




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (10): 83-97.DOI: 10.13304/j.nykjdb.2024.0365
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Zhenwei ZHANG1( ), Xiangshu DONG2, Jing YANG1, Xuejun LI1, Meijun QI1, Kuaile JIANG1, Yonglin YANG1, Butian WANG1, Xuedong SHI1, Junchao QIU1, Zhihua CHEN1, Yu GE1(
), Xiangshu DONG2, Jing YANG1, Xuejun LI1, Meijun QI1, Kuaile JIANG1, Yonglin YANG1, Butian WANG1, Xuedong SHI1, Junchao QIU1, Zhihua CHEN1, Yu GE1( )
)
Received:2024-05-08
Accepted:2024-06-21
Online:2024-10-15
Published:2024-10-18
Contact:
Yu GE
张振伟1( ), 董相书2, 杨婧1, 李学俊1, 杞美军1, 蒋快乐1, 杨永林1, 王步天1, 施学东1, 邱俊超1, 陈治华1, 葛宇1(
), 董相书2, 杨婧1, 李学俊1, 杞美军1, 蒋快乐1, 杨永林1, 王步天1, 施学东1, 邱俊超1, 陈治华1, 葛宇1( )
)
通讯作者:
葛宇
作者简介:张振伟E-mail:1010967244@qq.com;
基金资助:CLC Number:
Zhenwei ZHANG, Xiangshu DONG, Jing YANG, Xuejun LI, Meijun QI, Kuaile JIANG, Yonglin YANG, Butian WANG, Xuedong SHI, Junchao QIU, Zhihua CHEN, Yu GE. Genome-wide Identification and Expression Analysis of Key Chlorophyll Synthesis Related Gene CaPOR in Coffea arabica[J]. Journal of Agricultural Science and Technology, 2024, 26(10): 83-97.
张振伟, 董相书, 杨婧, 李学俊, 杞美军, 蒋快乐, 杨永林, 王步天, 施学东, 邱俊超, 陈治华, 葛宇. 小粒咖啡叶绿素合成基因CaPOR的全基因组鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(10): 83-97.
基因名称 Gene name | 正向引物序列 Forward primer sequence (5’-3’) | 反向引物序列 Revers primer sequence (5’-3’) |
|---|---|---|
| 24S | GCAGACAAGGCGGTGACTAT | CGGATCTTCTTGGCCCTGTT |
| CaPOR5 | GAAGTCACGCAGGGGATTGA | TCGAATGTGGTCCCCATGAC |
| CaPOR6 | TTAATTCGTCTGGCCGTCCC | GCTTTAACCGTCTGGCAAGC |
| CaPOR7 | GAAGTCACGCAGGGGATTGA | TCGAATGTGGTCCCCATGAC |
| CaPOR8 | TTAATTCGTCTGGCCGTCCC | GCTTTAACCGTCTGGCAAGC |
| CaPOR19 | GGCAGTGCGAAATGTGGATG | CGGAAAGGGCTGTAGCTTGA |
| CaPOR20 | GGCAGTGCGAAATGTGGATG | GCACGGAAAGGGCTTGATTG |
| CaPOR21 | TCAAGCTACAGCCCTTTCCG | CAGCACCCTGTGGAATGGAT |
Table 1 Primers of real-time fluorescence quantitative PCR amplification
基因名称 Gene name | 正向引物序列 Forward primer sequence (5’-3’) | 反向引物序列 Revers primer sequence (5’-3’) |
|---|---|---|
| 24S | GCAGACAAGGCGGTGACTAT | CGGATCTTCTTGGCCCTGTT |
| CaPOR5 | GAAGTCACGCAGGGGATTGA | TCGAATGTGGTCCCCATGAC |
| CaPOR6 | TTAATTCGTCTGGCCGTCCC | GCTTTAACCGTCTGGCAAGC |
| CaPOR7 | GAAGTCACGCAGGGGATTGA | TCGAATGTGGTCCCCATGAC |
| CaPOR8 | TTAATTCGTCTGGCCGTCCC | GCTTTAACCGTCTGGCAAGC |
| CaPOR19 | GGCAGTGCGAAATGTGGATG | CGGAAAGGGCTGTAGCTTGA |
| CaPOR20 | GGCAGTGCGAAATGTGGATG | GCACGGAAAGGGCTTGATTG |
| CaPOR21 | TCAAGCTACAGCCCTTTCCG | CAGCACCCTGTGGAATGGAT |
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaACSF1 | XP_027071950.1 | 428 | 49 460.89 | 9.07 | 40.60 | 75.98 | -0.340 |
| CaACSF2 | XP_027071221.1 | 427 | 49 387.84 | 8.91 | 38.47 | 76.39 | -0.332 |
| CaCAO1 | XP_027093472.1 | 535 | 60 461.69 | 8.48 | 41.80 | 85.07 | -0.279 |
| CaCAO2 | XP_027091395.1 | 535 | 60 451.67 | 8.14 | 42.68 | 85.25 | -0.267 |
Table 2 Physicochemical properties of protein encoded by genes related to chlorophyll synthesis of Coffea arabica
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaACSF1 | XP_027071950.1 | 428 | 49 460.89 | 9.07 | 40.60 | 75.98 | -0.340 |
| CaACSF2 | XP_027071221.1 | 427 | 49 387.84 | 8.91 | 38.47 | 76.39 | -0.332 |
| CaCAO1 | XP_027093472.1 | 535 | 60 461.69 | 8.48 | 41.80 | 85.07 | -0.279 |
| CaCAO2 | XP_027091395.1 | 535 | 60 451.67 | 8.14 | 42.68 | 85.25 | -0.267 |
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaCHLD1 | XP_027123386.1 | 763 | 83 861.49 | 6.18 | 46.55 | 88.64 | -0.324 |
| CaCHLD2 | XP_027079730.1 | 688 | 75 126.52 | 5.47 | 41.55 | 91.06 | -0.281 |
| CaCHLG1 | XP_027068575.1 | 380 | 40 758.37 | 7.58 | 22.00 | 111.16 | 0.386 |
| CaCHLG2 | XP_027072610.1 | 380 | 40 774.41 | 7.58 | 22.50 | 111.42 | 0.386 |
| CaCHLH1 | XP_027069536.1 | 1 382 | 153 535.80 | 5.74 | 39.12 | 88.81 | -0.271 |
| CaCHLH2 | XP_027069535.1 | 1 382 | 153 367.80 | 5.85 | 37.72 | 89.44 | -0.258 |
| CaCHLH3 | XP_027066034.1 | 1 155 | 127 594.20 | 6.53 | 38.64 | 92.68 | -0.180 |
| CaCHLI1 | XP_027078841.1 | 429 | 46 808.50 | 6.10 | 40.88 | 96.99 | -0.197 |
| CaCHLI2 | XP_027075032.1 | 429 | 46 886.54 | 6.10 | 42.42 | 96.55 | -0.198 |
| CaCHLM1 | XP_027123226.1 | 276 | 29 896.53 | 8.27 | 40.56 | 95.11 | -0.106 |
| CaCHLM2 | XP_027087377.1 | 321 | 34 625.96 | 8.57 | 38.44 | 90.90 | -0.058 |
| CaDVR1 | XP_027116070.1 | 366 | 40 327.44 | 5.26 | 24.61 | 93.20 | -0.061 |
| CaDVR2 | XP_027119467.1 | 415 | 45 457.33 | 6.21 | 25.54 | 89.98 | -0.061 |
| CaDVR3 | XP_027119468.1 | 415 | 45 562.42 | 7.52 | 26.73 | 88.82 | -0.092 |
| CaDVR4 | XP_027116071.1 | 366 | 40 320.50 | 5.74 | 24.87 | 92.16 | -0.067 |
| CaGSA1 | XP_027127512.1 | 477 | 50 838.38 | 5.91 | 33.78 | 81.82 | 0.020 |
| CaGSA2 | XP_027087467.1 | 477 | 50 854.42 | 5.83 | 32.56 | 83.04 | 0.032 |
| CaGSA3 | XP_027098002.1 | 479 | 52 160.00 | 8.74 | 31.67 | 90.19 | -0.096 |
| CaGSA4 | XP_027121611.1 | 479 | 52 206.09 | 8.70 | 30.44 | 90.19 | -0.090 |
| CaGSA5 | XP_027126106.1 | 441 | 48 043.70 | 8.26 | 25.68 | 90.00 | -0.008 |
| CaGSA6 | XP_027091382.1 | 484 | 52 975.94 | 8.27 | 32.52 | 88.04 | -0.093 |
| CaGSA7 | XP_027094060.1 | 523 | 57 122.38 | 6.84 | 50.47 | 87.00 | -0.115 |
| CaGSA8 | XP_027077773.1 | 475 | 52 482.79 | 8.13 | 37.84 | 91.20 | -0.136 |
| CaGSA9 | XP_027074140.1 | 475 | 52 471.77 | 8.44 | 38.41 | 90.17 | -0.152 |
| CaGSA10 | XP_027126874.1 | 468 | 51 593.81 | 8.31 | 38.51 | 91.30 | -0.129 |
| CaGSA11 | XP_027090984.1 | 430 | 46 240.96 | 7.18 | 37.03 | 91.91 | 0.036 |
| CaGSA12 | XP_027087761.1 | 457 | 49 018.23 | 6.57 | 38.55 | 96.50 | 0.039 |
| CaGSA13 | XP_027087466.1 | 461 | 49 307.69 | 6.09 | 37.98 | 100.09 | 0.133 |
| CaHEMA1 | XP_027112743.1 | 571 | 61 845.26 | 8.44 | 42.32 | 95.36 | -0.058 |
| CaHEMA2 | XP_027106141.1 | 571 | 61 889.21 | 8.44 | 42.85 | 94.85 | -0.077 |
| CaHEMA3 | XP_027089895.1 | 556 | 61 269.68 | 9.06 | 38.49 | 90.27 | -0.163 |
| CaHEMA4 | XP_027094467.1 | 555 | 61 104.53 | 9.20 | 38.37 | 90.43 | -0.178 |
| CaHEMB1 | XP_027081050.1 | 429 | 46 855.52 | 6.36 | 50.45 | 86.67 | -0.240 |
| CaHEMB2 | XP_027084484.1 | 429 | 46 697.38 | 6.36 | 50.45 | 85.78 | -0.239 |
| CaHEMC1 | XP_027061577.1 | 382 | 41 061.84 | 6.76 | 34.87 | 90.73 | -0.134 |
| CaHEMC2 | XP_027064519.1 | 383 | 41 150.93 | 6.80 | 33.88 | 91.51 | -0.127 |
| CaHEMD1 | XP_027089872.1 | 299 | 31 787.38 | 8.50 | 51.65 | 94.95 | 0.040 |
Table 2 Physicochemical properties of protein encoded by genes related to chlorophyll synthesis of Coffea arabica
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaCHLD1 | XP_027123386.1 | 763 | 83 861.49 | 6.18 | 46.55 | 88.64 | -0.324 |
| CaCHLD2 | XP_027079730.1 | 688 | 75 126.52 | 5.47 | 41.55 | 91.06 | -0.281 |
| CaCHLG1 | XP_027068575.1 | 380 | 40 758.37 | 7.58 | 22.00 | 111.16 | 0.386 |
| CaCHLG2 | XP_027072610.1 | 380 | 40 774.41 | 7.58 | 22.50 | 111.42 | 0.386 |
| CaCHLH1 | XP_027069536.1 | 1 382 | 153 535.80 | 5.74 | 39.12 | 88.81 | -0.271 |
| CaCHLH2 | XP_027069535.1 | 1 382 | 153 367.80 | 5.85 | 37.72 | 89.44 | -0.258 |
| CaCHLH3 | XP_027066034.1 | 1 155 | 127 594.20 | 6.53 | 38.64 | 92.68 | -0.180 |
| CaCHLI1 | XP_027078841.1 | 429 | 46 808.50 | 6.10 | 40.88 | 96.99 | -0.197 |
| CaCHLI2 | XP_027075032.1 | 429 | 46 886.54 | 6.10 | 42.42 | 96.55 | -0.198 |
| CaCHLM1 | XP_027123226.1 | 276 | 29 896.53 | 8.27 | 40.56 | 95.11 | -0.106 |
| CaCHLM2 | XP_027087377.1 | 321 | 34 625.96 | 8.57 | 38.44 | 90.90 | -0.058 |
| CaDVR1 | XP_027116070.1 | 366 | 40 327.44 | 5.26 | 24.61 | 93.20 | -0.061 |
| CaDVR2 | XP_027119467.1 | 415 | 45 457.33 | 6.21 | 25.54 | 89.98 | -0.061 |
| CaDVR3 | XP_027119468.1 | 415 | 45 562.42 | 7.52 | 26.73 | 88.82 | -0.092 |
| CaDVR4 | XP_027116071.1 | 366 | 40 320.50 | 5.74 | 24.87 | 92.16 | -0.067 |
| CaGSA1 | XP_027127512.1 | 477 | 50 838.38 | 5.91 | 33.78 | 81.82 | 0.020 |
| CaGSA2 | XP_027087467.1 | 477 | 50 854.42 | 5.83 | 32.56 | 83.04 | 0.032 |
| CaGSA3 | XP_027098002.1 | 479 | 52 160.00 | 8.74 | 31.67 | 90.19 | -0.096 |
| CaGSA4 | XP_027121611.1 | 479 | 52 206.09 | 8.70 | 30.44 | 90.19 | -0.090 |
| CaGSA5 | XP_027126106.1 | 441 | 48 043.70 | 8.26 | 25.68 | 90.00 | -0.008 |
| CaGSA6 | XP_027091382.1 | 484 | 52 975.94 | 8.27 | 32.52 | 88.04 | -0.093 |
| CaGSA7 | XP_027094060.1 | 523 | 57 122.38 | 6.84 | 50.47 | 87.00 | -0.115 |
| CaGSA8 | XP_027077773.1 | 475 | 52 482.79 | 8.13 | 37.84 | 91.20 | -0.136 |
| CaGSA9 | XP_027074140.1 | 475 | 52 471.77 | 8.44 | 38.41 | 90.17 | -0.152 |
| CaGSA10 | XP_027126874.1 | 468 | 51 593.81 | 8.31 | 38.51 | 91.30 | -0.129 |
| CaGSA11 | XP_027090984.1 | 430 | 46 240.96 | 7.18 | 37.03 | 91.91 | 0.036 |
| CaGSA12 | XP_027087761.1 | 457 | 49 018.23 | 6.57 | 38.55 | 96.50 | 0.039 |
| CaGSA13 | XP_027087466.1 | 461 | 49 307.69 | 6.09 | 37.98 | 100.09 | 0.133 |
| CaHEMA1 | XP_027112743.1 | 571 | 61 845.26 | 8.44 | 42.32 | 95.36 | -0.058 |
| CaHEMA2 | XP_027106141.1 | 571 | 61 889.21 | 8.44 | 42.85 | 94.85 | -0.077 |
| CaHEMA3 | XP_027089895.1 | 556 | 61 269.68 | 9.06 | 38.49 | 90.27 | -0.163 |
| CaHEMA4 | XP_027094467.1 | 555 | 61 104.53 | 9.20 | 38.37 | 90.43 | -0.178 |
| CaHEMB1 | XP_027081050.1 | 429 | 46 855.52 | 6.36 | 50.45 | 86.67 | -0.240 |
| CaHEMB2 | XP_027084484.1 | 429 | 46 697.38 | 6.36 | 50.45 | 85.78 | -0.239 |
| CaHEMC1 | XP_027061577.1 | 382 | 41 061.84 | 6.76 | 34.87 | 90.73 | -0.134 |
| CaHEMC2 | XP_027064519.1 | 383 | 41 150.93 | 6.80 | 33.88 | 91.51 | -0.127 |
| CaHEMD1 | XP_027089872.1 | 299 | 31 787.38 | 8.50 | 51.65 | 94.95 | 0.040 |
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaHEMD2 | XP_027095285.1 | 298 | 31 672.25 | 8.50 | 51.43 | 94.63 | 0.035 |
| CaHEMD3 | XP_027095286.1 | 210 | 22 397.50 | 5.11 | 39.93 | 94.29 | 0.052 |
| CaHEMD4 | XP_027089873.1 | 210 | 22 425.56 | 5.11 | 39.52 | 95.19 | 0.064 |
| CaHEME1.1 | XP_027062275.1 | 417 | 46 084.19 | 8.15 | 36.79 | 92.78 | -0.067 |
| CaHEME1.2 | XP_027065112.1 | 417 | 46 138.19 | 7.71 | 39.01 | 92.78 | -0.063 |
| CaHEME2.1 | XP_027104195.1 | 322 | 35 712.15 | 6.34 | 35.46 | 98.04 | -0.028 |
| CaHEME2.2 | XP_027110652.1 | 322 | 35 698.13 | 6.34 | 36.83 | 98.04 | -0.028 |
| CaHEME2.3 | XP_027110651.1 | 391 | 43 011.53 | 7.66 | 39.26 | 95.93 | 0.005 |
| CaHEME2.4 | XP_027104194.1 | 391 | 43 021.57 | 7.66 | 38.06 | 95.68 | 0.002 |
| CaHEMF | XP_027089153.1 | 414 | 45 918.50 | 6.43 | 60.99 | 65.34 | -0.453 |
| CaHEMG1.1 | XP_027099809.1 | 537 | 57 776.53 | 9.18 | 34.91 | 89.70 | -0.055 |
| CaHEMG1.2 | XP_027118697.1 | 537 | 57 869.83 | 9.30 | 35.53 | 91.34 | -0.085 |
| CaHEMG2.1 | XP_027102156.1 | 547 | 60 822.73 | 9.27 | 44.05 | 80.59 | -0.247 |
| CaPOR1 | XP_027103747.1 | 315 | 34 094.78 | 9.30 | 30.85 | 85.81 | -0.223 |
| CaPOR2 | XP_027116958.1 | 316 | 34 222.91 | 9.30 | 32.14 | 85.54 | -0.233 |
| CaPOR3 | XP_027116956.1 | 316 | 34 210.94 | 9.21 | 31.21 | 87.69 | -0.192 |
| CaPOR4 | XP_027109629.1 | 315 | 34 082.81 | 9.21 | 29.92 | 87.97 | -0.181 |
| CaPOR5 | XP_027109627.1 | 317 | 34 699.37 | 8.64 | 25.94 | 87.10 | -0.232 |
| CaPOR6 | XP_027109628.1 | 318 | 34 827.50 | 8.64 | 27.24 | 86.82 | -0.242 |
| CaPOR7 | XP_027100939.1 | 317 | 34 683.31 | 8.72 | 26.03 | 87.10 | -0.242 |
| CaPOR8 | XP_027103745.1 | 318 | 34 811.44 | 8.72 | 27.33 | 86.82 | -0.253 |
| CaPOR9 | XP_027080713.1 | 281 | 30 655.05 | 9.72 | 22.66 | 89.82 | -0.058 |
| CaPOR10 | XP_027080712.1 | 320 | 34 887.64 | 8.53 | 21.52 | 87.41 | -0.120 |
| CaPOR11 | XP_027080711.1 | 320 | 34 977.81 | 8.53 | 22.18 | 88.94 | -0.087 |
| CaPOR12 | XP_027103746.1 | 328 | 35 973.78 | 7.15 | 29.99 | 86.31 | -0.133 |
| CaPOR13 | XP_027081909.1 | 327 | 35 741.75 | 8.70 | 28.48 | 88.65 | -0.111 |
| CaPOR14 | XP_027085131.1 | 319 | 34 875.72 | 8.93 | 29.78 | 86.90 | -0.150 |
| CaPOR15 | XP_027071003.1 | 319 | 34 998.86 | 9.03 | 29.99 | 86.90 | -0.177 |
| CaPOR16 | XP_027071002.1 | 298 | 32 852.52 | 9.60 | 33.56 | 96.24 | -0.215 |
| CaPOR17 | XP_027122441.1 | 257 | 27 948.33 | 8.43 | 26.40 | 109.26 | 0.178 |
| CaPOR18 | XP_027122442.1 | 315 | 33 872.97 | 8.40 | 24.28 | 104.00 | 0.125 |
| CaPOR19 | XP_027082984.1 | 259 | 28 369.46 | 8.86 | 40.58 | 84.32 | -0.171 |
| CaPOR20 | XP_027082982.1 | 314 | 34 362.38 | 8.63 | 35.92 | 88.82 | -0.162 |
| CaPOR21 | XP_027082983.1 | 316 | 34 612.63 | 8.62 | 35.76 | 88.26 | -0.168 |
| CaPOR22 | XP_027082980.1 | 320 | 34 901.06 | 9.21 | 30.59 | 96.41 | -0.034 |
| CaPOR23 | XP_027110265.1 | 347 | 38 111.97 | 9.27 | 28.13 | 100.98 | 0.018 |
| CaPOR24 | XP_027079820.1 | 366 | 40 399.48 | 9.08 | 42.62 | 95.49 | -0.064 |
Table 2 Physicochemical properties of protein encoded by genes related to chlorophyll synthesis of Coffea arabica
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaHEMD2 | XP_027095285.1 | 298 | 31 672.25 | 8.50 | 51.43 | 94.63 | 0.035 |
| CaHEMD3 | XP_027095286.1 | 210 | 22 397.50 | 5.11 | 39.93 | 94.29 | 0.052 |
| CaHEMD4 | XP_027089873.1 | 210 | 22 425.56 | 5.11 | 39.52 | 95.19 | 0.064 |
| CaHEME1.1 | XP_027062275.1 | 417 | 46 084.19 | 8.15 | 36.79 | 92.78 | -0.067 |
| CaHEME1.2 | XP_027065112.1 | 417 | 46 138.19 | 7.71 | 39.01 | 92.78 | -0.063 |
| CaHEME2.1 | XP_027104195.1 | 322 | 35 712.15 | 6.34 | 35.46 | 98.04 | -0.028 |
| CaHEME2.2 | XP_027110652.1 | 322 | 35 698.13 | 6.34 | 36.83 | 98.04 | -0.028 |
| CaHEME2.3 | XP_027110651.1 | 391 | 43 011.53 | 7.66 | 39.26 | 95.93 | 0.005 |
| CaHEME2.4 | XP_027104194.1 | 391 | 43 021.57 | 7.66 | 38.06 | 95.68 | 0.002 |
| CaHEMF | XP_027089153.1 | 414 | 45 918.50 | 6.43 | 60.99 | 65.34 | -0.453 |
| CaHEMG1.1 | XP_027099809.1 | 537 | 57 776.53 | 9.18 | 34.91 | 89.70 | -0.055 |
| CaHEMG1.2 | XP_027118697.1 | 537 | 57 869.83 | 9.30 | 35.53 | 91.34 | -0.085 |
| CaHEMG2.1 | XP_027102156.1 | 547 | 60 822.73 | 9.27 | 44.05 | 80.59 | -0.247 |
| CaPOR1 | XP_027103747.1 | 315 | 34 094.78 | 9.30 | 30.85 | 85.81 | -0.223 |
| CaPOR2 | XP_027116958.1 | 316 | 34 222.91 | 9.30 | 32.14 | 85.54 | -0.233 |
| CaPOR3 | XP_027116956.1 | 316 | 34 210.94 | 9.21 | 31.21 | 87.69 | -0.192 |
| CaPOR4 | XP_027109629.1 | 315 | 34 082.81 | 9.21 | 29.92 | 87.97 | -0.181 |
| CaPOR5 | XP_027109627.1 | 317 | 34 699.37 | 8.64 | 25.94 | 87.10 | -0.232 |
| CaPOR6 | XP_027109628.1 | 318 | 34 827.50 | 8.64 | 27.24 | 86.82 | -0.242 |
| CaPOR7 | XP_027100939.1 | 317 | 34 683.31 | 8.72 | 26.03 | 87.10 | -0.242 |
| CaPOR8 | XP_027103745.1 | 318 | 34 811.44 | 8.72 | 27.33 | 86.82 | -0.253 |
| CaPOR9 | XP_027080713.1 | 281 | 30 655.05 | 9.72 | 22.66 | 89.82 | -0.058 |
| CaPOR10 | XP_027080712.1 | 320 | 34 887.64 | 8.53 | 21.52 | 87.41 | -0.120 |
| CaPOR11 | XP_027080711.1 | 320 | 34 977.81 | 8.53 | 22.18 | 88.94 | -0.087 |
| CaPOR12 | XP_027103746.1 | 328 | 35 973.78 | 7.15 | 29.99 | 86.31 | -0.133 |
| CaPOR13 | XP_027081909.1 | 327 | 35 741.75 | 8.70 | 28.48 | 88.65 | -0.111 |
| CaPOR14 | XP_027085131.1 | 319 | 34 875.72 | 8.93 | 29.78 | 86.90 | -0.150 |
| CaPOR15 | XP_027071003.1 | 319 | 34 998.86 | 9.03 | 29.99 | 86.90 | -0.177 |
| CaPOR16 | XP_027071002.1 | 298 | 32 852.52 | 9.60 | 33.56 | 96.24 | -0.215 |
| CaPOR17 | XP_027122441.1 | 257 | 27 948.33 | 8.43 | 26.40 | 109.26 | 0.178 |
| CaPOR18 | XP_027122442.1 | 315 | 33 872.97 | 8.40 | 24.28 | 104.00 | 0.125 |
| CaPOR19 | XP_027082984.1 | 259 | 28 369.46 | 8.86 | 40.58 | 84.32 | -0.171 |
| CaPOR20 | XP_027082982.1 | 314 | 34 362.38 | 8.63 | 35.92 | 88.82 | -0.162 |
| CaPOR21 | XP_027082983.1 | 316 | 34 612.63 | 8.62 | 35.76 | 88.26 | -0.168 |
| CaPOR22 | XP_027082980.1 | 320 | 34 901.06 | 9.21 | 30.59 | 96.41 | -0.034 |
| CaPOR23 | XP_027110265.1 | 347 | 38 111.97 | 9.27 | 28.13 | 100.98 | 0.018 |
| CaPOR24 | XP_027079820.1 | 366 | 40 399.48 | 9.08 | 42.62 | 95.49 | -0.064 |
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaPOR25 | XP_027079819.1 | 365 | 40 271.35 | 9.08 | 41.53 | 95.75 | -0.054 |
| CaPOR26 | XP_027082977.1 | 283 | 31 052.72 | 9.44 | 35.50 | 98.59 | 0.010 |
| CaPOR27 | XP_027082976.1 | 324 | 35 711.92 | 9.18 | 40.51 | 91.51 | -0.087 |
| CaPOR28 | XP_027082979.1 | 276 | 30 315.94 | 9.41 | 34.75 | 98.26 | 0.011 |
| CaPOR29 | XP_027082978.1 | 300 | 33 206.13 | 9.56 | 42.10 | 96.90 | -0.054 |
| CaPOR30 | XP_027121849.1 | 348 | 37 884.57 | 9.44 | 30.21 | 96.75 | -0.021 |
| CaPOR31 | XP_027125881.1 | 341 | 37 129.59 | 9.39 | 31.24 | 94.46 | 0.018 |
| CaPOR32 | XP_027122872.1 | 342 | 37 258.71 | 9.30 | 32.13 | 94.18 | 0.007 |
| CaPOR33 | XP_027110266.1 | 271 | 29 703.21 | 9.20 | 37.08 | 100.07 | 0.030 |
| CaPOR34 | XP_027110268.1 | 329 | 35 780.08 | 9.21 | 32.38 | 95.53 | 0.034 |
| CaPOR35 | XP_027103749.1 | 330 | 35 909.20 | 9.10 | 33.30 | 95.24 | 0.024 |
| CaPOR36 | XP_027103748.1 | 369 | 40 863.41 | 9.21 | 37.16 | 94.88 | -0.094 |
| CaPOR37 | XP_027110267.1 | 407 | 45 157.59 | 5.70 | 33.75 | 91.57 | -0.186 |
| CaPOR38 | XP_027116957.1 | 409 | 45 329.77 | 5.70 | 33.63 | 91.37 | -0.183 |
Table 2 Physicochemical properties of protein encoded by genes related to chlorophyll synthesis of Coffea arabica
基因名称 Gene name | 蛋白序列ID Protein sequence ID | 氨基酸数量 Number of amino acid/aa | 相对分子量 Molecular weight/Da | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| CaPOR25 | XP_027079819.1 | 365 | 40 271.35 | 9.08 | 41.53 | 95.75 | -0.054 |
| CaPOR26 | XP_027082977.1 | 283 | 31 052.72 | 9.44 | 35.50 | 98.59 | 0.010 |
| CaPOR27 | XP_027082976.1 | 324 | 35 711.92 | 9.18 | 40.51 | 91.51 | -0.087 |
| CaPOR28 | XP_027082979.1 | 276 | 30 315.94 | 9.41 | 34.75 | 98.26 | 0.011 |
| CaPOR29 | XP_027082978.1 | 300 | 33 206.13 | 9.56 | 42.10 | 96.90 | -0.054 |
| CaPOR30 | XP_027121849.1 | 348 | 37 884.57 | 9.44 | 30.21 | 96.75 | -0.021 |
| CaPOR31 | XP_027125881.1 | 341 | 37 129.59 | 9.39 | 31.24 | 94.46 | 0.018 |
| CaPOR32 | XP_027122872.1 | 342 | 37 258.71 | 9.30 | 32.13 | 94.18 | 0.007 |
| CaPOR33 | XP_027110266.1 | 271 | 29 703.21 | 9.20 | 37.08 | 100.07 | 0.030 |
| CaPOR34 | XP_027110268.1 | 329 | 35 780.08 | 9.21 | 32.38 | 95.53 | 0.034 |
| CaPOR35 | XP_027103749.1 | 330 | 35 909.20 | 9.10 | 33.30 | 95.24 | 0.024 |
| CaPOR36 | XP_027103748.1 | 369 | 40 863.41 | 9.21 | 37.16 | 94.88 | -0.094 |
| CaPOR37 | XP_027110267.1 | 407 | 45 157.59 | 5.70 | 33.75 | 91.57 | -0.186 |
| CaPOR38 | XP_027116957.1 | 409 | 45 329.77 | 5.70 | 33.63 | 91.37 | -0.183 |
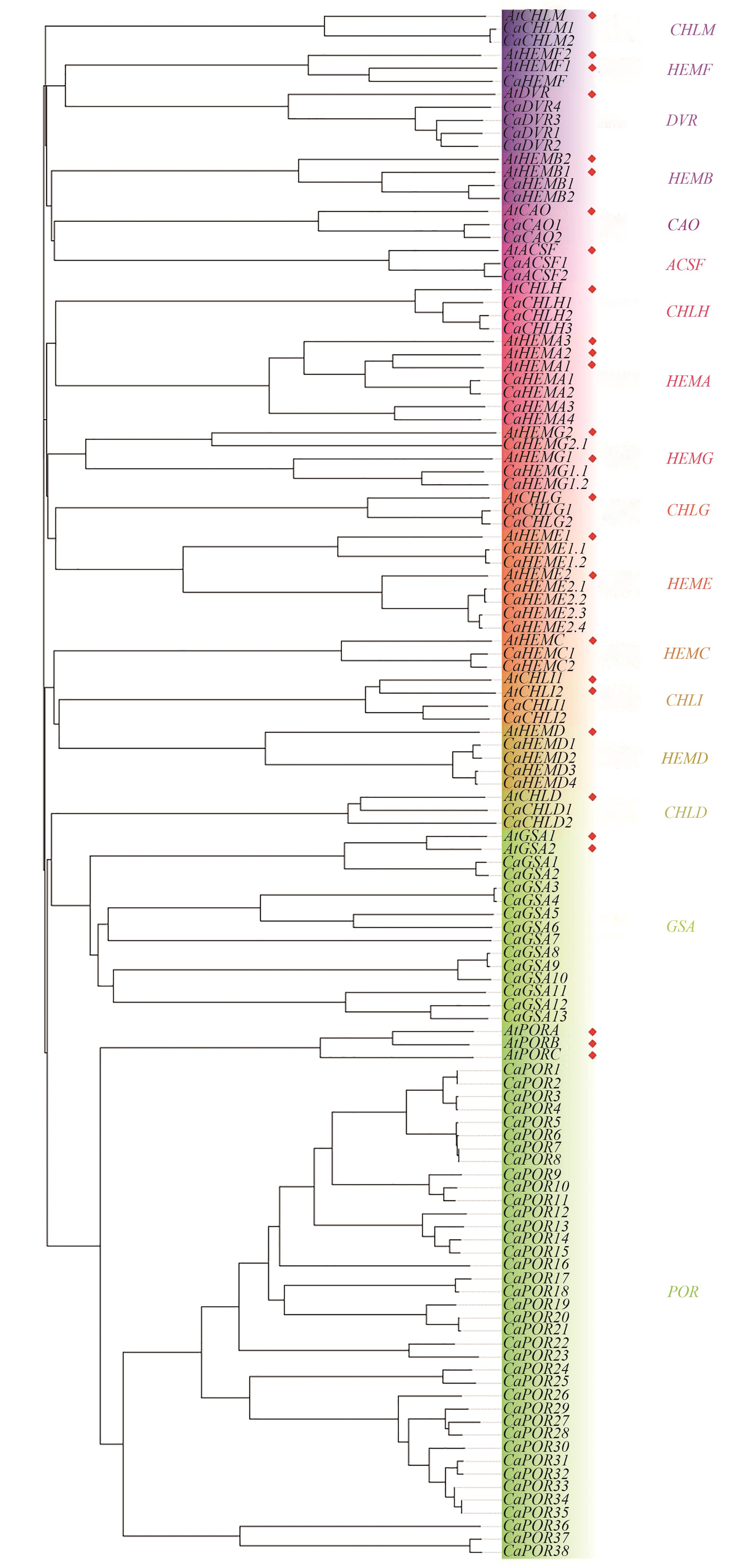
Fig. 1 Phylogenetic trees of genes related to chlorophyll synthesis of Arabidopsis thalian and Coffea arabicaNote:At—Arabidopsis thaliana; Ca—Coffea arabica; red diamond marks are genes from Arabidopsis thaliana; different colours represent different genes family.
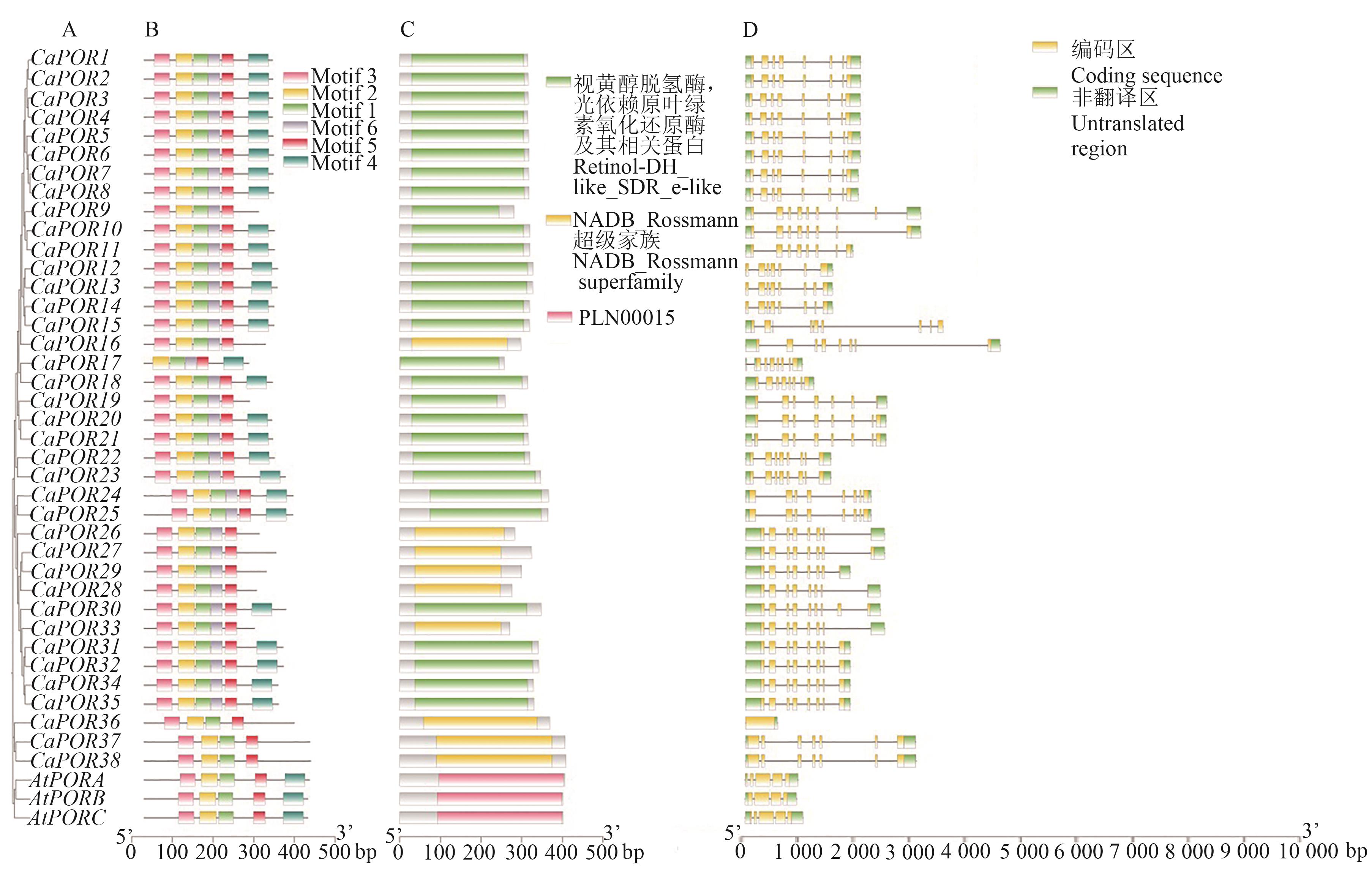
Fig. 4 Conserved motif, domain and gene structure of POR gene families in Coffea arabica and Arabidopsis thalianaA: Phylogenetic tree; B: Conserved motif; C: Domains; D: Gene structure
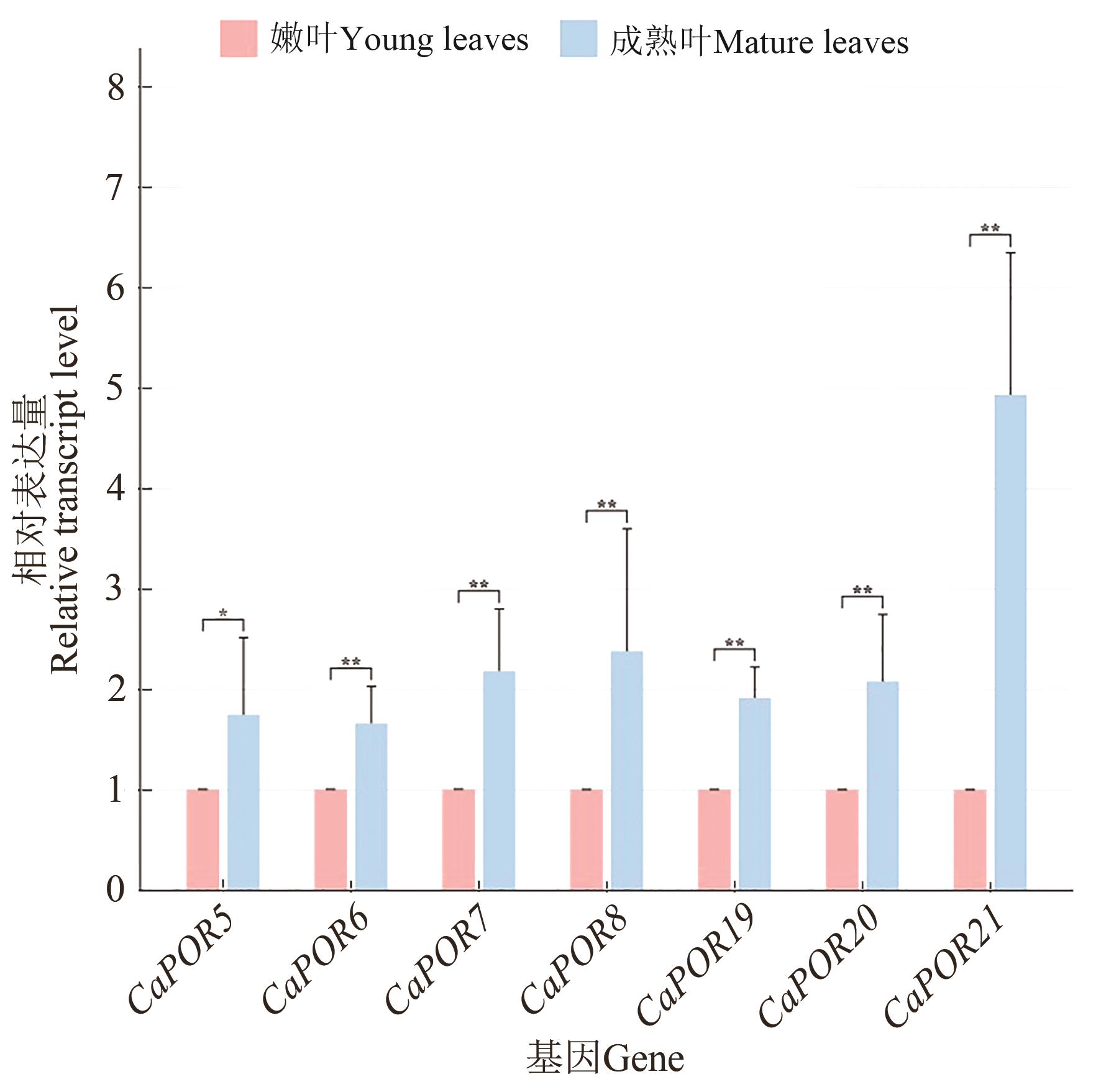
Fig. 12 Relative expression level of CaPOR gene in young and mature leaves of Coffea arabicaNote:* and ** indicate significant difference at P<0.05 and P<0.01 levels, respectively.
| 1 | 闫林,黄丽芳,王晓阳,等.基于ISSR标记的咖啡资源遗传多样性分析[J].热带作物学报,2019,40(2):300-307. |
| YAN L, HUANG L F, WANG X Y,et al..Genetic diversity of coffee germplasms by ISSR markers [J].Chin.J.Trop.Crops,2019,40(2):300-307. | |
| 2 | GE Y, WANG B, SHI X, et al.. Multi-omics analyses unravel genetic relationship of Chinese coffee germplasm resources [J/OL]. Forests, 2024, 15(1):163 [2024-04-10]. . |
| 3 | 闫林,黄丽芳,王晓阳,等.咖啡种质资源遗传多样性的ISSR分析[J].南方农业学报,2019,50(3):491-499.. |
| YAN L, HUANG L F, WANG X Y, et al.. Genetic diversity of coffee gemeplasms by ISSR analysis [J]. J. South. Agric., 2019, 50(3):491-499. | |
| 4 | 胡荣锁,董文江,宗迎,等.5个产区咖啡果皮成分分析与营养评价[J].热带作物学报,2018,39(5):987-992. |
| HU R S, DONG W J, ZONG Y, et al.. Analysis and evaluation of nutritional components of coffee peel from five different growing regions [J]. Chin. J. Trop. Crops, 2018, 39(5):987-992. | |
| 5 | 张修德.苹果叶绿素合成关键酶基因MdHEMA1克隆和功能验证[D].北京:中国农业科学院,2017. |
| ZHANG X D. Gene cloning and functional identification of MdHEMA1 encoding the key enzyme for chlorophyll biosynthesis in apple [D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. | |
| 6 | 王平荣,张帆涛,高家旭,等.高等植物叶绿素生物合成的研究进展[J].西北植物学报,2009,29(3):629-636. |
| WANG P R, ZHANG F T, GAO J X,et al..An overview of chlorophyll biosynthesis in higher plants [J].Acta Bot.Bor-Occid. Sin., 2009,29(3):629-636. | |
| 7 | 李佳佳,于旭东,蔡泽坪,等.高等植物叶绿素生物合成研究进展[J].分子植物育种,2019,17(18):6013-6019. |
| LI J J, YU X D, CAI Z P,et al..An overview of chlorophyll biosynthesis in higher plants [J]. Mol. Plant Breed.,2019,17(18):6013-6019. | |
| 8 | BEALE S I.Green genes gleaned [J].Trends Plant Sci.,2005,10(7):309-312. |
| 9 | ZENG Z Q, LIN T Z, ZHAO J Y, et al.. OsHemA gene,encoding glutamyl-tRNA reductase (GluTR) is essential for chlorophyll biosynthesis in rice (Oryza sativa) [J].J.Integr.Agric.,2020,19(3):612-623. |
| 10 | XU D D, SUN D, DIAO Y L, et al.. Fast mapping of a chlorophyll b synthesis-deficiency gene in barley (Hordeum vulgare L.) via bulked-segregant analysis with reduced-representation sequencing [J]. Crop J., 2019,7(1):58-64. |
| 11 | GABRUK M, MYSLIWA-KURDZIEL B.Light-dependent protochlorophyllide oxidoreductase:phylogeny,regulation,and catalytic properties [J]. Biochemistry, 2015, 54(34):5255-5262. |
| 12 | BUHR F, LAHROUSSI A, SPRINGER A,et al..NADPH:protochlorophyllide oxidoreductase B (PORB) action in Arabidopsis thaliana revisited through transgenic expression of engineered barley PORB mutant proteins [J]. Plant Mol. Biol., 2017,94(1):45-59. |
| 13 | LIANG M T, GU D C, LIE Z Y,et al..Regulation of chlorophyll biosynthesis by light-dependent acetylation of NADPH:protochlorophyll oxidoreductase A in Arabidopsis [J/OL].Plant Sci.,2023,330:111641 [2024-04-10]. . |
| 14 | ZHANG J, SUI C, LIU H,et al..Effect of chlorophyll biosynthesis-related genes on the leaf color in Hosta (Hosta plantaginea Aschers) and tobacco (Nicotiana tabacum L.) [J/OL].BMC Plant Biol.,2021,21(1):45 [2024-04-10]. . |
| 15 | CHEN J, WU S, DONG F,et al..Mechanism underlying the shading-induced chlorophyll accumulation in tea leaves [J/OL].Plant Sci.,2021,12:779819 [2024-04-10]. . |
| 16 | TIAN Y Y, WANG H Y, ZHANG Z Q,et al..An RNA-seq analysis reveals differential transcriptional responses to different light qualities in leaf color of Camellia sinensis cv.huangjinya [J]. J. Plant Growth Regul.,2022,41(2):612-627. |
| 17 | ABDELAZIZ M E, ATIA M A M, ABDELSATTAR M, et al.. Unravelling the role of piriformospora indica in combating water deficiency by modulating physiological performance and chlorophyll metabolism-related genes in cucumis sativus [J/OL]. Horticulturae, 2021, 7(10):399 [2024-04-10]. . |
| 18 | 李亚麒,严炜,娄予强,等.高产小粒咖啡叶绿体基因组密码子偏好性分析[J].南方农业学报,2023,54(8):2330-2339. |
| LI Y Q, YAN W, LOU Y Q,et al.. Codon usage bias in the chloroplast genome of high production Coffea arabica L. [J].J.South. Agric., 2023,54(8):2330-2339. | |
| 19 | CHEN C J, WU Y, LI J W,et al..TBtools-II:a “one for all,all for one” bioinformatics platform for biological big-data mining [J].Mol. Plant, 2023,16(11):1733-1742. |
| 20 | XIE J M, CHEN Y R, CAI G J,et al..Tree visualization by one table (tvBOT):a web application for visualizing,modifying and annotating phylogenetic trees [J].Nucl. Acids Res.,2023,51(1):587-592. |
| 21 | 郭睿,李汶东,陈大福,等.意大利蜜蜂幼虫肠道内球囊菌及其纯培养的高表达基因差异分析[J].微生物学通报,2018,45(2):368-375. |
| GUO R, LI W D, CHEN D F,et al..Highly-expressed gene differences between Ascosphaera apis stressing the larval gut of Apis mellifera ligustica and the pure culture of Ascosphaera apis [J].Microbiol. China,2018,45(2):368-375. | |
| 22 | MARCHLER-BAUER A, BO Y, HAN L,et al..CDD/SPARCLE:functional classification of proteins via subfamily domain architectures [J]. Nucl. Acids Res., 2017,45(1):200-203. |
| 23 | LU S, WANG J, CHITSAZ F,et al..CDD/SPARCLE:the conserved domain database in 2020 [J]. Nucl. Acids Res., 2020,48(1):265-268. |
| 24 | WANG J Y, CHITSAZ F, DERBYSHIRE M K,et al..The conserved domain database in 2023 [J]. Nucl. Acids Res., 2023,51(1):384-388. |
| 25 | LESCOT M, DÉHAIS P, THIJS G,et al..PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucl. Acids Res.,2002,30(1):325-327. |
| 26 | 黄思琦,张麒功,叶泽霖,等.5种柏科植物叶绿体基因组密码子偏好性分析[J].福建农林大学学报(自然科学版),2024,53(2):214-220. |
| HUANG S Q, ZHANG Q G, YE Z L, et al.. Codon bias analysis of chloroplast genomes of 5 Cupressaceae plants [J]. J. Fujian Agric. For. Univ. (Nat. Sci.), 2024, 53(2):214-220. | |
| 27 | 毛立彦,黄秋伟,龙凌云,等.7种睡莲属植物叶绿体基因组密码子偏好性分析[J].西北林学院学报,2022,37(2):98-107. |
| MAO L Y, HUANG Q W, LONG L Y,et al..Comparative analysis of codon usage bias in chloroplast genomes of seven Nymphaea species [J]. J. Northwest For. Univ., 2022,37(2):98-107. | |
| 28 | 罗永坚,王茹,赵仁菲,等.珙桐叶绿体基因组同义密码子使用偏好性分析[J].北京林业大学学报,2024,46(3):8-16. |
| LUO Y J, WANG R, ZHAO R F, et al.. Analysis of synonymous codon usage bias in the chloroplast genome of Davidia involucrata [J]. J. Beijing For. Univ., 2024, 46(3):8-16. | |
| 29 | FREITAS N C, BARRETO H G, FERNANDES-BRUM C N,et al..Validation of reference genes for qPCR analysis of Coffea arabica L.somatic embryogenesis-related tissues [J].Plant Cell Tissue Organ Cult. (PCTOC), 2017,128(3):663-678. |
| 30 | AROCHO A, CHEN B, LADANYI M,et al..Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts [J].Diagn. Mol. Pathol., 2006,15(1):56-61. |
| 31 | NIE L, ZHENG Y, ZHANG L,et al..Characterization and transcriptomic analysis of a novel yellow-green leaf Wucai (Brassica campestris L.) germplasm [J/OL].BMC Genomics,2021,22(1):258 [2024-04-10]. . |
| 32 | NGUYEN M K, SHIH T H, LIN S H, et al.. Transcription profile analysis of chlorophyll biosynthesis in leaves of wild-type and chlorophyll b-deficient rice (Oryza sativa L.) [J/OL]. Agriculture, 2021, 11(5):401 [2024-04-10]. . |
| 33 | KIM D H, LIM S H, LEE J Y.Expression of RsPORB is associated with radish root color [J/OL].Plants (Basel),2023,12(11):2214 [2024-04-10]. . |
| 34 | SAKURABA Y, RAHMAN M L, CHO S H,et al..The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions [J]. Plant J., 2013,74(1):122-133. |
| 35 | LI C F, XU Y X, MA J Q,et al..Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis) [J/OL].BMC Plant Biol.,2016,16(1):195 [2024-04-10]. . |
| 36 | 张巧丽,陈笛,宋艳萍,等.番茄果实叶绿素代谢转录调控网络研究进展[J].园艺学报,2023,50(9):2031-2047. |
| ZHANG Q L, CHEN D, SONG Y P,et al..Review on transcriptional regulation of chlorophyll metabolism network in tomato fruits [J]. Acta Hortic. Sin., 2023,50(9):2031-2047. | |
| 37 | ZHAO Y, XIE Q, YANG Q,et al.. Genome-wide identification and evolutionary analysis of the NRAMP gene family in the AC genomes of Brassica species [J/OL].BMC Plant Biol.,2024,24(1):311 [2024-04-10]. . |
| 38 | LI D, LI H, FENG H,et al.. Unveiling kiwifruit TCP genes:evolution,functions,and expression insights [J/OL].Plant Signal Behav.,2024,19(1):2338985 [2024-04-10]. . |
| 39 | 常箫月,王晨舒,李维蛟,等.滇重楼WRKY基因家族的鉴定及密码子偏好性分析[J/OL].分子植物育种,2024:1-11[2024-04-10].. |
| CHANG X Y, WANG C S, LI W J, et al.. Identification of WRKY gene family and analysis of codon bias in Paris polyphylla var. Yunnanensis [J/OL]. Mol. Plant Breeding., 2024:1-11 [2024-04-10]. . | |
| 40 | XIAO M K, HU X, LI Y Q,et al..Comparative analysis of codon usage patterns in the chloroplast genomes of nine forage legumes [J]. Physiol. Mol. Biol. Plants, 2024,30(2):153-166. |
| [1] | Ziqin LI, Jiaqiang WANG, Zhen LI, Deqiu ZOU, Xiaogong ZHANG, Xiaoyu LUO, Weiyang LIU. Estimation of Chlorophyll Density of Cotton Leaves Based on Spectral Index [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 103-111. |
| [2] | Fulin ZHANG, Rui XI, Yuxiang LIU, Zhaolong CHEN, Qinghui YU, Ning LI. Genome-wide Identification and Expression Analysis of Tomato BURP Structural Domain Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 51-62. |
| [3] | Yike XU, Shuang LI, Changle LIU, Peiwen KOU, Xiaochun SUN, Wenjing HUANG. Study on Agronomic Characters and Photosynthetic Physiological Characteristics of Pinellia ternata from Different Producing Areas [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 77-89. |
| [4] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [5] | Yurong DENG, Lian HAN, Jinlong WANG, Xinghan WEI, Xudong WANG, Ying ZHAO, Xiaohong WEI, Chaozhou LI. Identification of SOD Family Genes in Chenopodium quinoa and Their Response to Mixed Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 28-39. |
| [6] | Shengyan YANG, Man CAO, Baoshi GUO, Chao YANG, Zhixia HOU. Effects of Different Iron Environments on the Growth and Leaf Chlorophyll Fluorescence Characteristics of Blueberry [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 52-62. |
| [7] | Rui TIAN, Hua ZHANG, Meihong HUANG, Zhenqi SHAO, Xihuan LI, Caiying ZHANG. Mining of Candidate Genes and Genetic Loci Conferring Drought Tolerance in Soybean [J]. Journal of Agricultural Science and Technology, 2023, 25(9): 69-82. |
| [8] | Yingxue ZHU, Qi WANG, Xianfa MA, Yusheng JIAO, Jinxu GAO, Weijia MAO, Jia FU, Xuedong SUN, Ye YUAN. Leaf Color Characteristic Value and Nitrogen Diagnosis Model of Tobacco During Growth Period [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 54-62. |
| [9] | Lifang HUANG, Yuzhou LONG, Jinqin LI, Yunping DONG, Xiaoyang WANG, Peng CHEN, Xianwen WANG, Lin YAN. Physiological and Biochemical Characteristics of Coffea arabica Seedling Under Low Temperature Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(2): 60-67. |
| [10] | Peixin LIANG, Rong TANG, Jianguo LIU. Effects of Mixed Saline-alkali Stress on Photosynthetic Physiology and Yield in Cyperus esculentus L. [J]. Journal of Agricultural Science and Technology, 2023, 25(12): 195-204. |
| [11] | Wengyou TIAN, Hao LIU, Chaolin GAN, Liufen WU, Ai LI, Lifang YANG, Ying GAO. Photosynthetic Response and Spectral Characteristics of Cherry Rootstocks Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2022, 24(3): 77-83. |
| [12] | Jinfeng ZHAO, Aili YU, Yanfang LI, Yanwei DU, Gaohong WANG, Zhenhua WANG. Response Characteristics of SiCBL3 to Abiotic Stresses in Foxtail Millet [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 68-75. |
| [13] | LIU Zhengwen, WANG Xingfen, MENG Chengsheng, ZHANG Yan, SUN Zhengwen, WU Liqiang, MA Zhiying, ZHANG Guiyin. Genome-Wide Identification and Analysis of GH9 Gene Family in Gossypium barbadense L. [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 30-45. |
| [14] | PU Quanming, YANG Peng, YONG Lei, DENG Yuchuan, HE Zihan, LIN Bangmin, SHI Songmei, XIANG Chengyong, FANG Fang. Studies on Pigment Content and Photosyntheic Characteristics of Purple-red Leaf Color Mutant in Radish [J]. Journal of Agricultural Science and Technology, 2021, 23(8): 45-54. |
| [15] | CUI Jianghui§, YANG Puyuan§, CHANG Jinhua*. Identification and Expression Analysis Under Abiotic Stress of GRF Gene Family in Sorghum [J]. Journal of Agricultural Science and Technology, 2021, 23(4): 37-46. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号