




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (1): 78-88.DOI: 10.13304/j.nykjdb.2022.0622
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Xiangwu LI1,2( ), Ziyang LIU2,3, Yujun XU2, Jianbo ZHU1(
), Ziyang LIU2,3, Yujun XU2, Jianbo ZHU1( ), Yanmin WU2,3(
), Yanmin WU2,3( )
)
Received:2022-07-23
Accepted:2022-09-28
Online:2024-01-15
Published:2024-01-08
Contact:
Jianbo ZHU,Yanmin WU
李相吴1,2( ), 刘自扬2,3, 徐玉俊2, 祝建波1(
), 刘自扬2,3, 徐玉俊2, 祝建波1( ), 吴燕民2,3(
), 吴燕民2,3( )
)
通讯作者:
祝建波,吴燕民
作者简介:李相吴 E-mail:1003786451@qq.com;
基金资助:CLC Number:
Xiangwu LI, Ziyang LIU, Yujun XU, Jianbo ZHU, Yanmin WU. Explore of Molecular Mechanism on Fungal Elicitors Regulating Shikonin Synthesis[J]. Journal of Agricultural Science and Technology, 2024, 26(1): 78-88.
李相吴, 刘自扬, 徐玉俊, 祝建波, 吴燕民. 真菌诱导子调控紫草素合成的分子机制探究[J]. 中国农业科技导报, 2024, 26(1): 78-88.
处理 Treatment | 样品编号 Sample ID | 总碱基数 Base number | GC含量 GC content/% | Q30/% |
|---|---|---|---|---|
| F20 | F20-1 | 6 293 089 734 | 43.61 | 92.39 |
| F20-2 | 6 422 595 670 | 43.56 | 93.06 | |
| F20-3 | 7 834 038 640 | 43.62 | 92.46 | |
| R20 | R20-1 | 6 872 640 242 | 43.61 | 93.02 |
| R20-2 | 7 140 087 048 | 43.71 | 93.05 | |
| R20-3 | 6 946 752 472 | 43.66 | 92.70 | |
| CK | CK-1 | 7 792 349 390 | 43.66 | 92.82 |
| CK-2 | 6 870 409 240 | 43.68 | 92.81 | |
| CK-3 | 5 930 234 288 | 43.68 | 92.44 |
Table 1 sample sequencing data evaluation
处理 Treatment | 样品编号 Sample ID | 总碱基数 Base number | GC含量 GC content/% | Q30/% |
|---|---|---|---|---|
| F20 | F20-1 | 6 293 089 734 | 43.61 | 92.39 |
| F20-2 | 6 422 595 670 | 43.56 | 93.06 | |
| F20-3 | 7 834 038 640 | 43.62 | 92.46 | |
| R20 | R20-1 | 6 872 640 242 | 43.61 | 93.02 |
| R20-2 | 7 140 087 048 | 43.71 | 93.05 | |
| R20-3 | 6 946 752 472 | 43.66 | 92.70 | |
| CK | CK-1 | 7 792 349 390 | 43.66 | 92.82 |
| CK-2 | 6 870 409 240 | 43.68 | 92.81 | |
| CK-3 | 5 930 234 288 | 43.68 | 92.44 |
处理 Treatment | 样品编号 Sample ID | Read数 Reads number | 匹配Read数 Mapped reads number | 匹配率 Mapped ratio/% |
|---|---|---|---|---|
| F20 | F20-1 | 21 096 427 | 15 558 872 | 73.75 |
| F20-2 | 21 504 765 | 16 195 509 | 75.31 | |
| F20-3 | 26 236 242 | 19 430 135 | 74.06 | |
| R20 | R20-1 | 23 005 518 | 17 277 331 | 75.10 |
| R20-2 | 23 913 025 | 17 644 691 | 73.79 | |
| R20-3 | 23 283 171 | 17 129 464 | 73.57 | |
| CK | CK-1 | 26 093 101 | 19 723 787 | 75.59 |
| CK-2 | 23 036 789 | 17 376 776 | 75.43 | |
| CK-3 | 19 879 659 | 14 903 996 | 74.97 |
Table 2 Comparison of sequencing data with assembly results
处理 Treatment | 样品编号 Sample ID | Read数 Reads number | 匹配Read数 Mapped reads number | 匹配率 Mapped ratio/% |
|---|---|---|---|---|
| F20 | F20-1 | 21 096 427 | 15 558 872 | 73.75 |
| F20-2 | 21 504 765 | 16 195 509 | 75.31 | |
| F20-3 | 26 236 242 | 19 430 135 | 74.06 | |
| R20 | R20-1 | 23 005 518 | 17 277 331 | 75.10 |
| R20-2 | 23 913 025 | 17 644 691 | 73.79 | |
| R20-3 | 23 283 171 | 17 129 464 | 73.57 | |
| CK | CK-1 | 26 093 101 | 19 723 787 | 75.59 |
| CK-2 | 23 036 789 | 17 376 776 | 75.43 | |
| CK-3 | 19 879 659 | 14 903 996 | 74.97 |
| 数据库名称Database name | 被注释到的差异基因数量 Annotated number of DGEs | |
|---|---|---|
| F20/CK | R20/CK | |
| COG | 517 | 317 |
| GO | 1 276 | 785 |
| KEGG | 1 063 | 651 |
| KOG | 758 | 439 |
| Pfam | 1 294 | 775 |
| Swiss-Prot | 1 188 | 731 |
| NR | 1 555 | 943 |
Table 3 Numbers of functional annotated DEGs
| 数据库名称Database name | 被注释到的差异基因数量 Annotated number of DGEs | |
|---|---|---|
| F20/CK | R20/CK | |
| COG | 517 | 317 |
| GO | 1 276 | 785 |
| KEGG | 1 063 | 651 |
| KOG | 758 | 439 |
| Pfam | 1 294 | 775 |
| Swiss-Prot | 1 188 | 731 |
| NR | 1 555 | 943 |
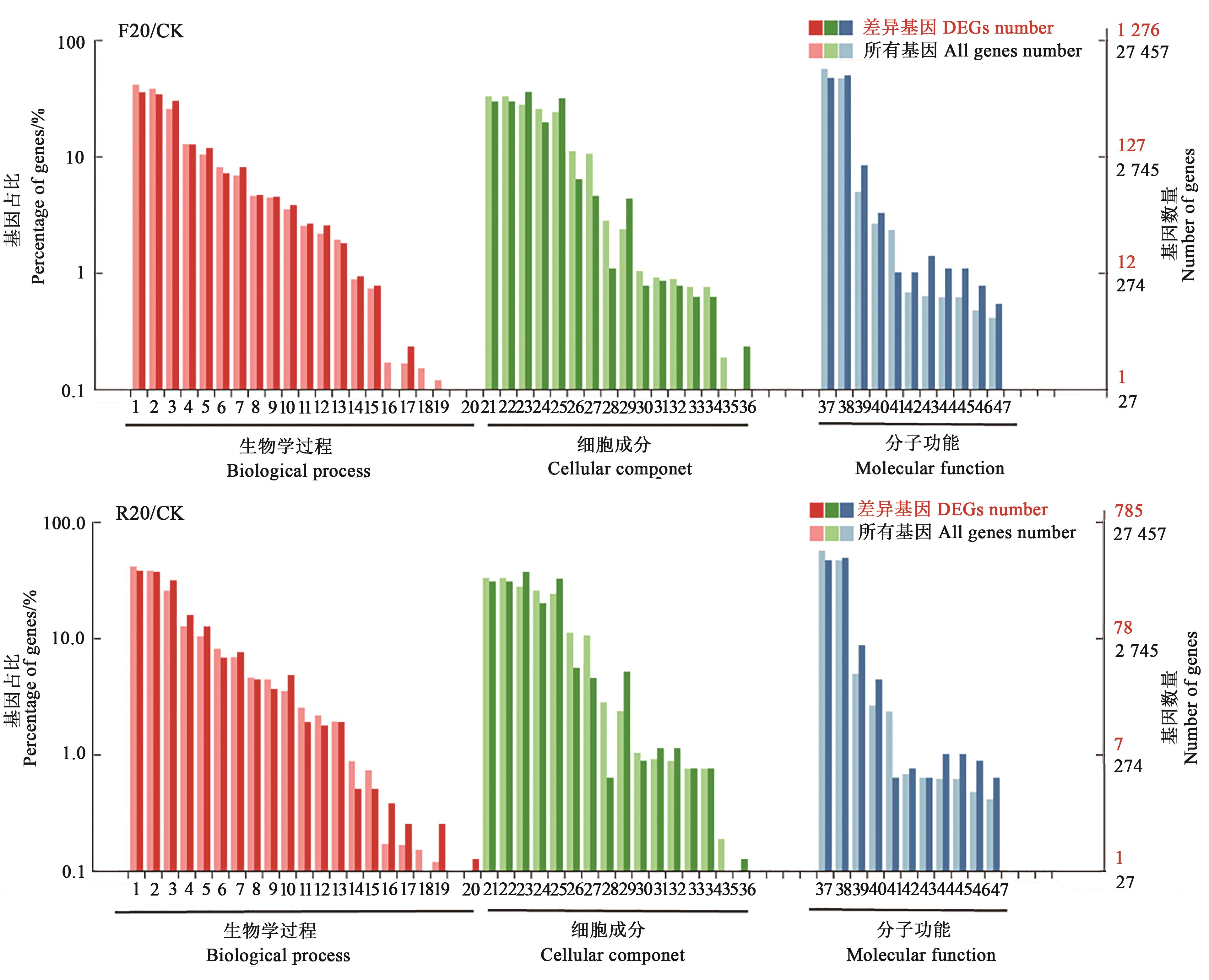
Fig. 2 GO functional annotation of differential expressed genesNote:1—Cellular process; 2—Metabolic process; 3—Single-organism process; 4—Biological regulation; 5—Response to stimulus; 6—Cellular component organization or biogenesis; 7—Localization; 8—Developmental process; 9—Multicellular organismal process; 10—Signaling; 11—Reproductive process; 12—Multi-organism process; 13—Reproduction; 14—Growth; 15—Immune system process; 16—Rhythmic process; 17—Locomotion; 18—Biological adhesion; 19—Detoxification; 20—Cell aggregation; 21—Cell; 22—Cell part; 23—membrane; 24—Organelle; 25—Membrane part; 26—Organelle part; 27—Macromolecular complex; 28—Membrane-enclosed lumen; 29—Extracellular region; 30—Extracellular region part; 31—Cell junction; 32—Symplast; 33—Other organism; 34—Other organism part; 35—Supramolecular complex; 36—Nucleoid; 37—Binding; 38—Catalytic activity; 39—Transporter activity; 40—Nucleic acid binding transcription factor activity; 41—Structural molecule activity; 42—Electron carrier activity; 43—Antioxidant activity; 44—Molecular transducer activity; 45—Signal transducer activity; 46—Transcription factor activity protein binding; 47—Molecular function regulator.
家族 Family | 基因ID Gene ID | log2FC | |
|---|---|---|---|
| F20/CK | R20/CK | ||
| TRAF | c65742.graph_c0 | 2.51 | 2.01 |
| Tify | c54302.graph_c0 | 1.59 | 0.96 |
| SRS | c54737.graph_c1 | -4.53 | -2.95 |
| MYB-related | c58018.graph_c0 | 4.26 | 2.80 |
| MYB | c62808.graph_c0 | 1.59 | 1.15 |
| LOB | c48779.graph_c0 | 1.71 | 1.28 |
| HB-other | c47461.graph_c0 | -2.71 | -1.85 |
| HB-BELL | c58247.graph_c0 | 1.70 | 1.54 |
| GARP-G2-like | c65836.graph_c0 | -1.95 | -1.23 |
| GARP-G2-like | c53563.graph_c1 | -1.78 | -1.82 |
| C2H2 | c56397.graph_c0 | -2.06 | -1.52 |
| C2C2-YABBY | c28651.graph_c0 | -3.50 | -2.26 |
| C2C2-Dof | c53061.graph_c0 | 1.71 | 1.50 |
| bHLH | c59392.graph_c0 | 2.16 | 1.43 |
| bHLH | c54786.graph_c0 | 1.65 | 1.66 |
| bHLH | c53656.graph_c0 | -6.60 | -3.19 |
| bHLH | c47522.graph_c0 | -2.99 | -1.71 |
| AP2/ERF-ERF | c60825.graph_c0 | -1.78 | -1.17 |
| AP2/ERF-ERF | c57468.graph_c0 | 2.29 | 1.93 |
| AP2/ERF-ERF | c55718.graph_c0 | -1.68 | -1.33 |
Table 4 Some differentially expressed transcription factor (top 20)
家族 Family | 基因ID Gene ID | log2FC | |
|---|---|---|---|
| F20/CK | R20/CK | ||
| TRAF | c65742.graph_c0 | 2.51 | 2.01 |
| Tify | c54302.graph_c0 | 1.59 | 0.96 |
| SRS | c54737.graph_c1 | -4.53 | -2.95 |
| MYB-related | c58018.graph_c0 | 4.26 | 2.80 |
| MYB | c62808.graph_c0 | 1.59 | 1.15 |
| LOB | c48779.graph_c0 | 1.71 | 1.28 |
| HB-other | c47461.graph_c0 | -2.71 | -1.85 |
| HB-BELL | c58247.graph_c0 | 1.70 | 1.54 |
| GARP-G2-like | c65836.graph_c0 | -1.95 | -1.23 |
| GARP-G2-like | c53563.graph_c1 | -1.78 | -1.82 |
| C2H2 | c56397.graph_c0 | -2.06 | -1.52 |
| C2C2-YABBY | c28651.graph_c0 | -3.50 | -2.26 |
| C2C2-Dof | c53061.graph_c0 | 1.71 | 1.50 |
| bHLH | c59392.graph_c0 | 2.16 | 1.43 |
| bHLH | c54786.graph_c0 | 1.65 | 1.66 |
| bHLH | c53656.graph_c0 | -6.60 | -3.19 |
| bHLH | c47522.graph_c0 | -2.99 | -1.71 |
| AP2/ERF-ERF | c60825.graph_c0 | -1.78 | -1.17 |
| AP2/ERF-ERF | c57468.graph_c0 | 2.29 | 1.93 |
| AP2/ERF-ERF | c55718.graph_c0 | -1.68 | -1.33 |
| 1 | 国家药典委员会.中华人民共和国药典-一部[M].北京:中国医药科技出版社,2020:355-356. |
| National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China-part1 [M]. Beijing: Chinese Medicine Science and Technology Press, 2020:355-356. | |
| 2 | WANG Y, ZHU Y, XIAO L, et al... Meroterpenoids isolated from Arnebia euchroma (Royle) Johnst. and their cytotoxic activity in human hepatocellular carcinoma cells [J]. Fitoterapia, 2018, 131(11):236-244. |
| 3 | CRUICKSHANK I, PERRIN D R. The isolation and partial characterization of monilicolin A, a polypeptide with phaseollin-inducing activity from Monilinia fructicola [J]. Life Sci., 1968, 7(10):449-458. |
| 4 | OKSMAN-CALDENTEY K, VERPOORTE R, VAN DER HEIJDEN R, et al... Engineering the plant cell factory for secondary metabolite production [J]. Transgenic Res., 2000, 9(4):323-343. |
| 5 | WANG Y, DAI C C, CAO J L, et al.. Comparison of the effects of fungal endophyte Gilmaniella sp. and its elicitor on atractylodes lancea plantlets [J]. World J. Microbiol. Biotechnol., 2011, 28(2):575-584. |
| 6 | ARGHAVANI P, HAGHBEEN K, MOUSAVI A. Enhancement of shikalkin production in Arnebia euchroma callus by a fungal elicitor, Rhizoctonia solani [J]. Iranian J. Biotechnol., 2015, 13(4):10-16. |
| 7 | 晏琼,胡宗定,吴建勇.生物与非生物诱导子协同作用对丹参毛状根培养生产丹参酮的影响[J].中国中药杂志,2006(3):188-191. |
| YAN Q, HU Z D, WU J Y. Synergistic effects of biotic and abiotic elicitors on the production of tanshinones in Salvia miltiorrhiza hairy root culture [J]. China J. Chin. Materia Medica, 2006(3):188-191. | |
| 8 | 田佩雯.白及内生真菌诱导子对宿主生长和主要活性物质的影响及调控[D].南宁:广西大学,2019. |
| TIAN P W. Effects and regulation of endophytic fungal elicitors from Bletilla striata on host growthand main substances [D]. Nanning: Guangxi University, 2019. | |
| 9 | 饶龙兵,杨汉波,郭洪英,等.不同倍性桤木属植物的转录组测序和分析[J].分子植物育种.2016,14(11):3047-3055. |
| RAO L B, YANG H B, GUO H Y, et al.. Analysis on transcriptome sequenced for alnus plants with different ploidy [J]. Mol. Plant Breed., 2016, 14(11):3047-3055. | |
| 10 | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 [J/OL]. Genome Biol., 2014, 15(12):550 [2022-06-02]. |
| 11 | GAISSER S, HEIDE L. Inhibition and regulation of shikonin biosynthesis in suspension cultures of Lithospermum [J]. Phytochemistry, 1996, 41(4):1065-1072. |
| 12 | SYKŁOWSKA-BARANEK K, PIETROSIUK A, NALIWAJSKI M R, et al.. Effect of L-phenylalanine on PAL activity and production of naphthoquinone pigments in suspension cultures of Arnebia euchroma (Royle) Johnst [J]. In Vitro Cellular Dev. Biol. Plant, 2012, 48(5):555-564. |
| 13 | YAZAKI K, KUNIHISA M, FUJISAKI T, et al.. Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon: cloning and characterization of a key enzyme in shikonin biosynthesis [J]. J. Biol. Chem., 2002, 277(8):6240-6246. |
| 14 | WANG S, WANG R S, LIU T G, et al.. CYP76B74 catalyzes the 3’’-hydroxylation of geranylhydroquinone in shikonin biosynthesis [J]. Plant Physiol. (Bethesda), 2019, 179(2):402-414. |
| 15 | 梁玖雯,李锬,王瑞杉,等.新疆紫草中2条CYP450基因的干扰毛状根体系的建立及其影响研究[J].中国中药杂志,2020,45(14):3422-3431. |
| LIANG J W, LI T, WANG R S, et al.. Establishment of RNA interfered hairy root system of two CYP450 genes in Arnebia euchroma and its influence [J]. China J. Chin. Materia Medica, 2020, 45(14):3422-3431. | |
| 16 | LIAO M, ZENG C, LIANG F. Two new dimeric naphthoquinones from Arnebia euchroma [J]. Phytochem. Letters, 2020, 37(1):106-109. |
| 17 | YAZAKI K, MATSUOKA H, SHIMOMURA K, et al.. A novel dark-inducible protein, LeDI-2, and its involvement in root-specific secondary metabolism in Lithospermum erythrorhizon [J]. Plant Physiol. (Bethesda), 2001, 125(3):1831-1841. |
| 18 | YAMAMURA Y N C U, SAHIN F P, NAGATSU A, et al.. Molecular cloning and characterization of a cDNA encoding a novel apoplastic protein preferentially expressed in a shikonin-producing callus strain of Lithospermum erythrorhizon [J]. Plant Cell Physiol., 2003, 44(4):437-446. |
| 19 | SAHA S, PAL D. Elicitor Signal Transduction Leading to the Production of Plant Secondary Metabolites [M]. Cham: Springer International Publishing, 2020:1-39. |
| 20 | 周雅涵.水杨酸、膜醭毕赤酵母、壳寡糖诱导柑橘果实抗病性及其生物学机制研究[D].重庆:西南大学,2017. |
| ZHOU Y H. Salicylic acid, Pichia membranaefaciens and oligochitosan induced disease resistance of citrus fruit and the possible biological mechanisms involved [D]. Chongqing: Southwest University, 2017. | |
| 21 | 瞿巾卓.酿酒葡萄细胞对内生真菌诱导子的代谢响应与机制[D].昆明:云南大学,2020. |
| JU J Z. Metabolic response and mechanism of Wine grape cells to elicitors from fungal endophytes [D]. Kunming: Yunnan University, 2020 | |
| 22 | 张明菊,朱莉,夏启中.植物激素对胁迫反应调控的研究进展[J].湖北大学学报(自然科学版).2021,43(3):242-253. |
| ZHANG M J, ZHU L, XIA Q ZAND. Research progress on the regulation of plant hormones to stress responses [J]. J. Hubei Univ., 2021, 43(3):242-253. | |
| 23 | SHAH L, ALI A, ZHU Y L, et al.. Wheat defense response to Fusarium head blight and possibilities of its improvement [J]. Physiol. Mol. Plant Pathol., 2017, 98(2):9-17. |
| 24 | NEMESIO-GORRIZ M, BLAIR P B, DALMAN K, et al.. Identification of Norway spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway [J/OL]. Front. Plant Sci., 2017, 8:305 [2022-06-02]. . |
| 25 | GRAEFF M, STRAUB D, EGUEN T, et al.. Microprotein-mediated recruitment of constans into a topless trimeric complex represses flowering in Arabidopsis [J/OL]. PLoS Genet., 2016, 12(3):e1005959 [2022-06-02]. . |
| 26 | DENG B, HUANG Z, GE F, et al.. An AP2/ERF family transcription factor PnERF1 raised the biosynthesis of saponins in panax notoginseng [J]. J. Plant Growth Regul., 2017, 36(3):691-701. |
| 27 | GAUTAM J K, GIRI M K, SINGH D, et al.. MYC2 influences salicylic acid biosynthesis and defense against bacterial pathogens in Arabidopsis thaliana [J]. Physiol. Plantarum., 2021, 173(4):2248-2261. |
| 28 | KAZAN K, MANNERS J M. MYC2: the master in action [J]. Mol. Plant, 2013, 6(3):686-703. |
| 29 | WANG F, ZHU H, CHEN D, et al.. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana [J]. Plant Cell. Tissue Organ Cult., 2016, 125(2):387-398. |
| 30 | ZHANG M, LI S T, NIE L, et al.. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis [J]. Plant Mol. Biol., 2015, 89(4-5):463-473. |
| 31 | EL-SAYED A S A, MOHAMED N Z, SAFAN S, et al.. Restoring the taxol biosynthetic machinery of aspergillus terreus by Podocarpus gracilior pilger microbiome, with retrieving the ribosome biogenesis proteins of WD40 superfamily [J]. Sci. Rep., 2019, 9(1):11512-11534. |
| 32 | HAN Z, YANG T, GUO Y, et al.. The transcription factor PagLBD3 contributes to the regulation of secondary growth in Populus [J]. J. Exp. Bot., 2021, 72(20):7092-7106. |
| 33 | TANG X M, WANG X, HUANG Y, et al... Natural variations of TFIIAγ gene and LOB1 promoter contribute to citrus canker disease resistance in Atalantia buxifolia [J]. PLoS Genetics. 2021, 17(1):e1009316 [2022-06-02]. . |
| 34 | HAO H, LEI C, DONG Q, et al... Effects of exogenous methyl jasmonate on the biosynthesis of shikonin derivatives in callus tissues of Arnebia euchroma [J]. Appl. Biochem. Biotechnol., 2014, 173(8):2198-2210. |
| 35 | WANG C G, WU J Y, MEI X G. Enhancement of taxol production and excretion in Taxus chinensis cell culture by fungal elicitation and medium renewal [J]. Appl. Microbiol. Biotechnol., 2001, 55(4):404-410. |
| [1] | Xiaoran WANG, Xiaoyu LI, Hui SUN, Haidong YU, Yongchun SHI. Transcriptome Analysis of Tobacco Leaves Under Boron Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 53-64. |
| [2] | Yunsheng WANG, Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG. Effects of Overexpression of veA Gene on Secondary Metabolism of Eurotium cristatus [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 77-86. |
| [3] | Lan MA, Qing PENG, Xiaoqing XU, Shuo YANG, Yuwei ZHANG, Dandan TIAN, Linbo SHI, Bo SHI, Yu QIAO. Gene Expression in Escherichia coli O157∶H7 Biofilms [J]. Journal of Agricultural Science and Technology, 2023, 25(6): 71-88. |
| [4] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [5] | Ruifeng GUO, Yuemei REN, Zhong YANG, Guishan LIU, Guangbing REN, Shou ZHANG, Wenjuan ZHU. Transcriptomic Analysis of Mechanism of Foxtail Millet Male Infertility Induced by Glyphosate Ammonium Salt [J]. Journal of Agricultural Science and Technology, 2022, 24(10): 35-43. |
| [6] | LI Shuxin, ZHANG Hao, ZHENG Housheng, ZHENG Peihe, PANG Shifeng, XU Shiquan. Transcriptome Analysis of Phenotypic Differences Between Ermaya and Changbo of Forest Cultivated Panax ginseng [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 56-68. |
| [7] | LIU Yuan, ZHANG Xiuyan, XU Miaoyun, ZHENG Hongyan, ZOU Junjie, ZHANG Lan, WANG Lei. Global Small RNA Transcriptome Profiling of Rice Under Drought Stress [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 23-32. |
| [8] | ZHANG Wenyun1, ZHANG Jiancheng2, YAO Jingzhen2*. Comparative Transcriptome Analysis of Wheat Leaf in Response to Low Nitrogen Stress#br# [J]. Journal of Agricultural Science and Technology, 2020, 22(11): 26-34. |
| [9] | CHEN Cheng1, LIU Xiaofei2, LI Qiang3, WANG Jian1, FU Rongtao1, ZHANG Hong1, LU Daihua1*. Bioinformatic Analysis of Simple Sequence Repeat (SSR) Loci in Ganoderma oregonense Transcriptome [J]. Journal of Agricultural Science and Technology, 2018, 20(7): 48-55. |
| [10] | WANG Lin1, YANG Lijun2, ZHAO Jiajia2, HUANG Haitang2, XU Zicheng1*. Application of Omics Analysis Technology in Tobacco Research [J]. Journal of Agricultural Science and Technology, 2018, 20(7): 56-62. |
| [11] | HUANG Juan, DENG Jiao, CHEN Qingfu*. Transcriptome Analysis of Fagopyrum Root and Identification of Genes Involved in Flavonoid Biosynthesis [J]. Journal of Agricultural Science and Technology, 2017, 19(2): 9-19. |
| [12] | YUE Chun-jiang1§, CHEN Chuan-chuan1§, GUO Feng-xian1, LI Hua1, SUN Hong-bo1, PEI Dan-ning1, MA Xiao-qing1, CHEN Fu-xin1, YANG Huo-li1, LI Qin1, LIU Yue1,2*. Data Mining of Simple Sequence Repeats in Transcriptome Sequences of Mongolia Medicinal Plant Artemisia frigida Willd [J]. Journal of Agricultural Science and Technology, 2016, 18(6): 31-43. |
| [13] | LI Rui-yuan1,PAN Fan2, CHEN Qing-fu2, SHI Tao-xiong2*. Excavation and Polymorphism Analysis of EST-SSR from Transcriptome of Tartary Buckwheat [J]. , 2015, 17(4): 42-52. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号