




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (10): 98-109.DOI: 10.13304/j.nykjdb.2024.0022
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Wanjing XU1,2( ), Fang PENG1, Doudou ZHAO1,2, Jiaojiao LUO1, Shan TAO1, Hailang LIAO1, Changqing MAO1, Yu WU1, Xiu ZHU3, Zhengjun XU2(
), Fang PENG1, Doudou ZHAO1,2, Jiaojiao LUO1, Shan TAO1, Hailang LIAO1, Changqing MAO1, Yu WU1, Xiu ZHU3, Zhengjun XU2( ), Chao ZHANG1(
), Chao ZHANG1( )
)
Received:2024-01-09
Accepted:2024-03-11
Online:2024-10-15
Published:2024-10-18
Contact:
Zhengjun XU,Chao ZHANG
徐皖菁1,2( ), 彭芳1, 赵豆豆1,2, 罗姣姣1, 陶珊1, 廖海浪1, 毛常清1, 吴宇1, 朱秀3, 徐正君2(
), 彭芳1, 赵豆豆1,2, 罗姣姣1, 陶珊1, 廖海浪1, 毛常清1, 吴宇1, 朱秀3, 徐正君2( ), 张超1(
), 张超1( )
)
通讯作者:
徐正君,张超
作者简介:徐皖菁 E-mail:1315024678@qq.com
基金资助:CLC Number:
Wanjing XU, Fang PENG, Doudou ZHAO, Jiaojiao LUO, Shan TAO, Hailang LIAO, Changqing MAO, Yu WU, Xiu ZHU, Zhengjun XU, Chao ZHANG. Analysis of Response Mechanism of Ligusticum chuanxiong to Cadmium Stress Based on Transcriptome and Metabolome[J]. Journal of Agricultural Science and Technology, 2024, 26(10): 98-109.
徐皖菁, 彭芳, 赵豆豆, 罗姣姣, 陶珊, 廖海浪, 毛常清, 吴宇, 朱秀, 徐正君, 张超. 基于转录组和代谢组解析川芎对镉胁迫的响应机制[J]. 中国农业科技导报, 2024, 26(10): 98-109.
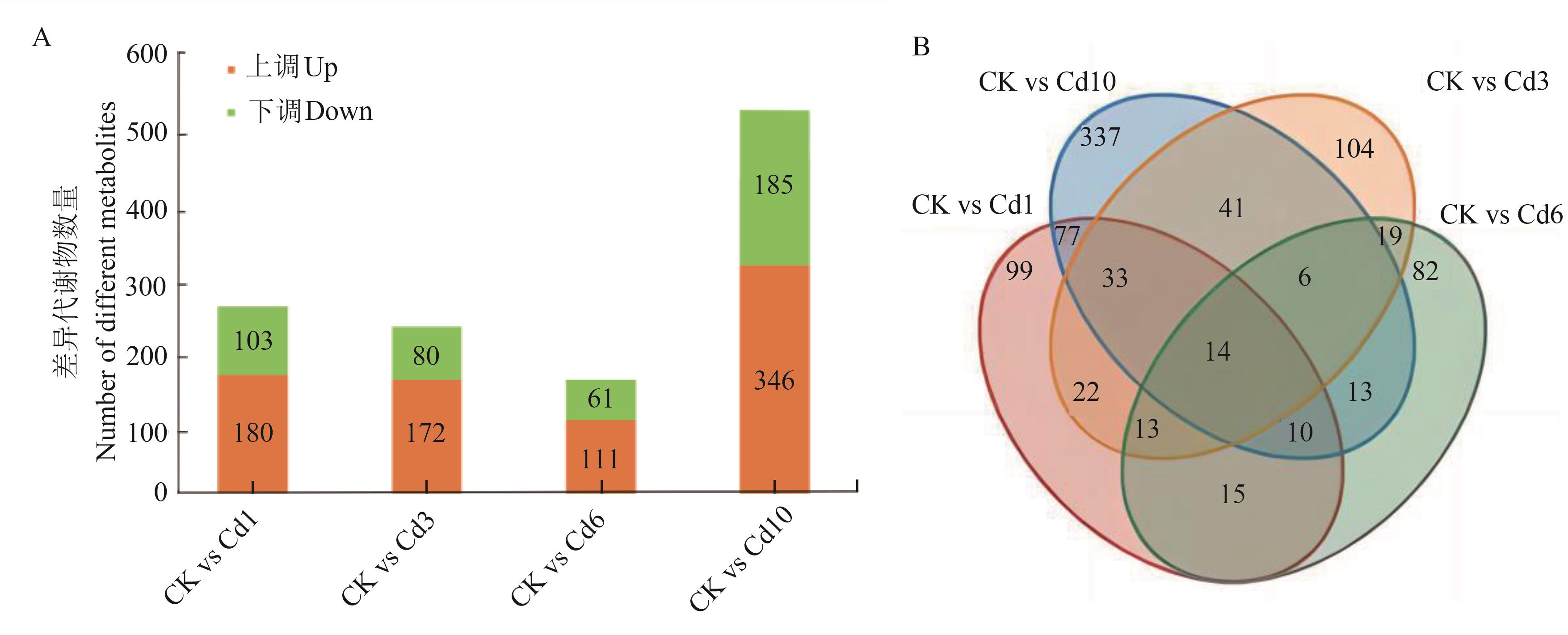
Fig. 1 Numbers of the up-regulated and down-regulated DAMs in pairwise comparisonsA:Number of differential accumulated metabolites in each group; B:Differential accumulated metabolite Venn diagram

Fig. 2 Partial least squares discriminant analysis orthogonal projection score of metabolites of Ligusticum chuanxiong treated with different levels of CdNote: TP—Score value of the predicted component;TO—Score value of the orthogonal component.
样品编号 Sample number | Clean reads数目 Number of clean reads | 总碱基数 Total base number/nt | GC含量 GC content/% | Q30/% |
|---|---|---|---|---|
| CK-1 | 29 678 610 | 8 835 337 288 | 43.62 | 93.76 |
| CK-2 | 24 429 573 | 7 276 365 402 | 43.79 | 93.26 |
| CK-3 | 19 868 507 | 5 905 914 150 | 44.19 | 93.33 |
| Cd1-1 | 20 935 573 | 6 215 766 952 | 44.79 | 93.93 |
| Cd1-2 | 25 071 449 | 7 465 487 210 | 43.97 | 93.66 |
| Cd1-3 | 21 681 836 | 6 460 887 440 | 43.92 | 93.61 |
| Cd3-1 | 19 483 124 | 5 787 607 824 | 43.61 | 93.79 |
| Cd3-2 | 20 832 819 | 6 215 838 452 | 43.87 | 93.05 |
| Cd3-3 | 21 394 788 | 6 382 109 148 | 43.90 | 93.59 |
| Cd6-1 | 20 610 107 | 6 120 785 704 | 43.97 | 93.70 |
| Cd6-2 | 20 640 197 | 6 157 201 056 | 43.79 | 93.78 |
| Cd6-3 | 22 624 009 | 6 720 661 646 | 43.88 | 93.87 |
| Cd10-1 | 19 376 308 | 5 766 904 348 | 43.94 | 93.65 |
| Cd10-2 | 19 731 742 | 5 880 325 626 | 43.67 | 93.46 |
| Cd10-3 | 21 006 650 | 6 270 393 312 | 43.95 | 93.36 |
| 合计 Total | 327 365 292 | 97 461 585 558 | — | — |
Table 1 Transcriptome sequencing product analysis
样品编号 Sample number | Clean reads数目 Number of clean reads | 总碱基数 Total base number/nt | GC含量 GC content/% | Q30/% |
|---|---|---|---|---|
| CK-1 | 29 678 610 | 8 835 337 288 | 43.62 | 93.76 |
| CK-2 | 24 429 573 | 7 276 365 402 | 43.79 | 93.26 |
| CK-3 | 19 868 507 | 5 905 914 150 | 44.19 | 93.33 |
| Cd1-1 | 20 935 573 | 6 215 766 952 | 44.79 | 93.93 |
| Cd1-2 | 25 071 449 | 7 465 487 210 | 43.97 | 93.66 |
| Cd1-3 | 21 681 836 | 6 460 887 440 | 43.92 | 93.61 |
| Cd3-1 | 19 483 124 | 5 787 607 824 | 43.61 | 93.79 |
| Cd3-2 | 20 832 819 | 6 215 838 452 | 43.87 | 93.05 |
| Cd3-3 | 21 394 788 | 6 382 109 148 | 43.90 | 93.59 |
| Cd6-1 | 20 610 107 | 6 120 785 704 | 43.97 | 93.70 |
| Cd6-2 | 20 640 197 | 6 157 201 056 | 43.79 | 93.78 |
| Cd6-3 | 22 624 009 | 6 720 661 646 | 43.88 | 93.87 |
| Cd10-1 | 19 376 308 | 5 766 904 348 | 43.94 | 93.65 |
| Cd10-2 | 19 731 742 | 5 880 325 626 | 43.67 | 93.46 |
| Cd10-3 | 21 006 650 | 6 270 393 312 | 43.95 | 93.36 |
| 合计 Total | 327 365 292 | 97 461 585 558 | — | — |
数据库 Database | Unigenes数量 Number of unigenes | |||
|---|---|---|---|---|
总量 Total | 300≤长度<1 000 bp 300≤Length<1 000 bp | 长度≥1 000 bp Length≥1 000 bp | 占比 Proportion/% | |
| COG | 11 717 | 4 635 | 7 082 | 32.25 |
| GO | 28 277 | 11 510 | 16 747 | 77.82 |
| KEGG | 23 357 | 9 252 | 14 105 | 64.28 |
| KOG | 21 487 | 8 946 | 12 541 | 59.13 |
| Pfam | 28 001 | 10 941 | 17 060 | 77.06 |
| Swiss-Prot | 22 042 | 8 924 | 13 118 | 60.66 |
| TrEMBL | 31 952 | 13 037 | 18 915 | 87.93 |
| EggNOG4.5 | 26 245 | 10 422 | 15 823 | 72.23 |
| NR | 34 643 | 14 628 | 20 015 | 95.34 |
总注释转录本 Total annotated transcript | 36 336 | 15 887 | 20 429 | 100.00 |
Table 2 Statistics of unigene annotation
数据库 Database | Unigenes数量 Number of unigenes | |||
|---|---|---|---|---|
总量 Total | 300≤长度<1 000 bp 300≤Length<1 000 bp | 长度≥1 000 bp Length≥1 000 bp | 占比 Proportion/% | |
| COG | 11 717 | 4 635 | 7 082 | 32.25 |
| GO | 28 277 | 11 510 | 16 747 | 77.82 |
| KEGG | 23 357 | 9 252 | 14 105 | 64.28 |
| KOG | 21 487 | 8 946 | 12 541 | 59.13 |
| Pfam | 28 001 | 10 941 | 17 060 | 77.06 |
| Swiss-Prot | 22 042 | 8 924 | 13 118 | 60.66 |
| TrEMBL | 31 952 | 13 037 | 18 915 | 87.93 |
| EggNOG4.5 | 26 245 | 10 422 | 15 823 | 72.23 |
| NR | 34 643 | 14 628 | 20 015 | 95.34 |
总注释转录本 Total annotated transcript | 36 336 | 15 887 | 20 429 | 100.00 |
分组 Group | 差异表达基因总数 Total number of differentially expressed genes | 上调 Up | 下调 Down |
|---|---|---|---|
| CK vs Cd1 | 2 385 | 2 247 | 138 |
| CK vs Cd3 | 5 703 | 4 205 | 1 498 |
| CK vs Cd6 | 286 | 257 | 29 |
| CK vs Cd10 | 195 | 150 | 45 |
| 合计 Total | 8 569 | 6 859 | 1 710 |
Table 3 Statistical results of differentially expressed genes
分组 Group | 差异表达基因总数 Total number of differentially expressed genes | 上调 Up | 下调 Down |
|---|---|---|---|
| CK vs Cd1 | 2 385 | 2 247 | 138 |
| CK vs Cd3 | 5 703 | 4 205 | 1 498 |
| CK vs Cd6 | 286 | 257 | 29 |
| CK vs Cd10 | 195 | 150 | 45 |
| 合计 Total | 8 569 | 6 859 | 1 710 |
| 1 | 王艺涵,赵佳琛,金艳,等.经典名方中川芎的本草考证[J].中国实验方剂学杂志,2022,28(10):262-274. |
| WANG Y H, ZHAO J C, JIN Y, et al.. Herbal textual research on Chuanxiong rhizoma in famous classical formulas [J]. Chin. J. Exp. Tradit. Med. Formulae, 2022,28(10):262-274. | |
| 2 | 廖雪梅,陈媛媛,陶珊,等.川芎重金属镉污染现状及降镉策略研究进展[J].河南农业科学,2020,49(10):1-11. |
| LIAO X M, CHEN Y Y, TAO S,et al..Research progress on present situation of heavy metal cadmium pollution and strategies to reduce cadmium in Ligusticum chuanxiong [J]. J.Henan Agric. Sci., 2020,49(10):1-11. | |
| 3 | 王晓亚,鲁建丽.川芎和抚芎的多糖和重金属含量分析[J].广东微量元素科学,2006,13(1):49-52. |
| WANG X Y, LU J L.Determination of polysaccharides and heavy mentals in Ligusticum chuanxiong hort and Ligusticum chuanxiong hort cv.Fuxiong [J].Guangdong Trace Elem.Sci.,2006,13(1):49-52. | |
| 4 | 何平,李佳颖,刘宇哲,等.土壤镉污染背景下川芎生态适宜区规划研究[J].中国中药杂志,2022,47(5):1196-1204. |
| HE P, LI J Y, LIU Y Z,et al..Planning of ecologically suitable areas for Ligusticum chuanxiong under background of soil cadmium pollution [J].China J.Chin.Mater.Med.,2022,47(5):1196-1204. | |
| 5 | 陈云子,刘薇,黎智,等.川芎镉污染现状调查及原因分析[J].中药与临床,2022,13(6):1-4, 13. |
| CHEN Y Z, LIU W, LI Z,et al..Investigation and analysis of cadmium pollution in Chuanxiong [J].Pharm.Clin.Chin.Mater.Med.,2022,13(6):1-4, 13. | |
| 6 | 李笑媛,陈润芍,许安妮,等.川芎对镉、铅及其复合处理的生理响应[J].应用与环境生物学报,2019,25(2):321-327. |
| LI X Y, CHEN R S, XU A N,et al..Physiological response to cadmium,lead,and their combination stress in Ligusticum chuanxiong Hort [J].Chin.J.Appl.Environ.Biol.,2019,25(2):321-327. | |
| 7 | GHATAK A, CHATURVEDI P, WECKWERTH W.Metabolomics in plant stress physiology [J].Adv Biochem. Eng. Biotechnol.,2018,164:187-236. |
| 8 | 罗庆.镉、铅胁迫下东南景天根系分泌物的代谢组学研究[D].沈阳:东北大学,2015. |
| LUO Q. Metabolomics study on root exudates of Sedum alfredii under Cd and Pb stress [D]. Shenyang: Northeastern University, 2015. | |
| 9 | 盛莎莎,刘荣鹏,王晓云,等.车前响应镉胁迫的生理学和代谢组学分析[J].植物科学学报,2023,41(2):234-244. |
| SHENG S S, LIU R P, WANG X Y,et al..Physiological and metabolomic analysis of Plantago asiatica L.in response to cadmium stress [J]. Plant Sci. J., 2023,41(2):234-244. | |
| 10 | 汪京超.基于转录组学的油菜镉胁迫响应机制研究[D].重庆:西南大学,2016. |
| WANG J C. Mechanism research on cadmium stress response of brassica based on transcriptomic [D]. Chongqing: Southwest University, 2016. | |
| 11 | 王书凤.高低镉积累油菜品种响应镉胁迫的分子机制研究[D].重庆:西南大学,2019. |
| WANG S F. Study on the molecular mechanisms of high/low cadmium accumulation rapeseed cultivars response to Cd stress [D]. Chongqing: Southwest University,2019. | |
| 12 | 吴迪,张燕燕,林楠,等.基于代谢组学和转录组学探究草珊瑚叶和根中黄酮类成分差异积累的转录调控机制[J].中国中药杂志,2023,48(21):5767-5778. |
| WU D, ZHANG Y Y, LIN N,et al..Transcriptional regulation mechanism of differential accumulation of flavonoids in leaves and roots of Sarcandra glabra based on metabonomics and transcriptomics [J].China J. Chin. Mater. Med.,2023,48(21):5767-5778. | |
| 13 | 刁瑞宁,杨莎,张佳蕾,等.基于转录组学和代谢组学解析氮调控花生结瘤固氮的机理[J].山东农业科学,2023,55(4):1-10. |
| DIAO R N, YANG S, ZHANG J L,et al.. Mechanism analysis of nitrogen regulation on peanut nodulating based on transcriptomes and metabonomics [J].Shandong Agric.Sci.,2023,55(4):1-10. | |
| 14 | 王婷婷,陈鸽,包悦琳,等.大豆根系响应早期缺铁胁迫的转录组分析[J].南京农业大学学报,2022,45(2):224-234. |
| WANG T T, CHEN G, BAO Y L, et al.. Transcriptome analysis of soybean roots in response to early iron deficiency stress [J].J.Nanjing Agric.Univ., 2022,45(2):224-234. | |
| 15 | 武丽娟,刘海学,张亚东,等.不同镉胁迫水平对水稻幼苗差异代谢物的影响[J/OL].分子植物育种,2022:1-21[2023-12-08]. . |
| WU L J, LIU H X, ZHANG Y D, et al.. Effects of different cadmium stress levels on differential metabolites in rice seedlings [J/OL]. Mol. Plant Breeding,2022:1-21 [2023-12-08]. . | |
| 16 | 宋佳阳,胡瑜,徐倩,等.不同秋眠级紫花苜蓿叶片响应渍水胁迫的代谢组学分析[J].草地学报,2023,31(11):3355-3363. |
| SONG J Y, HU Y, XU Q,et al..Metabolomic analysis on the response of leaves of alfalfa with different fall dormancy levels to waterlogging stress [J].Acta Agrestia Sin.,2023,31(11):3355-3363. | |
| 17 | 袁俊,盛莎莎,刘荣鹏,等.镉胁迫对丹参生理特性和代谢特征的影响[J].植物科学学报,2022,40(3):408-417. |
| YUAN J, SHENG S S, LIU R P,et al..Effects of cadmium on physiological characteristics and metabolic profiles of Salvia miltiorrhiza Bunge [J]. Plant Sci. J., 2022,40(3):408-417. | |
| 18 | STEPANSKY A, LEUSTEK T.Histidine biosynthesis in plants [J].Amino Acids,2006,30(2):127-142. |
| 19 | 马洪娜,吴依琳,马珊珊,等.朱砂根叶响应Ca2+胁迫的差异代谢物分析[J].植物生理学报,2023,59(9):1771-1782. |
| MA H N, WU Y L, MA S S, et al.. Analysis of differential metabolites in Ardisia crenata leaves in response to Ca2+ stress [J]. Plant Physiol. J., 2023, 59(9):1771-1782. | |
| 20 | HO C L, SAITO K. Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana [J]. Amino Acids, 2001,20(3):243-259. |
| 21 | WADITEE R, BHUIYAN N H, HIRATA E,et al..Metabolic engineering for betaine accumulation in microbes and plants [J]. J.Biol. Chem.,2007,282(47):34185-34193. |
| 22 | ROS R, CASCALES-MIÑANA B, SEGURA J, et al.. Serine biosynthesis by photorespiratory and nonphotorespiratory pathways: an interesting interplay with unknown regulatory networks [J]. Plant Biol., 2013,15:707-712. |
| 23 | PENG H, HE X, GAO J,et al..Transcriptomic changes during maize roots development responsive to Cadmium (Cd) pollution using comparative RNAseq-based approach [J].Biochem. Biophys. Res. Commun., 2015,464(4):1040-1047. |
| 24 | 张尉欣,刘阳,刘帅,等.镉胁迫下菜心的转录组分析[J].深圳大学学报(理工版),2018,35(5):543-550. |
| ZHANG Y X, LIU Y, LIU S,et al..Transcriptome analysis of Brassica rapa ssp.chinensis var. parachinensis under cadmium stress [J]. J. Shenzhen Univ. (Sci.Eng.), 2018,35(5):543-550. | |
| 25 | ARBONA V, MANZI M, OLLAS C D,et al..Metabolomics as a tool to investigate abiotic stress tolerance in plants [J].Int. J. Mol. Sci., 2013,14(3):4885-4911. |
| 26 | BHEEMAREDDY B R, PULIPETA M, IYER P, et al.. Effect of the total galactose content on complement-dependent cytotoxicity of the therapeutic anti-CD20 IgG1 antibodies under temperature stress conditions [J/OL]. J. Carbohydr. Chem., 2019,38(1):1541995 [2023-12-08]. . |
| 27 | NÄGELE T, STUTZ S, HÖRMILLER I I,et al..Identification of a metabolic bottleneck for cold acclimation in Arabidopsis thaliana [J]. Plant J., 2012,72(1):102-114. |
| 28 | 王丽华,李改玲,李晶,等.外源糖对盐胁迫下小黑麦幼苗糖代谢的影响[J].麦类作物学报,2017,37(4):548-553. |
| WANG L H, LI G L, LI J, et al.. Effect of exogenous sugar on the sugar metabolism in Triticale seedling under salt stress [J]. J.Triticeae Crops,2017,37(4):548-553. | |
| 29 | LI C, LIU Y, TIAN J,et al.. Changes in sucrose metabolism in maize varieties with different cadmium sensitivities under cadmium stress [J/OL].PLoS One,2020,15(12):e0243835 [2023-12-08]. . |
| 30 | 陈文玲,张晴晴,唐韶华,等.甘油-3-磷酸酰基转移酶在植物脂质代谢、生长及逆境反应中的作用[J].植物生理学报,2018,54(5):725-735. |
| CHEN W L, ZHANG Q Q, TANG S H,et al..Glycerol-3-phosphate acyltransferase in lipid metabolism,growth and response to stresses in plants [J].Plant Physiol.J.,2018,54(5):725-735. | |
| 31 | 汪怡文.转录组和代谢组整合分析冬小麦镉胁迫响应的关键代谢通路[D].武汉:华中农业大学, 2022. |
| WANG Y W. Integrated transcriptome and metabonomic analysis of key metabolic pathways in response to cadmium stress in winter wheat [D].Wuhan:Huazhong Agricultural University, 2022. | |
| 32 | 钱禛锋,谷书杰,赵雪婷,等.基于转录组的蔗茅CML基因家族鉴定及冷胁迫表达分析[J].农业生物技术学报,2022,30(5):885-895. |
| QIAN Z F, GU S J, ZHAO X T,et al.. Identification and cold stress expression analysis of CML gene family in Erianthus fulvus based on transcriptome [J].J.Agric.Biotechnol.,2022,30(5):885-895. | |
| 33 | MAGNAN F, RANTY B, CHARPENTEAU M,et al.. Mutations in AtCML9,a calmodulin-like protein from Arabidopsis thaliana,alter plant responses to abiotic stress and abscisic acid [J]. Plant J., 2008,56(4):575-589. |
| 34 | HIRAI M Y, SAWADA Y, KANAYA S,et al.. Toward genome-wide metabolotyping and elucidation of metabolic system:metabolic profiling of large-scale bioresources [J]. J. Plant Res., 2010,123(3):291-298. |
| 35 | 赵云霞,魏艳玲,黄先忠.新疆无苞芥类钙调素蛋白基因OpCML13的克隆与分析[J].西北植物学报,2014,34(3):431-437. |
| ZHAO Y X, WEI Y L, HUANG X Z. Clone and expression analysis of calmodulin-like protein gene OpCML13 from Olimarabidopsis pumila of Xinjiang [J]. Acta Bot.Bor-Occid. Sin., 2014,34(3):431-437. | |
| 36 | VANDERBELD B, SNEDDEN W A. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37,CML38 and CML39 [J].Plant Mol. Biol., 2007,64(6):683-697. |
| [1] | Qian ZHANG, Lina MEN, Yiran LI, Qiao LIU, Angie DENG, Xiaowen HU, Yuhong ZHANG, Zhiwei ZHANG, Wei ZHANG. Differential Expression Paradigm of Chemoreceptor Genes Between Males and Females at Different Developmental Stages of Carposina sasakii Matsumura [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 151-162. |
| [2] | Yiwei LU, Xueyan XIA, Yu ZHAO, Jihan CUI, Meng LIU, Meihong HUANG, Cheng CHU, Jianjun LIU, Shunguo LI. Transcriptome Profiling and Gene Mining of Millet Response to Potassium Deficiency Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 30-44. |
| [3] | Jiarui XU, Yiru WANG, Shaogeng ZHAO, Kun LI, Jun ZHENG. Functional Study and Transcriptome Analysis of Corn Gene ZmCCoAOMT1 Involved in Lignin Synthesis Pathway [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 30-43. |
| [4] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [5] | Wei JI, Ying FAN, Jiaxing HUANG, Huipeng YANG, Jin XU, Xiaoying LI, Yueqin GUO, Yueguo WU, Jilian LI, Jun YAO. Transcriptome Analysis of Stigma Response Mechanism of Blueberry with Different Pollination Intensity [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 71-82. |
| [6] | Xiangwu LI, Ziyang LIU, Yujun XU, Jianbo ZHU, Yanmin WU. Explore of Molecular Mechanism on Fungal Elicitors Regulating Shikonin Synthesis [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 78-88. |
| [7] | Xiaoran WANG, Xiaoyu LI, Hui SUN, Haidong YU, Yongchun SHI. Transcriptome Analysis of Tobacco Leaves Under Boron Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 53-64. |
| [8] | Xiang WU, Juan LI, Yan CAO, Yanrong CHENG, Xuyu YAN, Ling LI. Research Advances on Plant Root Exudates in Response to Cadmium Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 12-20. |
| [9] | Yunsheng WANG, Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG. Effects of Overexpression of veA Gene on Secondary Metabolism of Eurotium cristatus [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 77-86. |
| [10] | Yunfei LIU, Fengjie WEI, Maolin XIA, Zhaojin YU, Hao XIA, Chunyu YI, Jianbo CHANG, Xiaoming JI. Alleviative Effect of New Composite Hydrogels on Cadmium Stress Tobacco Seedlings [J]. Journal of Agricultural Science and Technology, 2023, 25(3): 188-197. |
| [11] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [12] | LI Shuxin, ZHANG Hao, ZHENG Housheng, ZHENG Peihe, PANG Shifeng, XU Shiquan. Transcriptome Analysis of Phenotypic Differences Between Ermaya and Changbo of Forest Cultivated Panax ginseng [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 56-68. |
| [13] | LIU Yuan, ZHANG Xiuyan, XU Miaoyun, ZHENG Hongyan, ZOU Junjie, ZHANG Lan, WANG Lei. Global Small RNA Transcriptome Profiling of Rice Under Drought Stress [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 23-32. |
| [14] | HAO Zhenggang, ZHAO Huijun, WEI Yuqing*, ZENG Zhouqi, WANG Zhiheng. Physiological and Biochemical Responses of Sweet Sorghum to Cadmium Stress and Its Cadmium Accumulation [J]. Journal of Agricultural Science and Technology, 2021, 23(1): 30-42. |
| [15] | LIU Shidou1, ZHU Xinping1,2*, ZHAO Yi1, WANG Boyan1, HAN Yaoguang1, YANG Beibei1, LU Zhi1, JIA Hongtao1,2. Cotton Stalk Biochar Retarding Stress of Cadmium on Growth of Rice [J]. Journal of Agricultural Science and Technology, 2020, 22(4): 139-146. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号