













Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (4): 22-35.DOI: 10.13304/j.nykjdb.2024.0620
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Bei MA1,2( ), Jie GONG2(
), Jie GONG2( ), Yinke DU2,3, Yuwei GAN1,2, Rong CHENG2,3, Bo ZHU1, Lixia YI1(
), Yinke DU2,3, Yuwei GAN1,2, Rong CHENG2,3, Bo ZHU1, Lixia YI1( ), Jinxiu MA2(
), Jinxiu MA2( ), Shiqing GAO2(
), Shiqing GAO2( )
)
Received:2024-08-02
Accepted:2024-09-18
Online:2025-04-15
Published:2025-04-15
Contact:
Lixia YI,Jinxiu MA,Shiqing GAO
马蓓1,2( ), 公杰2(
), 公杰2( ), 杜银柯2,3, 甘雨薇1,2, 程蓉2,3, 朱波1, 易丽霞1(
), 杜银柯2,3, 甘雨薇1,2, 程蓉2,3, 朱波1, 易丽霞1( ), 马锦绣2(
), 马锦绣2( ), 高世庆2(
), 高世庆2( )
)
通讯作者:
易丽霞,马锦绣,高世庆
作者简介:马蓓 E-mail:18034656852@163.com基金资助:CLC Number:
Bei MA, Jie GONG, Yinke DU, Yuwei GAN, Rong CHENG, Bo ZHU, Lixia YI, Jinxiu MA, Shiqing GAO. Identification and Expression Analysis of TaINP1 Gene Related to Pollen Pore Development in Wheat[J]. Journal of Agricultural Science and Technology, 2025, 27(4): 22-35.
马蓓, 公杰, 杜银柯, 甘雨薇, 程蓉, 朱波, 易丽霞, 马锦绣, 高世庆. 小麦花粉孔发育相关TaINP1基因鉴定及表达分析[J]. 中国农业科技导报, 2025, 27(4): 22-35.
基因名称 Gene name | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaINP2.2 | TCGTAGATGGGATAATGGGGC | CAAGCCACAGGAAGCACCTT |
| TaINP2.5 | ATTGGGTTTCAGACGGAGCA | CAAGGGCTTGTCAGACCTGT |
| TaINP2.8 | CCGCTGGAATCTTGTCCGTTA | TTCCTTGCAGCCTCACGATT |
| TaINP3.2 | TCCATGAGTGGAAATGGGGC | ACTACGCAGCTCAGTATCGC |
| TaINP3.7 | TTTTAAAGCTTGAACCCCTAACTGA | ACCCAGCCAATGTTTCTGCC |
| TaINP3.12 | TTTTAAAGCTTGAACCCCTAACTGA | CCGATAGACCCAGCCAACG |
| TaINP3.4 | GCAGTTGATCCTCAGGCACT | AGGGTTTGTAGCCGCAGATT |
| TaINP3.9 | CTCAGCACACCATCCCAACA | TCTTCCACATTCCCGACAGC |
| TaINP3.14 | CAGGCTGATAATCTTCGTCTGC | GTGCAGATTGGCGAGTGGTT |
| TaINP4.1 | CCAAAAGGTGGTATTGCGGA | ACTGCTGCTCTGTTAGAGGC |
| TaINP4.4 | ACTCCAGAAAGCTCGACAGC | CAGAGAGAACTGAGGGCACG |
| 18s | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
Table 1 Fluorescence quantification primers
基因名称 Gene name | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaINP2.2 | TCGTAGATGGGATAATGGGGC | CAAGCCACAGGAAGCACCTT |
| TaINP2.5 | ATTGGGTTTCAGACGGAGCA | CAAGGGCTTGTCAGACCTGT |
| TaINP2.8 | CCGCTGGAATCTTGTCCGTTA | TTCCTTGCAGCCTCACGATT |
| TaINP3.2 | TCCATGAGTGGAAATGGGGC | ACTACGCAGCTCAGTATCGC |
| TaINP3.7 | TTTTAAAGCTTGAACCCCTAACTGA | ACCCAGCCAATGTTTCTGCC |
| TaINP3.12 | TTTTAAAGCTTGAACCCCTAACTGA | CCGATAGACCCAGCCAACG |
| TaINP3.4 | GCAGTTGATCCTCAGGCACT | AGGGTTTGTAGCCGCAGATT |
| TaINP3.9 | CTCAGCACACCATCCCAACA | TCTTCCACATTCCCGACAGC |
| TaINP3.14 | CAGGCTGATAATCTTCGTCTGC | GTGCAGATTGGCGAGTGGTT |
| TaINP4.1 | CCAAAAGGTGGTATTGCGGA | ACTGCTGCTCTGTTAGAGGC |
| TaINP4.4 | ACTCCAGAAAGCTCGACAGC | CAGAGAGAACTGAGGGCACG |
| 18s | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP1.4 | TraesCS1B02G127400 | 1B | 333 | 36 564.21 | 6.14 | 54.41 | 83.60 | -0.416 | 细胞核Nucleus |
| TaINP6.2 | TraesCS6A02G248300 | 6A | 232 | 25 301.97 | 9.61 | 67.01 | 93.15 | -0.088 | 细胞核Nucleus |
| TaINP3.6 | TraesCS3B02G120900 | 3B | 260 | 27 937.63 | 5.43 | 54.74 | 86.58 | -0.181 | 细胞核Nucleus |
| TaINP2.4 | TraesCS2A02G526300 | 2A | 419 | 46 270.21 | 6.24 | 59.92 | 71.89 | -0.536 | 细胞核Nucleus |
| TaINP3.3 | TraesCS3A02G306200 | 3A | 296 | 31 824.06 | 5.63 | 49.12 | 88.34 | -0.210 | 细胞核Nucleus |
| TaINP3.11 | TraesCS3D02G105800 | 3D | 260 | 27 976.71 | 5.46 | 55.33 | 85.85 | -0.202 | 细胞核Nucleus |
| TaINP1.9 | TraesCS1D02G400300 | 1D | 226 | 2 449.19 | 5.65 | 46.52 | 98.14 | -0.078 | 线粒体Mitochondria |
| TaINP1.6 | TraesCS1B02G420300 | 1B | 226 | 24 449.82 | 5.65 | 47.19 | 96.86 | -0.098 | 线粒体Mitochondria |
| TaINP5.3 | TraesCS5B02G265300 | 5B | 525 | 58 620.02 | 6.58 | 70.75 | 76.63 | -0.560 | 细胞核Nucleus |
| TaINP4.2 | TraesCS4A02G183400 | 4A | 539 | 58 911.63 | 6.44 | 54.62 | 69.13 | -0.568 | 细胞核Nucleus |
| TaINP5.2 | TraesCS5A02G265600 | 5A | 518 | 57 856.21 | 6.51 | 68.93 | 77.30 | -0.550 | 细胞核Nucleus |
| TaINP1.1 | TraesCS1A02G096300 | 1A | 335 | 36 675.16 | 5.71 | 57.88 | 82.27 | -0.438 | 细胞核Nucleus |
| TaINP1.8 | TraesCS1D02G276100 | 1D | 481 | 53 546.29 | 5.95 | 62.72 | 74.39 | -0.503 | 细胞核Nucleus |
| TaINP4.1 | TraesCS4A02G126300 | 4A | 335 | 37 349.28 | 7.87 | 51.81 | 84.00 | -0.491 | 细胞核Nucleus |
| TaINP7.2 | TraesCS7B02G114300 | 7B | 448 | 48 464.72 | 7.36 | 56.89 | 82.01 | -0.285 | 细胞核Nucleus |
| TaINP1.5 | TraesCS1B02G285800 | 1B | 526 | 58 065.31 | 5.79 | 61.88 | 73.23 | -0.471 | 细胞核Nucleus |
| TaINP6.4 | TraesCS6B02G276400 | 6B | 232 | 25 314.02 | 9.46 | 68.12 | 92.72 | -0.100 | 叶绿体Chloroplast |
| TaINP4.5 | TraesCS4D02G129900 | 4D | 537 | 58 671.34 | 6.41 | 54.16 | 68.47 | -0.567 | 细胞核Nucleus |
| TaINP3.13 | TraesCS3D02G295800 | 3D | 292 | 31 490.63 | 5.37 | 49.44 | 89.55 | -0.191 | 叶绿体Chloroplast |
| TaINP3.9 | TraesCS3B02G365100 | 3B | 477 | 51 918.14 | 6.78 | 50.58 | 75.68 | -0.494 | 细胞核Nucleus |
| TaINP4.3 | TraesCS4B02G135000 | 4B | 566 | 6 217.82 | 6.40 | 56.40 | 66.70 | -0.614 | 细胞核Nucleus |
| TaINP2.9 | TraesCS2D02G529000 | 2D | 425 | 46 843.83 | 5.86 | 57.18 | 73.39 | -0.507 | 细胞核Nucleus |
| TaINP5.1 | TraesCS5A02G174200 | 5A | 467 | 51 251.99 | 7.02 | 53.54 | 77.41 | -0.473 | 细胞核Nucleus |
| TaINP2.6 | TraesCS2B02G512700 | 2B | 240 | 26 178.71 | 9.20 | 44.06 | 86.33 | -0.215 | 细胞质Cytoplasm |
Table 2 Basic physical and chemical properties of TaINP1 gene family in wheat
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP1.4 | TraesCS1B02G127400 | 1B | 333 | 36 564.21 | 6.14 | 54.41 | 83.60 | -0.416 | 细胞核Nucleus |
| TaINP6.2 | TraesCS6A02G248300 | 6A | 232 | 25 301.97 | 9.61 | 67.01 | 93.15 | -0.088 | 细胞核Nucleus |
| TaINP3.6 | TraesCS3B02G120900 | 3B | 260 | 27 937.63 | 5.43 | 54.74 | 86.58 | -0.181 | 细胞核Nucleus |
| TaINP2.4 | TraesCS2A02G526300 | 2A | 419 | 46 270.21 | 6.24 | 59.92 | 71.89 | -0.536 | 细胞核Nucleus |
| TaINP3.3 | TraesCS3A02G306200 | 3A | 296 | 31 824.06 | 5.63 | 49.12 | 88.34 | -0.210 | 细胞核Nucleus |
| TaINP3.11 | TraesCS3D02G105800 | 3D | 260 | 27 976.71 | 5.46 | 55.33 | 85.85 | -0.202 | 细胞核Nucleus |
| TaINP1.9 | TraesCS1D02G400300 | 1D | 226 | 2 449.19 | 5.65 | 46.52 | 98.14 | -0.078 | 线粒体Mitochondria |
| TaINP1.6 | TraesCS1B02G420300 | 1B | 226 | 24 449.82 | 5.65 | 47.19 | 96.86 | -0.098 | 线粒体Mitochondria |
| TaINP5.3 | TraesCS5B02G265300 | 5B | 525 | 58 620.02 | 6.58 | 70.75 | 76.63 | -0.560 | 细胞核Nucleus |
| TaINP4.2 | TraesCS4A02G183400 | 4A | 539 | 58 911.63 | 6.44 | 54.62 | 69.13 | -0.568 | 细胞核Nucleus |
| TaINP5.2 | TraesCS5A02G265600 | 5A | 518 | 57 856.21 | 6.51 | 68.93 | 77.30 | -0.550 | 细胞核Nucleus |
| TaINP1.1 | TraesCS1A02G096300 | 1A | 335 | 36 675.16 | 5.71 | 57.88 | 82.27 | -0.438 | 细胞核Nucleus |
| TaINP1.8 | TraesCS1D02G276100 | 1D | 481 | 53 546.29 | 5.95 | 62.72 | 74.39 | -0.503 | 细胞核Nucleus |
| TaINP4.1 | TraesCS4A02G126300 | 4A | 335 | 37 349.28 | 7.87 | 51.81 | 84.00 | -0.491 | 细胞核Nucleus |
| TaINP7.2 | TraesCS7B02G114300 | 7B | 448 | 48 464.72 | 7.36 | 56.89 | 82.01 | -0.285 | 细胞核Nucleus |
| TaINP1.5 | TraesCS1B02G285800 | 1B | 526 | 58 065.31 | 5.79 | 61.88 | 73.23 | -0.471 | 细胞核Nucleus |
| TaINP6.4 | TraesCS6B02G276400 | 6B | 232 | 25 314.02 | 9.46 | 68.12 | 92.72 | -0.100 | 叶绿体Chloroplast |
| TaINP4.5 | TraesCS4D02G129900 | 4D | 537 | 58 671.34 | 6.41 | 54.16 | 68.47 | -0.567 | 细胞核Nucleus |
| TaINP3.13 | TraesCS3D02G295800 | 3D | 292 | 31 490.63 | 5.37 | 49.44 | 89.55 | -0.191 | 叶绿体Chloroplast |
| TaINP3.9 | TraesCS3B02G365100 | 3B | 477 | 51 918.14 | 6.78 | 50.58 | 75.68 | -0.494 | 细胞核Nucleus |
| TaINP4.3 | TraesCS4B02G135000 | 4B | 566 | 6 217.82 | 6.40 | 56.40 | 66.70 | -0.614 | 细胞核Nucleus |
| TaINP2.9 | TraesCS2D02G529000 | 2D | 425 | 46 843.83 | 5.86 | 57.18 | 73.39 | -0.507 | 细胞核Nucleus |
| TaINP5.1 | TraesCS5A02G174200 | 5A | 467 | 51 251.99 | 7.02 | 53.54 | 77.41 | -0.473 | 细胞核Nucleus |
| TaINP2.6 | TraesCS2B02G512700 | 2B | 240 | 26 178.71 | 9.20 | 44.06 | 86.33 | -0.215 | 细胞质Cytoplasm |
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP3.4 | TraesCS3A02G334100 | 3A | 475 | 51 766.93 | 6.53 | 52.32 | 74.36 | -0.514 | 细胞核Nucleus |
| TaINP4.4 | TraesCS4B02G178600 | 4B | 334 | 37 268.18 | 7.87 | 50.04 | 82.22 | -0.513 | 细胞核Nucleus |
| TaINP4.5 | TraesCS3B02G220400 | 3B | 327 | 36 381.83 | 7.79 | 59.51 | 76.51 | -0.622 | 细胞核Nucleus |
| TaINP3.5 | TraesCS3A02G372400 | 3A | 557 | 60 348.69 | 6.31 | 59.06 | 70.09 | -0.472 | 细胞核Nucleus |
| TaINP6.6 | TraesCS6D02G230400 | 6D | 232 | 25 274.05 | 9.97 | 65.71 | 96.08 | -0.029 | 叶绿体Chloroplast |
| TaINP1.3 | TraesCS1A02G392200 | 1A | 226 | 24 449.82 | 5.47 | 43.61 | 98.14 | -0.073 | 线粒体Mitochondria |
| TaINP6.3 | TraesCS6B02G193200 | 6B | 458 | 50 063.37 | 7.30 | 52.50 | 76.05 | -0.464 | 细胞核Nucleus |
| TaINP3.14 | TraesCS3D02G327600 | 3D | 477 | 51 979.22 | 6.53 | 53.94 | 75.47 | -0.495 | 细胞核Nucleus |
| TaINP1.2 | TraesCS1A02G276600 | 1A | 481 | 53 549.34 | 6.29 | 63.78 | 73.97 | -0.512 | 细胞核Nucleus |
| TaINP1.7 | TraesCS1D02G105300 | 1D | 338 | 37 042.89 | 6.56 | 58.57 | 84.97 | -0.371 | 细胞核Nucleus |
| TaINP3.12 | TraesCS3D02G194800 | 3D | 327 | 36 386.85 | 8.49 | 59.18 | 76.51 | -0.626 | 细胞核Nucleus |
| TaINP2.1 | TraesCS2A02G010800 | 2A | 198 | 21 663.49 | 9.56 | 54.16 | 83.54 | -0.271 | 细胞核Nucleus |
| TaINP3.1 | TraesCS3A02G103500 | 3A | 260 | 27 872.59 | 5.60 | 50.35 | 85.85 | -0.191 | 叶绿体Chloroplast |
| TaINP4.6 | TraesCS4D02G180200 | 4D | 329 | 36 682.46 | 7.87 | 52.80 | 81.40 | -0.527 | 胞核Nucleus |
| TaINP5.5 | TraesCS5D02G273500 | 5D | 517 | 57 757.09 | 6.55 | 68.94 | 77.43 | -0.544 | 细胞核Nucleus |
| TaINP2.8 | TraesCS2D02G096800 | 2D | 294 | 32 644.01 | 6.01 | 60.35 | 78.10 | -0.431 | 细胞核Nucleus |
| TaINP3.2 | TraesCS3A02G190700 | 3A | 350 | 38 979.69 | 6.36 | 59.18 | 75.11 | -0.647 | 细胞核Nucleus |
Table 2 Basic physical and chemical properties of TaINP1 gene family in wheat
基因名称 Gene name | 基因号 Gene ID | 染色体Chromosome | 蛋白长度Number of amino acid/aa | 分子量Molecular weight/Da | 等电点 pI | 不稳定指数Instability index | 脂肪系数Aliphatic index | 总亲水性指数 GRAVY | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| TaINP3.4 | TraesCS3A02G334100 | 3A | 475 | 51 766.93 | 6.53 | 52.32 | 74.36 | -0.514 | 细胞核Nucleus |
| TaINP4.4 | TraesCS4B02G178600 | 4B | 334 | 37 268.18 | 7.87 | 50.04 | 82.22 | -0.513 | 细胞核Nucleus |
| TaINP4.5 | TraesCS3B02G220400 | 3B | 327 | 36 381.83 | 7.79 | 59.51 | 76.51 | -0.622 | 细胞核Nucleus |
| TaINP3.5 | TraesCS3A02G372400 | 3A | 557 | 60 348.69 | 6.31 | 59.06 | 70.09 | -0.472 | 细胞核Nucleus |
| TaINP6.6 | TraesCS6D02G230400 | 6D | 232 | 25 274.05 | 9.97 | 65.71 | 96.08 | -0.029 | 叶绿体Chloroplast |
| TaINP1.3 | TraesCS1A02G392200 | 1A | 226 | 24 449.82 | 5.47 | 43.61 | 98.14 | -0.073 | 线粒体Mitochondria |
| TaINP6.3 | TraesCS6B02G193200 | 6B | 458 | 50 063.37 | 7.30 | 52.50 | 76.05 | -0.464 | 细胞核Nucleus |
| TaINP3.14 | TraesCS3D02G327600 | 3D | 477 | 51 979.22 | 6.53 | 53.94 | 75.47 | -0.495 | 细胞核Nucleus |
| TaINP1.2 | TraesCS1A02G276600 | 1A | 481 | 53 549.34 | 6.29 | 63.78 | 73.97 | -0.512 | 细胞核Nucleus |
| TaINP1.7 | TraesCS1D02G105300 | 1D | 338 | 37 042.89 | 6.56 | 58.57 | 84.97 | -0.371 | 细胞核Nucleus |
| TaINP3.12 | TraesCS3D02G194800 | 3D | 327 | 36 386.85 | 8.49 | 59.18 | 76.51 | -0.626 | 细胞核Nucleus |
| TaINP2.1 | TraesCS2A02G010800 | 2A | 198 | 21 663.49 | 9.56 | 54.16 | 83.54 | -0.271 | 细胞核Nucleus |
| TaINP3.1 | TraesCS3A02G103500 | 3A | 260 | 27 872.59 | 5.60 | 50.35 | 85.85 | -0.191 | 叶绿体Chloroplast |
| TaINP4.6 | TraesCS4D02G180200 | 4D | 329 | 36 682.46 | 7.87 | 52.80 | 81.40 | -0.527 | 胞核Nucleus |
| TaINP5.5 | TraesCS5D02G273500 | 5D | 517 | 57 757.09 | 6.55 | 68.94 | 77.43 | -0.544 | 细胞核Nucleus |
| TaINP2.8 | TraesCS2D02G096800 | 2D | 294 | 32 644.01 | 6.01 | 60.35 | 78.10 | -0.431 | 细胞核Nucleus |
| TaINP3.2 | TraesCS3A02G190700 | 3A | 350 | 38 979.69 | 6.36 | 59.18 | 75.11 | -0.647 | 细胞核Nucleus |

Fig. 1 Phylogenetic tree of INP1 genes among different speciesNote:Ta—Triticum aestivum L.; Os—Oryza sativa L.; Zm—Zea mays L.; At—Arabidopsis thaliana; Hv—Hordeum vulgare L.; Pp—Physcomitrium patens L..

Fig. 3 Collinear analysis of INP1 genesA: Collinear analysis of TaINP1 genes in wheat among A, B, D chromosomes; B: Collinear analysis between TaINP1 genes in wheat and rice, Arabidopsis and maize; Ta—Triticum aestivum L.; Zm—Zea mays L.; Os—Oryza sativa L.; At—Arabidopsis thaliana
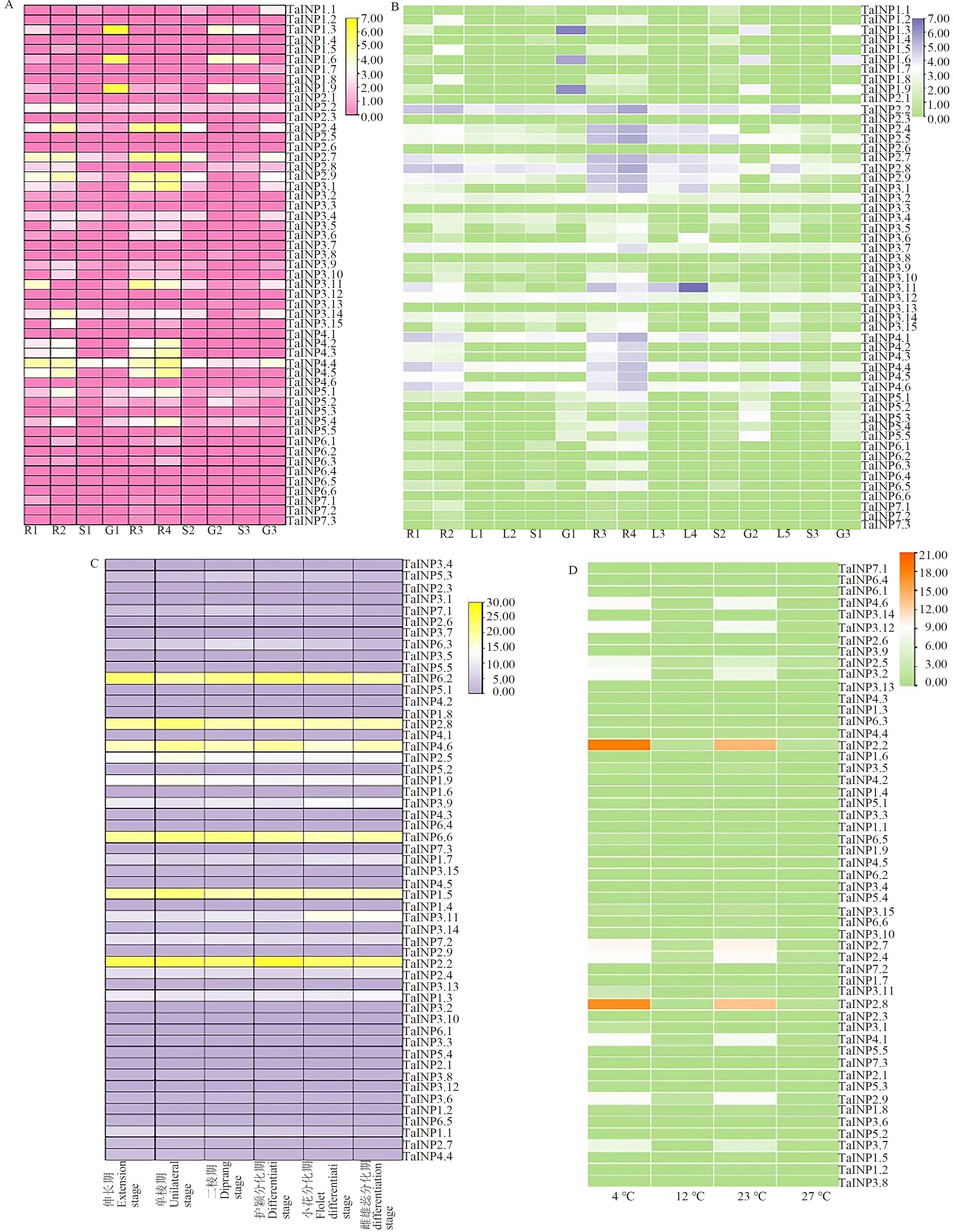
Fig. 4 Expression heat map of TaINP1 genes in wheatA:Expression of different tissues based on Wheatomics database; B: Expression of early panicle with different stages based on Wheat Expression Browser; C:Expression of young spikes in different stages D: Expression of different temperature treatments. R1—Roots of Azhurnaya in vegetative growth period; R2—Roots of Azhurnaya in reproductive growth period; R3—Roots of Chinese spring in vegetative growth period; R4—Roots of Chinese spring in reproductive growth period; L1—Leaves of Azhurnaya in vegetative growth period; L2—Leaves of Azhurnaya in reproductive growth period; L3—Leaves of Chinese spring in vegetative growth period; L4—Leaves of Chinese spring in reproductive growth period; L5—Leaves of other germplasms; S1—Spike of Azhurnaya;S2—Spike of Chinese spring; G1—Grains of Azhurnaya;G2—Grains of Chinese spring; S3—Spike of other germplasms; G3—Grains of other germplasms

Fig. 5 Expression analysis of TaINP1 genes in different tissues of wheatNote:Different lowercase letters indicate significant differences between different tissues at P<0.05 level.
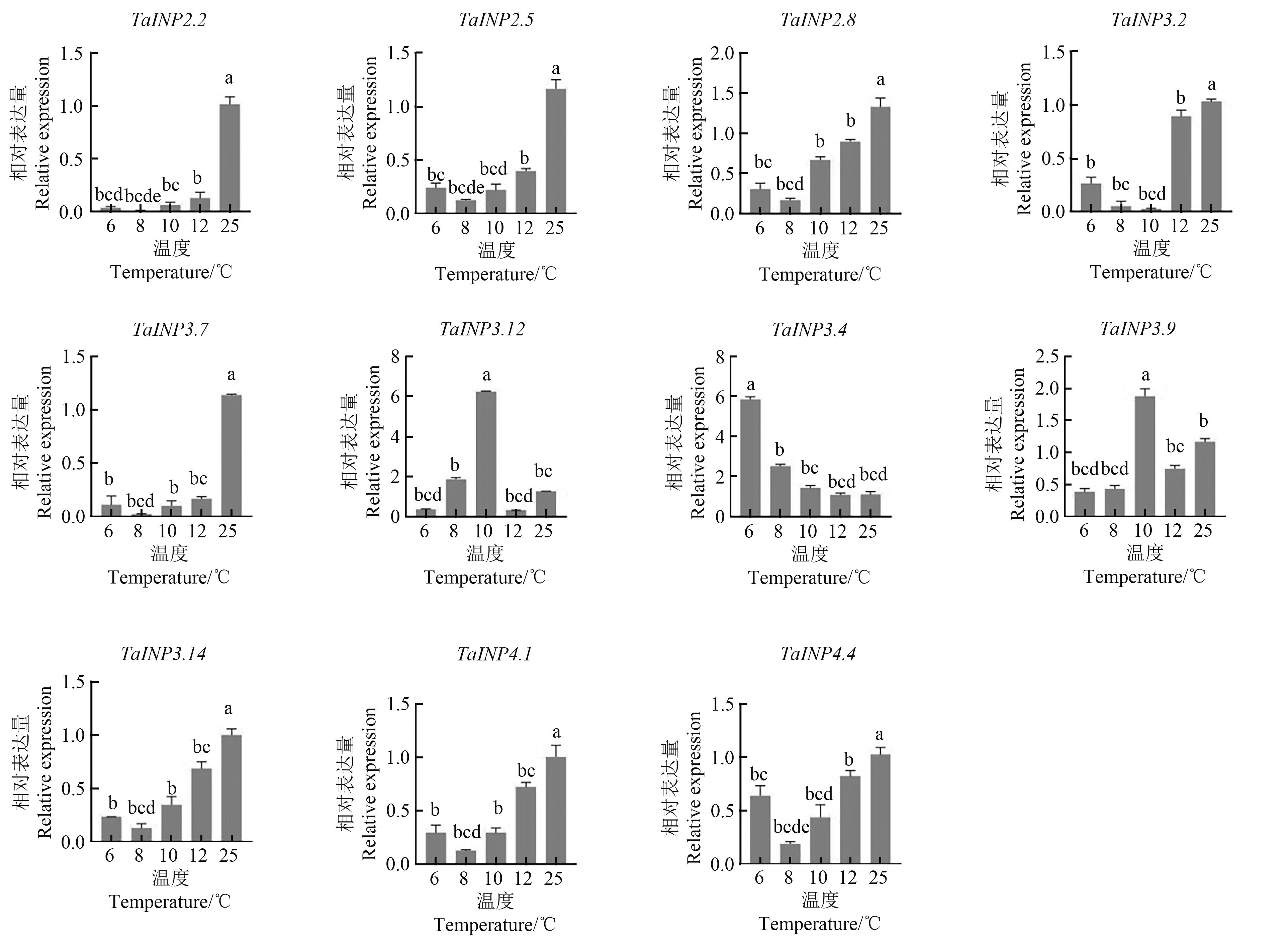
Fig. 6 Expression of TaINP1 genes in pollen under low temperature treatmentNote:Different lowercase letters indicate significant differences between different temperatures at P<0.05 level.
| 1 | HUANG J B, YANG L, YANG L, et al.. Stigma receptors control intraspecies and interspecies barriers in Brassicaceae [J].Nature, 2023,614(7947):303-308. |
| 2 | YU X B, ZHANG X C, ZHAO P, et al.. Fertilized egg cells secrete endopeptidases to avoid polytubey [J]. Nature, 2021,592:433-437. |
| 3 | ZHANG Z B, SUN M K, XIONG T, et al.. Development and genetic regulation of pollen intine in Arabidopsis and rice [J/OL].Gene, 2024,893:147936[2024-07-20].. |
| 4 | SHI J X, CUI M H, YANG L, et al.. Genetic and biochemical mechanisms of pollen wall development [J]. Trends Plant Sci., 2015,20(11):741-753. |
| 5 | LI F S, PHYO P, JACOBOWITZ J, et al.. The molecular structure of plant sporopollenin [J]. Nat. Plants, 2019,5(1):41-46. |
| 6 | QUILICHINI T D, GRIENENBERGER E, DOUGLAS C J. The biosynthesis,composition and assembly of the outer pollen wall:a tough case to crack [J]. Phytochemistry, 2015,113:170-182. |
| 7 | FURNESS C A, RUDALL P J. Pollen aperture evolution:a crucial factor for eudicot success? [J]. Trends Plant Sci., 2004,9(3):154-158. |
| 8 | EDLUND A F, SWANSON R, PREUSS D. Pollen and stigma structure and function:the role of diversity in pollination [J].Plant Cell, 2004,16(Sl):S84-S97. |
| 9 | ZHANG X, ZHAO G C, TAN Q, et al.. Rice pollen aperture formation is regulated by the interplay between OsINP1 and OsDAF1 [J]. Nat. Plants, 2020,6(4):394-403. |
| 10 | DOBRITSA A A, REEDER S H. Formation of pollen apertures in Arabidopsis requires an interplay between male meiosis,development of INP1-decorated plasma membrane domains,and the callose wall [J/OL]. Plant Signal. Behav., 2017,12(12):e1393136 [2024-07-20]. . |
| 11 | DOBRITSA A A, COERPER D. The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine [J]. Plant cell, 2012,24(11):4452-4464. |
| 12 | LI P, BEN-MENNI SCHULER S, REEDER S H, et al.. INP1 involvement in pollen aperture formation is evolutionarily conserved and may require species-specific partners [J]. J. Exp.Bot., 2018,69(5):983-996. |
| 13 | DOBRITSA A A, KIRKPATRICK A B, REEDER S H, et al.. Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition [J]. Plant Physiol., 2018,176(1):326-339. |
| 14 | DE SOUSA T, RIBEIRO M, SABENÇA C, et al.. The 10,000-year success story of wheat! [J/OL]. Foods, 2021,10(9):2124 [2024-07-20]. . |
| 15 | CARRILLO-BARRAL N, DEL CARMEN RODRÍGUEZ-GACIO M, MATILLA A J. Delay of germination-1 (DOG1):a key to understanding seed dormancy [J/OL]. Plants (Basel), 2020,9(4):480 [2024-07-20]. . |
| 16 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE,a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002,30(1):325-327. |
| 17 | ZHANG J P, QI Y N, WANG L M, et al.. Genomic comparison and population diversity analysis provide insights into the domestication and improvement of flax [J/OL]. iScience, 2020,23(4):100967 [2024-07-20]. . |
| 18 | SONG S Q, TANG C F, JIANG X C, et al.. Research progress on DOG1, a key regulator of seed dormancy and germination [J]. Plant Sci., 2024, 42(2): 254-266. |
| 19 | 崔荣秀,张议文,陈晓倩,等.植物bZIP参与胁迫应答调控的最新研究进展[J].生物技术通报,2019,35(2):143-155. |
| CUI R X, ZHANG Y W, CHEN X Q, et al.. The latest research progress on the stress responses of bZIP involved in plants [J].Biotechnol. Bull., 2019,35(2):143-155. | |
| 20 | 杜朝金,张汉尧,罗心平,等.基因调控植物花器官发育的研究进展[J].植物遗传资源学报,2024,25(2):151-161. |
| DU C J, ZHANG H R, LUO X P, et al.. Progress in gene regulation of plant floral organ development [J]. J. Plant Genetic Resour., 2024, 25(2):151-161 | |
| 21 | WANG Z P, ZHANG Z B, ZHENG D Y, et al.. Efficient and genotype independent maize transformation using pollen transfected by DNA-coated magnetic nanoparticles [J]. J. Integr. Plant Biol., 2022,64(6):1145-1156. |
| 22 | WILKINS M R, GASTEIGER E, BAIROCH A, et al.. Protein identification and analysis tools in the ExPASy server [J]. Methods Mol. Biol., 1999,112:531-552. |
| [1] | Zhenyu XUE, Kangkang ZHANG, Yuanyuan ZHANG, Qiangqiang YAN, Lirong YAO, Hong ZHANG, Yaxiong MENG, Erjing SI, Baochun LI, Xiaole MA, Huajun WANG, Juncheng WANG. Screening and Functional Gene Detection of High-quality and Drought-resistant Wheat Germplasms [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 35-49. |
| [2] | Xianyin SUN, Qiuhuan MU, Yong MI, Guangde LYU, Xiaolei QI, Yingying SUN, Xundong YIN, Ruixia WANG, Ke WU, Zhaoguo QIAN, Yan ZHAO, Minggang GAO. Classification and Evaluation of New Wheat Lines Based on GT Biplot [J]. Journal of Agricultural Science and Technology, 2024, 26(7): 14-24. |
| [3] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [4] | Yangyang DU, Yuanyuan BAO, Xiangyu LIU, Xinyong ZHANG. Effects of Tartary Buckwheat Rotation on Enzyme Activities and Microorganisms in Rhizosphere Soil of Cultivated Potato in Yunnan Province [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 192-200. |
| [5] | Gang ZHAO, Shuying WANG, Shangzhong LI, Jianjun ZHANG, Yi DANG, Lei WANG, Xingmao LI, Wanli CHENG, Gang ZHOU, Shengli NI, Tinglu FAN. Effects of Precipitation on Yield and Water Consumption of Winter Wheat in Loess Plateau in Recent 40 Years [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 164-173. |
| [6] | Hong ZHANG, Weiguo LI, Xiaodong ZHANG, Bihui LU, Chengcheng ZHANG, Wei LI, Tinghuai MA. Extraction of Winter Wheat Planting Area Based on Fusion Features of HJ-1 and GF-1 Image [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 109-119. |
| [7] | Jingyun ZHANG, Feng GUAN, Bo SHI, Xinjian WAN. Effects of Wheat Root Exudates on Bitter Gourd Seeding Growth and Soil Environment [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 181-190. |
| [8] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [9] | Yan JIN, Quanhao SONG, Jiajing SONG, Liang CHEN, Lishang ZHAO, Jie CHEN, Dong BAI, Tongquan ZHU. Comprehensive Evaluation of 69 Wheat Germplasm Resources [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 33-45. |
| [10] | Hao WANG, Bing FANG, Dapeng YE, Limin XIE. Measurement and Analysis of Dielectric Properties of Wheat Bran at Microwave Frequency [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 115-121. |
| [11] | Haixia LIU, Yinhui ZHANG, Lei ZHUANG, Mengjiao GUO, Li ZHAO, Meijuan WU, Jian HOU, Tian LI, Hongxia LIU, Xueyong ZHANG, Chenyang HAO. Discovering of Candidate Genes for Wheat SDS-Sedimentation Value Using Association Study and Development of KASP Marker [J]. Journal of Agricultural Science and Technology, 2024, 26(12): 18-29. |
| [12] | Zhaoyang YANG, Peng XU, Tiejian YUAN, Xiaoqiong LI, Donggen PENG, Zhentao ZHANG, Junling YANG, Chuangchuang DING, Jizhou ZHU. Study on Low Temperature Drying Characteristics and Quality of Hybrid Broussonetia papyrifera [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 157-170. |
| [13] | Peng TIAN, Jie SONG, Xia LI, Yuzhi WANG, Bangbang WU. Analysis of Folate Content and Its Derivatives in Grains of Wheat with Different Grain Colors [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 56-65. |
| [14] | Jieru YUE, Zhilie QIN, Qiling HOU, Shaohua YUAN, Xiaocong HAO, Jifang YANG, Xiucheng BAI, Changping ZHAO, Fengting ZHANG, Hui SUN. Study on Stigma Exsertion Pattern of BS Type Photo-thermo Sensitive Male Sterile Lines of Wheat [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 22-29. |
| [15] | Yunhong WANG, Qi MIAO, Junchao LI, Hongye WANG, Jishi ZHANG, Zhenling CUI. Effect of Comprehensive Management Measures on Productivity of Medium and Low Yield Farmland in Coastal Saline Areas [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 163-172. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 63
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 299
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
 京公网安备11010802021197号
京公网安备11010802021197号