0.029 | 叶绿体Chloroplast |
| TaINP1.3 | TraesCS1A02G392200 | 1A | 226 | 24 449.82 | 5.47 | 43.61 | 98.14 | -0.073 | 线粒体Mitochondria |
| TaINP6.3 | TraesCS6B02G193200 | 6B | 458 | 50 063.37 | 7.30 | 52.50 | 76.05 | -0.464 | 细胞核Nucleus |
| TaINP3.14 | TraesCS3D02G327600 | 3D | 477 | 51 979.22 | 6.53 | 53.94 | 75.47 | -0.495 | 细胞核Nucleus |
| TaINP1.2 | TraesCS1A02G276600 | 1A | 481 | 53 549.34 | 6.29 | 63.78 | 73.97 | -0.512 | 细胞核Nucleus |
| TaINP1.7 | TraesCS1D02G105300 | 1D | 338 | 37 042.89 | 6.56 | 58.57 | 84.97 | -0.371 | 细胞核Nucleus |
| TaINP3.12 | TraesCS3D02G194800 | 3D | 327 | 36 386.85 | 8.49 | 59.18 | 76.51 | -0.626 | 细胞核Nucleus |
| TaINP2.1 | TraesCS2A02G010800 | 2A | 198 | 21 663.49 | 9.56 | 54.16 | 83.54 | -0.271 | 细胞核Nucleus |
| TaINP3.1 | TraesCS3A02G103500 | 3A | 260 | 27 872.59 | 5.60 | 50.35 | 85.85 | -0.191 | 叶绿体Chloroplast |
| TaINP4.6 | TraesCS4D02G180200 | 4D | 329 | 36 682.46 | 7.87 | 52.80 | 81.40 | -0.527 | 胞核Nucleus |
| TaINP5.5 | TraesCS5D02G273500 | 5D | 517 | 57 757.09 | 6.55 | 68.94 | 77.43 | -0.544 | 细胞核Nucleus |
| TaINP2.8 | TraesCS2D02G096800 | 2D | 294 | 32 644.01 | 6.01 | 60.35 | 78.10 | -0.431 | 细胞核Nucleus |
| TaINP3.2 | TraesCS3A02G190700 | 3A | 350 | 38 979.69 | 6.36 | 59.18 | 75.11 | -0.647 | 细胞核Nucleus |
表2 小麦 TaINP1基因家族成员的基本理化性质 (续表Continued)
Table 2 Basic physical and chemical properties of TaINP1 gene family in wheat
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP3.4 | TraesCS3A02G334100 | 3A | 475 | 51 766.93 | 6.53 | 52.32 | 74.36 | -0.514 | 细胞核Nucleus |
| TaINP4.4 | TraesCS4B02G178600 | 4B | 334 | 37 268.18 | 7.87 | 50.04 | 82.22 | -0.513 | 细胞核Nucleus |
| TaINP4.5 | TraesCS3B02G220400 | 3B | 327 | 36 381.83 | 7.79 | 59.51 | 76.51 | -0.622 | 细胞核Nucleus |
| TaINP3.5 | TraesCS3A02G372400 | 3A | 557 | 60 348.69 | 6.31 | 59.06 | 70.09 | -0.472 | 细胞核Nucleus |
| TaINP6.6 | TraesCS6D02G230400 | 6D | 232 | 25 274.05 | 9.97 | 65.71 | 96.08 | -0.029 | 叶绿体Chloroplast |
| TaINP1.3 | TraesCS1A02G392200 | 1A | 226 | 24 449.82 | 5.47 | 43.61 | 98.14 | -0.073 | 线粒体Mitochondria |
| TaINP6.3 | TraesCS6B02G193200 | 6B | 458 | 50 063.37 | 7.30 | 52.50 | 76.05 | -0.464 | 细胞核Nucleus |
| TaINP3.14 | TraesCS3D02G327600 | 3D | 477 | 51 979.22 | 6.53 | 53.94 | 75.47 | -0.495 | 细胞核Nucleus |
| TaINP1.2 | TraesCS1A02G276600 | 1A | 481 | 53 549.34 | 6.29 | 63.78 | 73.97 | -0.512 | 细胞核Nucleus |
| TaINP1.7 | TraesCS1D02G105300 | 1D | 338 | 37 042.89 | 6.56 | 58.57 | 84.97 | -0.371 | 细胞核Nucleus |
| TaINP3.12 | TraesCS3D02G194800 | 3D | 327 | 36 386.85 | 8.49 | 59.18 | 76.51 | -0.626 | 细胞核Nucleus |
| TaINP2.1 | TraesCS2A02G010800 | 2A | 198 | 21 663.49 | 9.56 | 54.16 | 83.54 | -0.271 | 细胞核Nucleus |
| TaINP3.1 | TraesCS3A02G103500 | 3A | 260 | 27 872.59 | 5.60 | 50.35 | 85.85 | -0.191 | 叶绿体Chloroplast |
| TaINP4.6 | TraesCS4D02G180200 | 4D | 329 | 36 682.46 | 7.87 | 52.80 | 81.40 | -0.527 | 胞核Nucleus |
| TaINP5.5 | TraesCS5D02G273500 | 5D | 517 | 57 757.09 | 6.55 | 68.94 | 77.43 | -0.544 | 细胞核Nucleus |
| TaINP2.8 | TraesCS2D02G096800 | 2D | 294 | 32 644.01 | 6.01 | 60.35 | 78.10 | -0.431 | 细胞核Nucleus |
| TaINP3.2 | TraesCS3A02G190700 | 3A | 350 | 38 979.69 | 6.36 | 59.18 | 75.11 | -0.647 | 细胞核Nucleus |

图1 不同物种INP1基因的系统进化树注:Ta—小麦;Os—水稻;Zm—玉米;At—拟南芥;Hv—大麦;Pp—帕滕斯藻。
Fig. 1 Phylogenetic tree of INP1 genes among different speciesNote:Ta—Triticum aestivum L.; Os—Oryza sativa L.; Zm—Zea mays L.; At—Arabidopsis thaliana; Hv—Hordeum vulgare L.; Pp—Physcomitrium patens L..

图3 INP1基因的共线性分析A:小麦TaINP1基因在A、B、D染色体的共线性分析;B:小麦TaINP1基因与水稻、拟南芥、玉米的共线性分析。Ta—小麦;Zm—玉米;Os—水稻;At—拟南芥
Fig. 3 Collinear analysis of INP1 genesA: Collinear analysis of TaINP1 genes in wheat among A, B, D chromosomes; B: Collinear analysis between TaINP1 genes in wheat and rice, Arabidopsis and maize; Ta—Triticum aestivum L.; Zm—Zea mays L.; Os—Oryza sativa L.; At—Arabidopsis thaliana
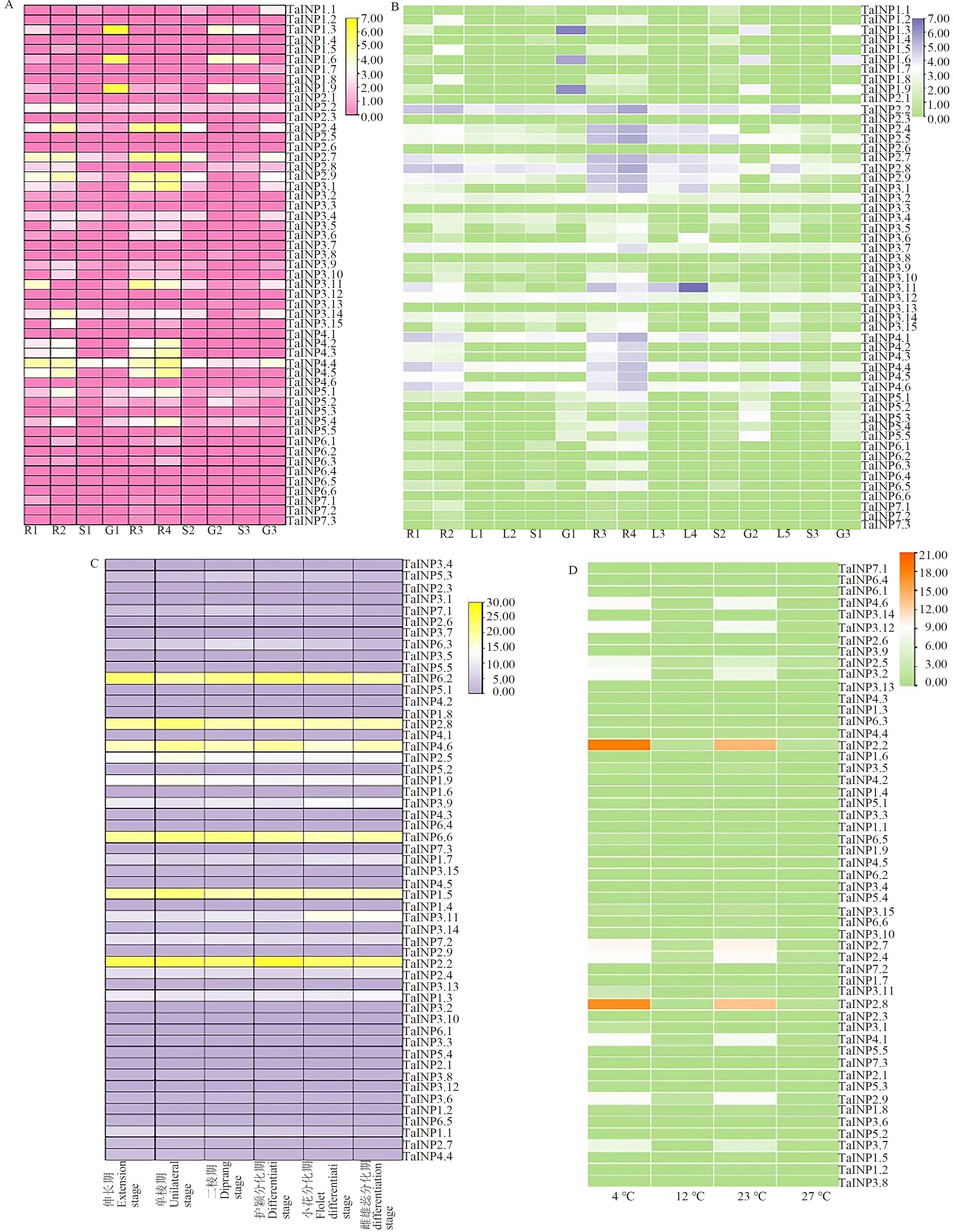
图4 小麦TaINP1基因表达热图A:基于Wheatomics数据库在不同组织中的表达;B: 基于Wheat Expression Browser数据库在不同组织中的表达;C:在幼穗不同发育时期的表达;D:在不同温度处理下的表达。R1—Azhurnaya营养生长期根;R2—Azhurnaya生殖生长期根;R3—中国春营养生长期根;R4—中国春生殖生长期根;L1—Azhurnaya营养生长期叶;L2—Azhurnaya生殖生长期叶;L3—中国春营养生长期叶;L4—中国春生殖生长期叶;L5—其他种质叶;S1—Azhurnaya穗;S2—中国春穗;G1—Azhurnaya籽粒;G2—中国春籽粒;S3—其他种质穗;G3—其他种质籽粒
Fig. 4 Expression heat map of TaINP1 genes in wheatA:Expression of different tissues based on Wheatomics database; B: Expression of early panicle with different stages based on Wheat Expression Browser; C:Expression of young spikes in different stages D: Expression of different temperature treatments. R1—Roots of Azhurnaya in vegetative growth period; R2—Roots of Azhurnaya in reproductive growth period; R3—Roots of Chinese spring in vegetative growth period; R4—Roots of Chinese spring in reproductive growth period; L1—Leaves of Azhurnaya in vegetative growth period; L2—Leaves of Azhurnaya in reproductive growth period; L3—Leaves of Chinese spring in vegetative growth period; L4—Leaves of Chinese spring in reproductive growth period; L5—Leaves of other germplasms; S1—Spike of Azhurnaya;S2—Spike of Chinese spring; G1—Grains of Azhurnaya;G2—Grains of Chinese spring; S3—Spike of other germplasms; G3—Grains of other germplasms

图5 TaINP1基因在小麦不同组织中的表达注:不同小写字母表示不同器官间在P<0.05水平差异显著。
Fig. 5 Expression analysis of TaINP1 genes in different tissues of wheatNote:Different lowercase letters indicate significant differences between different tissues at P<0.05 level.
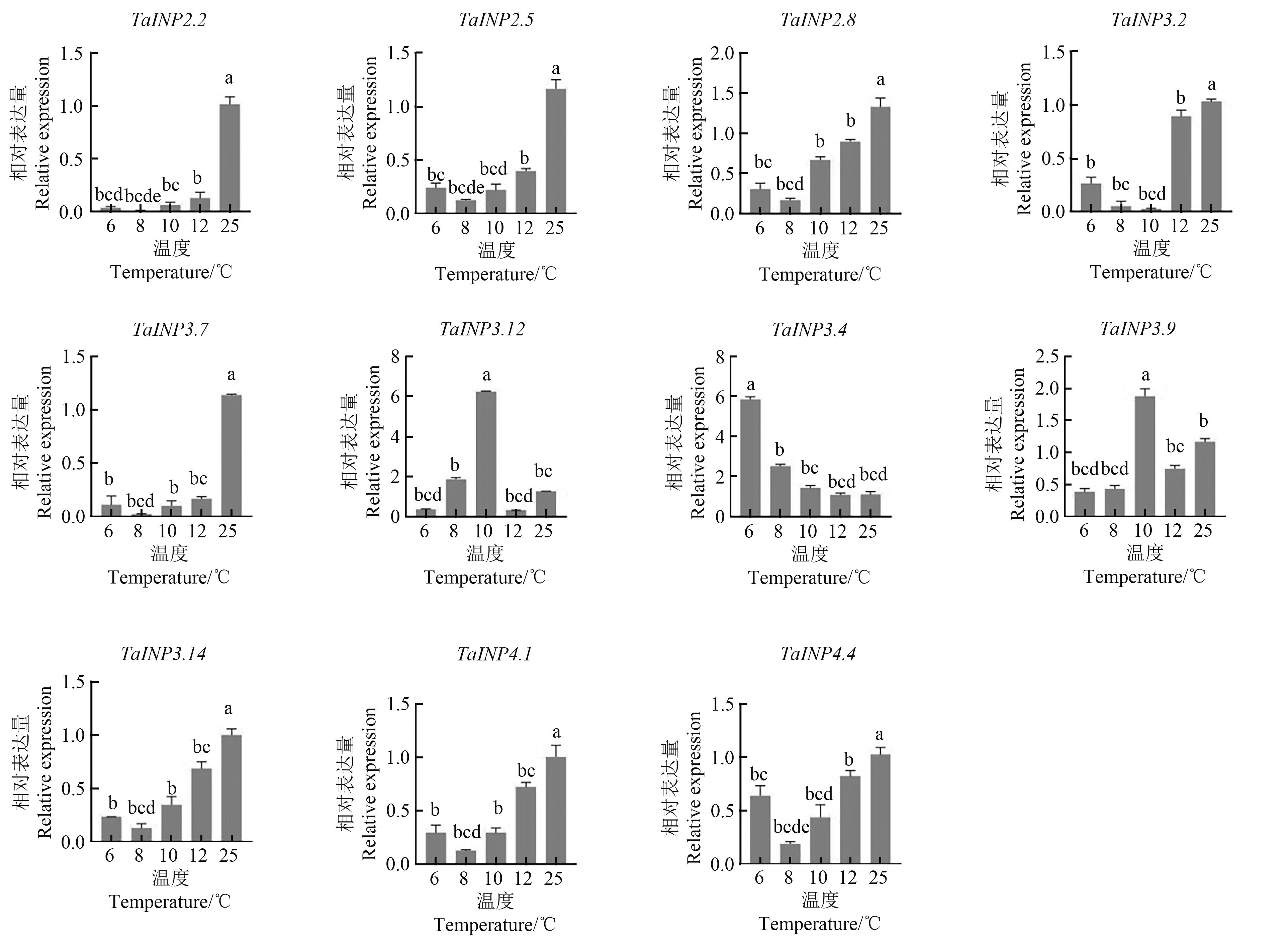
图6 TaINP1基因在低温处理下花粉中的表达注:不同小写字母表示不同温度间在P<0.05水平差异显著。
Fig. 6 Expression of TaINP1 genes in pollen under low temperature treatmentNote:Different lowercase letters indicate significant differences between different temperatures at P<0.05 level.
| 1 | HUANG J B, YANG L, YANG L, et al.. Stigma receptors control intraspecies and interspecies barriers in Brassicaceae [J].Nature, 2023,614(7947):303-308. |
| 2 | YU X B, ZHANG X C, ZHAO P, et al.. Fertilized egg cells secrete endopeptidases to avoid polytubey [J]. Nature, 2021,592:433-437. |
| 3 | ZHANG Z B, SUN M K, XIONG T, et al.. Development and genetic regulation of pollen intine in Arabidopsis and rice [J/OL].Gene, 2024,893:147936[2024-07-20].. |
| 4 | SHI J X, CUI M H, YANG L, et al.. Genetic and biochemical mechanisms of pollen wall development [J]. Trends Plant Sci., 2015,20(11):741-753. |
| 5 | LI F S, PHYO P, JACOBOWITZ J, et al.. The molecular structure of plant sporopollenin [J]. Nat. Plants, 2019,5(1):41-46. |
| 6 | QUILICHINI T D, GRIENENBERGER E, DOUGLAS C J. The biosynthesis,composition and assembly of the outer pollen wall:a tough case to crack [J]. Phytochemistry, 2015,113:170-182. |
| 7 | FURNESS C A, RUDALL P J. Pollen aperture evolution:a crucial factor for eudicot success? [J]. Trends Plant Sci., 2004,9(3):154-158. |
| 8 | EDLUND A F, SWANSON R, PREUSS D. Pollen and stigma structure and function:the role of diversity in pollination [J].Plant Cell, 2004,16(Sl):S84-S97. |
| 9 | ZHANG X, ZHAO G C, TAN Q, et al.. Rice pollen aperture formation is regulated by the interplay between OsINP1 and OsDAF1 [J]. Nat. Plants, 2020,6(4):394-403. |
| 10 | DOBRITSA A A, REEDER S H. Formation of pollen apertures in Arabidopsis requires an interplay between male meiosis,development of INP1-decorated plasma membrane domains,and the callose wall [J/OL]. Plant Signal. Behav., 2017,12(12):e1393136 [2024-07-20]. . |
| 11 | DOBRITSA A A, COERPER D. The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine [J]. Plant cell, 2012,24(11):4452-4464. |
| 12 | LI P, BEN-MENNI SCHULER S, REEDER S H, et al.. INP1 involvement in pollen aperture formation is evolutionarily conserved and may require species-specific partners [J]. J. Exp.Bot., 2018,69(5):983-996. |
| 13 | DOBRITSA A A, KIRKPATRICK A B, REEDER S H, et al.. Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition [J]. Plant Physiol., 2018,176(1):326-339. |
| 14 | DE SOUSA T, RIBEIRO M, SABENÇA C, et al.. The 10,000-year success story of wheat! [J/OL]. Foods, 2021,10(9):2124 [2024-07-20]. . |
| 15 | CARRILLO-BARRAL N, DEL CARMEN RODRÍGUEZ-GACIO M, MATILLA A J. Delay of germination-1 (DOG1):a key to understanding seed dormancy [J/OL]. Plants (Basel), 2020,9(4):480 [2024-07-20]. . |
| 16 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE,a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002,30(1):325-327. |
| 17 | ZHANG J P, QI Y N, WANG L M, et al.. Genomic comparison and population diversity analysis provide insights into the domestication and improvement of flax [J/OL]. iScience, 2020,23(4):100967 [2024-07-20]. . |
| 18 | SONG S Q, TANG C F, JIANG X C, et al.. Research progress on DOG1, a key regulator of seed dormancy and germination [J]. Plant Sci., 2024, 42(2): 254-266. |
| 19 | 崔荣秀,张议文,陈晓倩,等.植物bZIP参与胁迫应答调控的最新研究进展[J].生物技术通报,2019,35(2):143-155. |
| CUI R X, ZHANG Y W, CHEN X Q, et al.. The latest research progress on the stress responses of bZIP involved in plants [J].Biotechnol. Bull., 2019,35(2):143-155. | |
| 20 | 杜朝金,张汉尧,罗心平,等.基因调控植物花器官发育的研究进展[J].植物遗传资源学报,2024,25(2):151-161. |
| DU C J, ZHANG H R, LUO X P, et al.. Progress in gene regulation of plant floral organ development [J]. J. Plant Genetic Resour., 2024, 25(2):151-161 | |
| 21 | WANG Z P, ZHANG Z B, ZHENG D Y, et al.. Efficient and genotype independent maize transformation using pollen transfected by DNA-coated magnetic nanoparticles [J]. J. Integr. Plant Biol., 2022,64(6):1145-1156. |
| 22 | WILKINS M R, GASTEIGER E, BAIROCH A, et al.. Protein identification and analysis tools in the ExPASy server [J]. Methods Mol. Biol., 1999,112:531-552. |
| [1] | 薛振宇, 张康康, 张元元, 闫强强, 姚立蓉, 张宏, 孟亚雄, 司二静, 李葆春, 马小乐, 王化俊, 汪军成. 优质抗旱小麦种质的筛选及功能基因检测[J]. 中国农业科技导报, 2025, 27(1): 35-49. |
| [2] | 孙宪印, 牟秋焕, 米勇, 吕广德, 亓晓蕾, 孙盈盈, 尹逊栋, 王瑞霞, 吴科, 钱兆国, 赵岩, 高明刚. 基于GT双标图对小麦新品系的分类评价[J]. 中国农业科技导报, 2024, 26(7): 14-24. |
| [3] | 鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21. |
| [4] | 陈明迪, 胡桂花, 张海文, 王旺田. 水稻RR基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(5): 20-29. |
| [5] | 张绥林, 李洋, 李琰, 张赟齐, 齐建勋, 侯智霞. 核桃晚霜危害特性及影响机制研究进展[J]. 中国农业科技导报, 2024, 26(4): 18-26. |
| [6] | 赵刚, 王淑英, 李尚中, 张建军, 党翼, 王磊, 李兴茂, 程万莉, 周刚, 倪胜利, 樊廷录. 黄土旱塬区近40年降水对冬小麦耗水和产量的影响[J]. 中国农业科技导报, 2024, 26(3): 164-173. |
| [7] | 张宏, 李卫国, 张晓东, 卢必慧, 张琤琤, 李伟, 马廷淮. 基于HJ-1星和GF-1号影像融合特征提取冬小麦种植面积[J]. 中国农业科技导报, 2024, 26(2): 109-119. |
| [8] | 张景云, 关峰, 石博, 万新建. 小麦根系分泌物对苦瓜幼苗生长及土壤生物学环境的影响[J]. 中国农业科技导报, 2024, 26(2): 181-190. |
| [9] | 李双, 王爱英, 焦浈, 池青, 孙昊, 焦涛. 盐胁迫下不同抗性小麦幼苗生理生化特性及转录组分析[J]. 中国农业科技导报, 2024, 26(2): 20-32. |
| [10] | 苗宇, 王婕, 赵尧尧, 张丽佳, 刘美君. 低温胁迫后紫花苜蓿叶片光合作用的恢复特性研究[J]. 中国农业科技导报, 2024, 26(2): 80-89. |
| [11] | 刘海霞, 张寅辉, 庄蕾, 郭梦娇, 赵李, 吴美娟, 侯健, 李甜, 刘红霞, 张学勇, 郝晨阳. 基于关联分析挖掘小麦SDS沉降值相关候选基因及KASP标记开发[J]. 中国农业科技导报, 2024, 26(12): 18-29. |
| [12] | 杨朝阳, 徐鹏, 苑铁键, 李晓琼, 彭冬根, 张振涛, 杨俊玲, 丁闯闯, 朱纪洲. 杂交构树低温干燥特性及品质研究[J]. 中国农业科技导报, 2024, 26(11): 157-170. |
| [13] | 胡墨明, 孟琰珺, 苗淑媛, 胡西宁, 许晴, 马红珍, 王鹏飞, 刘国芹, 康国章. 小麦淀粉合成关键基因TaAGPL1功能标记的开发[J]. 中国农业科技导报, 2024, 26(11): 43-55. |
| [14] | 田鹏, 宋洁, 李霞, 王裕智, 武棒棒. 不同粒色小麦籽粒中叶酸及衍生物含量分析[J]. 中国农业科技导报, 2024, 26(11): 56-65. |
| [15] | 尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤, 黄耿青. 薰衣草 JAZ和MYC2-like基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(11): 79-90. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号