




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (8): 151-162.DOI: 10.13304/j.nykjdb.2023.0338
• ANIMAL AND PLANT HEALTH • Previous Articles
Qian ZHANG1,2( ), Lina MEN1(
), Lina MEN1( ), Yiran LI1, Qiao LIU1, Angie DENG3, Xiaowen HU1, Yuhong ZHANG2(
), Yiran LI1, Qiao LIU1, Angie DENG3, Xiaowen HU1, Yuhong ZHANG2( ), Zhiwei ZHANG1(
), Zhiwei ZHANG1( ), Wei ZHANG2
), Wei ZHANG2
Online:2024-08-15
Published:2024-08-12
Contact:
Yuhong ZHANG,Zhiwei ZHANG
About author:ZHANG Qian E-mail: 16635047582@163.com
张倩1,2( ), 门丽娜1(
), 门丽娜1( ), 李一然1, 刘巧1, 胡晓雯1, 张宇宏2(
), 李一然1, 刘巧1, 胡晓雯1, 张宇宏2( ), 张志伟1(
), 张志伟1( ), 张伟2
), 张伟2
通讯作者:
张宇宏,张志伟
基金资助:CLC Number:
Qian ZHANG, Lina MEN, Yiran LI, Qiao LIU, Angie DENG, Xiaowen HU, Yuhong ZHANG, Zhiwei ZHANG, Wei ZHANG. Differential Expression Paradigm of Chemoreceptor Genes Between Males and Females at Different Developmental Stages of Carposina sasakii Matsumura[J]. Journal of Agricultural Science and Technology, 2024, 26(8): 151-162.
张倩, 门丽娜, 李一然, 刘巧, 胡晓雯, 张宇宏, 张志伟, 张伟. 桃蛀果蛾雌雄虫不同发育时期嗅觉基因的表达水平差异[J]. 中国农业科技导报, 2024, 26(8): 151-162.
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| obp11 | AAAATGCTGTCCCTCCTGCC | GGCATAAGTCAGAGCCACCT |
| pbp2 | TGGATCCTGATGGCAAGCTG | GCCATCTTTGAAGCAGCGAG |
| pbp3 | AAACGCTGTCTCCTTGTCGG | TCCGACCTCCTTGCTTACCA |
| csp8 | GTCCGGCTCTGCAAGATGTA | AGAGGACGCCACAAATCTCG |
| or2 | CGCCTCTTCTGAACCGTCAT | CACGCTCACTCTACTCGCAT |
| or5 | GTCTCGCAGCTCCCATACAA | CATTACGGCCTCTACCGTGG |
| or14 | TGAGCATAAGATCCCGACGC | TGCTTTTTCTCGTTCACCTGT |
| or21 | AGCAGATGGAGAATCCAGCG | TCGCAACCATTCCCGTTGTA |
| or45 | GGAACAAACGCAGACCAACA | GGGATGAAAGGTGGCAGGAG |
| or48 | AAGCAGAGCAGTAAAGCGGA | TATGCCAGCGCCAAGAGATT |
| ir1 | ACGCTTTGTTGGAGTGACCA | ATATTCGGGCACGTCAGGTC |
| ir5 | CGGCAGAAAGGGAAGAGGTT | AAAGCCGGTGAGGACTAACG |
| ir7 | ACCCTGTTAGCCGCATCAAA | CCCAAGGGTCACAATCGACA |
Table 1 Primers used for RT?qPCR
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| obp11 | AAAATGCTGTCCCTCCTGCC | GGCATAAGTCAGAGCCACCT |
| pbp2 | TGGATCCTGATGGCAAGCTG | GCCATCTTTGAAGCAGCGAG |
| pbp3 | AAACGCTGTCTCCTTGTCGG | TCCGACCTCCTTGCTTACCA |
| csp8 | GTCCGGCTCTGCAAGATGTA | AGAGGACGCCACAAATCTCG |
| or2 | CGCCTCTTCTGAACCGTCAT | CACGCTCACTCTACTCGCAT |
| or5 | GTCTCGCAGCTCCCATACAA | CATTACGGCCTCTACCGTGG |
| or14 | TGAGCATAAGATCCCGACGC | TGCTTTTTCTCGTTCACCTGT |
| or21 | AGCAGATGGAGAATCCAGCG | TCGCAACCATTCCCGTTGTA |
| or45 | GGAACAAACGCAGACCAACA | GGGATGAAAGGTGGCAGGAG |
| or48 | AAGCAGAGCAGTAAAGCGGA | TATGCCAGCGCCAAGAGATT |
| ir1 | ACGCTTTGTTGGAGTGACCA | ATATTCGGGCACGTCAGGTC |
| ir5 | CGGCAGAAAGGGAAGAGGTT | AAAGCCGGTGAGGACTAACG |
| ir7 | ACCCTGTTAGCCGCATCAAA | CCCAAGGGTCACAATCGACA |
| Sample | ReadSum | BaseSum | GC/% | N/% | Q20/% | Cycle Q20/% | Q30/% |
|---|---|---|---|---|---|---|---|
| 1M | 34 008 437 | 8 564 069 432 | 45.38 | 0.00 | 91.76 | 99.60 | 85.84 |
| 2M | 31 812 028 | 8 012 439 316 | 44.85 | 0.00 | 91.98 | 99.60 | 86.20 |
| 3M | 31 090 051 | 7 829 911 646 | 45.09 | 0.00 | 91.84 | 99.60 | 86.03 |
| 1F | 31 277 367 | 7 875 117 126 | 45.14 | 0.00 | 91.85 | 99.60 | 85.99 |
| 2F | 25 584 959 | 6 443 563 720 | 45.03 | 0.00 | 92.56 | 99.60 | 86.86 |
| 3F | 25 864 927 | 6 513 965 846 | 44.78 | 0.00 | 91.67 | 99.60 | 85.55 |
Table 2 Statistical table of C. sasakii sequencing data
| Sample | ReadSum | BaseSum | GC/% | N/% | Q20/% | Cycle Q20/% | Q30/% |
|---|---|---|---|---|---|---|---|
| 1M | 34 008 437 | 8 564 069 432 | 45.38 | 0.00 | 91.76 | 99.60 | 85.84 |
| 2M | 31 812 028 | 8 012 439 316 | 44.85 | 0.00 | 91.98 | 99.60 | 86.20 |
| 3M | 31 090 051 | 7 829 911 646 | 45.09 | 0.00 | 91.84 | 99.60 | 86.03 |
| 1F | 31 277 367 | 7 875 117 126 | 45.14 | 0.00 | 91.85 | 99.60 | 85.99 |
| 2F | 25 584 959 | 6 443 563 720 | 45.03 | 0.00 | 92.56 | 99.60 | 86.86 |
| 3F | 25 864 927 | 6 513 965 846 | 44.78 | 0.00 | 91.67 | 99.60 | 85.55 |
| Length range/bp | Unigenes | Percentage of all unigenes/% |
|---|---|---|
| 200~300 | 36 325 | 42.48 |
| 301~500 | 22 047 | 25.79 |
| 501~1 000 | 12 611 | 14.75 |
| 1 001~2 000 | 7 243 | 8.47 |
| 2 000+ | 7 275 | 8.51 |
| Total number | 85 501 | |
| Total length/bp | 62 718 690 | |
| N50 length/bp | 1 515 | |
| Mean length/bp | 733.543 4 |
Table 3 Summary of data related to unigene
| Length range/bp | Unigenes | Percentage of all unigenes/% |
|---|---|---|
| 200~300 | 36 325 | 42.48 |
| 301~500 | 22 047 | 25.79 |
| 501~1 000 | 12 611 | 14.75 |
| 1 001~2 000 | 7 243 | 8.47 |
| 2 000+ | 7 275 | 8.51 |
| Total number | 85 501 | |
| Total length/bp | 62 718 690 | |
| N50 length/bp | 1 515 | |
| Mean length/bp | 733.543 4 |
| Annotated database | Unigenes | Percentage/% |
|---|---|---|
| COG annotation | 5 159 | 29.55 |
| GO annotation | 8 968 | 51.37 |
| KEGG annotation | 6 217 | 35.61 |
| KOG annotation | 9 969 | 57.11 |
| Pfam annotation | 12 127 | 69.47 |
| Swissprot_annotation | 8 825 | 50.55 |
| eggNOG annotation | 15 553 | 89.09 |
| Nr annotation | 16 753 | 95.97 |
| All annotated | 17 457 | 100.00 |
Table 4 Summary of database annotations
| Annotated database | Unigenes | Percentage/% |
|---|---|---|
| COG annotation | 5 159 | 29.55 |
| GO annotation | 8 968 | 51.37 |
| KEGG annotation | 6 217 | 35.61 |
| KOG annotation | 9 969 | 57.11 |
| Pfam annotation | 12 127 | 69.47 |
| Swissprot_annotation | 8 825 | 50.55 |
| eggNOG annotation | 15 553 | 89.09 |
| Nr annotation | 16 753 | 95.97 |
| All annotated | 17 457 | 100.00 |
| Olfactory gene | Stages team | Total | |||||
|---|---|---|---|---|---|---|---|
| 1M | 2M | 3M | 1F | 2F | 3F | ||
| obp | 38 | 38 | 34 | 39 | 39 | 37 | 40 |
| pbp | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| csp | 15 | 14 | 15 | 14 | 13 | 16 | 16 |
| or | 67 | 68 | 63 | 61 | 64 | 62 | 74 |
| orco | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ir | 11 | 12 | 10 | 11 | 14 | 12 | 14 |
| gr | 71 | 79 | 68 | 67 | 68 | 66 | 88 |
| ode | 6 | 4 | 5 | 4 | 5 | 5 | 6 |
Table 5 Differentially expressed genes related to olfaction in 3 stages of C. sasakii
| Olfactory gene | Stages team | Total | |||||
|---|---|---|---|---|---|---|---|
| 1M | 2M | 3M | 1F | 2F | 3F | ||
| obp | 38 | 38 | 34 | 39 | 39 | 37 | 40 |
| pbp | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| csp | 15 | 14 | 15 | 14 | 13 | 16 | 16 |
| or | 67 | 68 | 63 | 61 | 64 | 62 | 74 |
| orco | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ir | 11 | 12 | 10 | 11 | 14 | 12 | 14 |
| gr | 71 | 79 | 68 | 67 | 68 | 66 | 88 |
| ode | 6 | 4 | 5 | 4 | 5 | 5 | 6 |
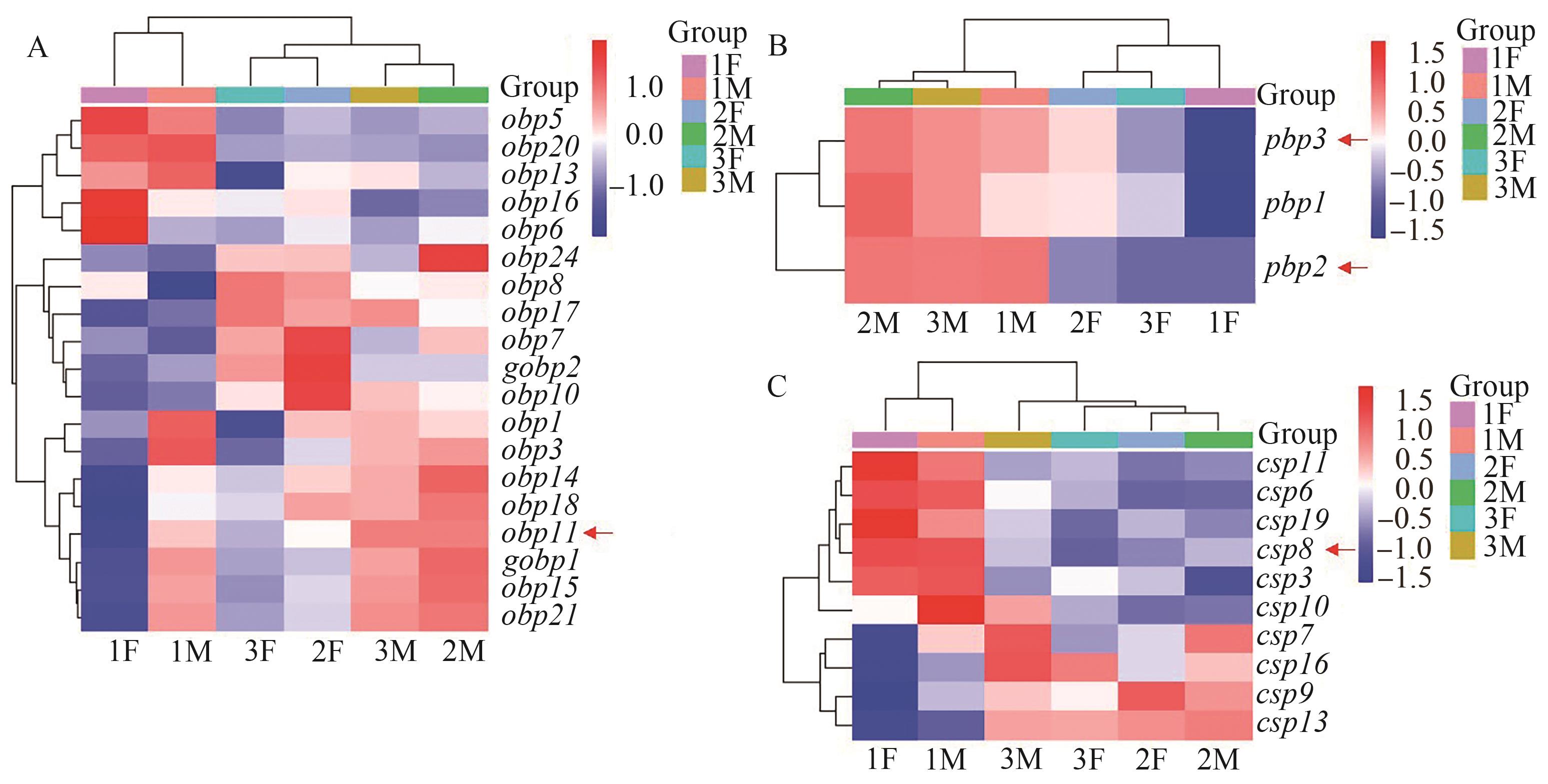
Fig. 2 Differential expression of semiochemical binding genesA: Differential expression of obps genes; B: Differential expression of pbps genes; C: Differential expression of csps genes. Red arrows show the interested differentially expressed genes

Fig. 3 Differential expression of semiochemical receptor genesA: Differential expression of ors genes; B: Differential expression of irs genes; C: Differential expression of grs genes. Red arrows show the interested differentially expressed genes
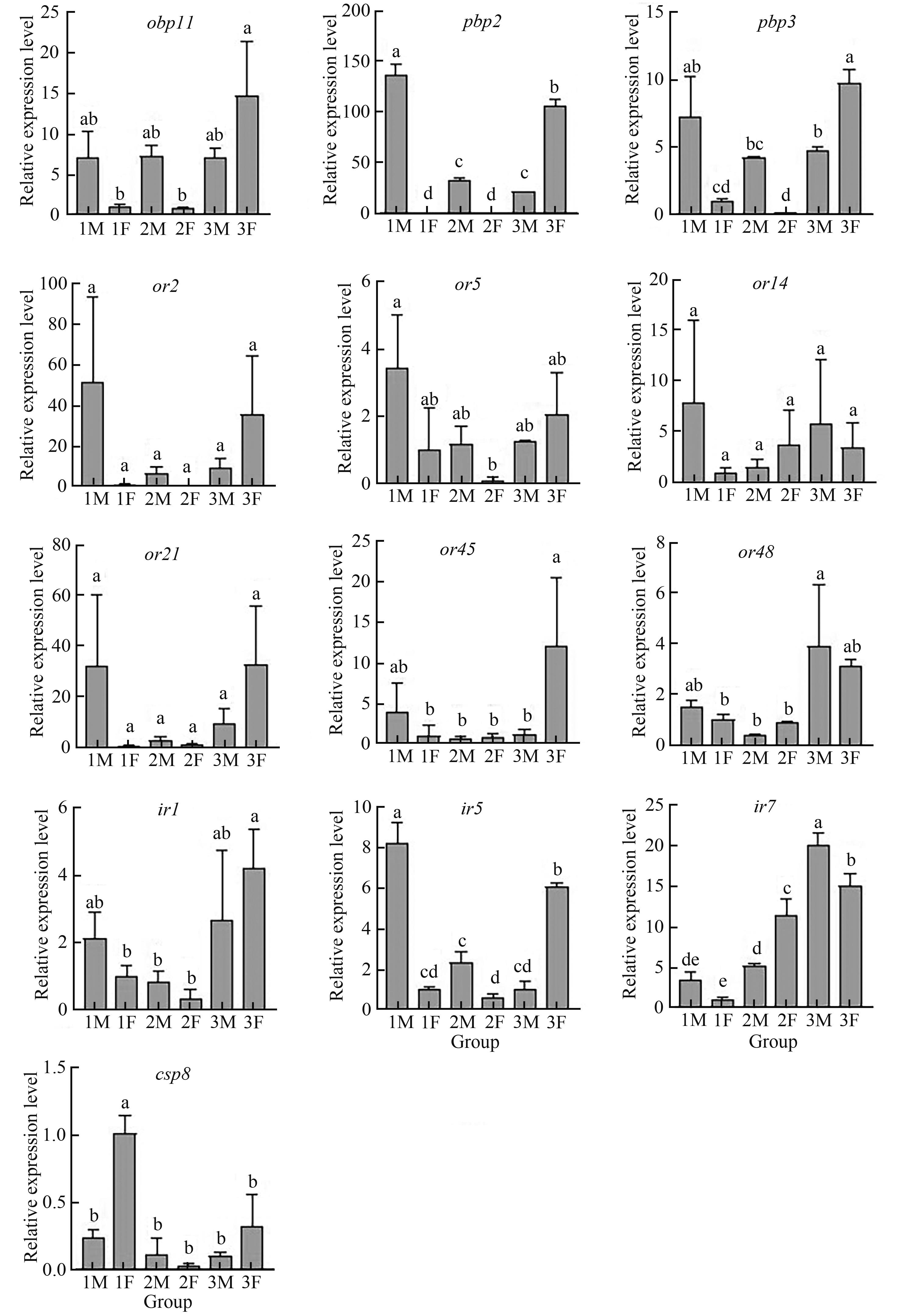
Fig. 4 Different sex and developmental stage expression profiles of 15 olfactory?related genes by RT?qPCRNote:Different lowercase letters indicate significant differences between different treatments at P<0.05 level.
| 1 | LIU B, ZHAN G P, REN L L, et al.. Toxicity of pure phosphine to Carposina sasakii Matsumura (Lepidoptera: Carposinadae) [J]. Plant Prot., 2016, 42( 6): 191- 196. |
| 2 | KIM D S, LEE J H, YIEM M S. Temperature-dependent development of Carposina sasakii (Lepidoptera: Carposinidae) and its stage emergence models [J]. Environ. Entomol., 2001, 30( 2): 298- 305. |
| 3 | HUA B Z, ZENG X H, ZHANG H. Influences of apple maturity on the development and diapause of Carposina sasakii Matsumura [J]. J. Northwest A&F Univ., 1996, 24( 6): 42- 45. |
| 4 | FENG D D, XUE Q Q, MEN L N, et al.. Demography and mass-rearing of Carposina sasakii Matsumura (Lepidoptera: Carposinidae) reared on golden delicious and red fuji apples in the laboratory [J]. J. Asia-Pac. Entomol., 2020, 23( 4): 1194- 1201. |
| 5 | ZHANG Z W, LI X W, XUE Y H, et al.. Increased trapping efficiency for the peach fruit moth Carposina sasakii (Matsumura) with synthetic sex pheromone [J]. Agric. For. Entomol., 2017, 19( 4): 424- 432. |
| 6 | YANG H B, DONG J F, SUN Y L, et al.. Antennal transcriptome analysis and expression profiles of putative chemosensory soluble proteins in Histia rhodope Cramer (Lepidoptera: Zygaenidae) [J/OL]. CBP, Part D: Genomics Proteomics, 2020, 33: 100654 [ 2023-03-25]. . |
| 7 | HU P, WANG J Z, CUI M M, et al.. Antennal transcriptome analysis of the asian longhorned beetle Anoplophora glabripennis [J/OL]. Sci. Rep., 2016, 6: 26652 [ 2023-03-25]. . |
| 8 | CHOO Y M, XU P X, HWANG J K, et al.. Reverse chemical ecology approach for the identification of an oviposition attractant for culex quinquefasciatus [J]. Proc. Natl. Acad. Sci. USA, 2018, 115: 714- 719. |
| 9 | KEPCHIA D, MOLIVER S, CHOHAN K, et al.. Inhibition of insect olfactory behavior by an airborne antagonist of the insect odorant receptor co-receptor subunit [J/OL]. PLoS One, 2017, 12( 8):e 0183009 [ 2023-03-25]. . |
| 10 | CAO S, SUN D D, LIU Y, et al.. Mutagenesis of odorant coreceptor orco reveals the distinct role of olfaction between sexes in Spodoptera frugiperda [J/OL]. J. Integr. Agric., 2022, 11: 004 [ 2023-03-25]. . |
| 11 | GRABHERR M G, HAAS B J, YASSOUR M, et al.. Full length transcriptome assembly from RNA seq data without a reference genome [J]. Nat. Biotechnol., 2011, 29: 644- 652. |
| 12 | TRAPNELL C, WILLIAMS B A, PERTEA G, et al.. Transcript assembly and quantification by RNA seq reveals unannotated transcripts and isoform switching during cell differentiation [J]. Nat. Biotechnol., 2010, 28( 5): 511- 515. |
| 13 | LI B, DEWEY C N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome [J/OL]. BMC Bioinf., 2011, 12: 323 [ 2023-03-25]. . |
| 14 | WANG E H, WANG X Q, LI H T, et al.. Effective extraction of total RNA from Carposina sasakii and the clone of the gene β⁃ actin [J]. J. Shenyang Agric. Univ., 2014, 45( 4): 403- 407. |
| 15 | SCHMITTGEN T D, LIVAK K J. Analyzing real-time PCR data by the comparative C(T) method [J]. Nat. Protocol, 2008, 3( 6): 1101- 1108. |
| 16 | LAN X N, XIANG S S, ZHU H. Research progress of the types and functions of insect antennal sensilla [J]. J. Environ. Entomol., 2023, 45( 5): 1197- 1216.. |
| 17 | CHENG J, WANG C Y, LYU Z H, et al.. Candidate olfactory genes identified in Heortia vitessoides (Lepidoptera: Crambidae) by antennal transcriptome analysis [J]. CBP, Part D: Genomics Proteomics, 2019, 29: 117- 130. |
| 18 | BRITO N F, MOREIRA M F, MELO A C. A look inside odorant-binding proteins in insect chemoreception [J]. J. Insect Physiol., 2016, 95: 51- 65. |
| 19 | LEAL W S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes [J]. Annu. Rev. Entomol., 2013, 58: 373- 391. |
| 20 | LIU N Y, ZHANG T, YE Z F, et al.. Identification and characterization of candidate chemosensory gene families from Spodoptera exigua developmental transcriptomes [J]. Int. J. Biol. Sci., 2015, 11( 9): 1036- 1048. |
| 21 | TIAN Z Q, SUN L N, LI Y Y, et al.. Antennal transcriptome analysis of the chemosensory gene families in Carposina sasakii (Lepidoptera: Carposinidae) [J/OL]. BMC Genomics, 2018, 19: 544 [ 2023-03-25]. . |
| 22 | ZHOU S S, SUN Z, MA W H, et al.. De novo analysis of the Nilaparvata lugens (Stål) antenna transcriptome and expression patterns of olfactory genes [J]. CBP, Part D: Genomics Proteomics, 2014, 9: 31- 39. |
| 23 | ANTONY B, JOHNY J, ALDOSARI S A. Silencing the odorant binding protein RferOBP1768 reduces the strong preference of palm weevil for the major aggregation pheromone compound ferrugineol [J/OL]. Front. Physiol., 2018, 9: 252 [ 2023-03-25]. . |
| 24 | ZHANG Y Y, GUO J M, WEI Z Q, et al.. Identification and sex expression profiles of olfactory-related genes in Mythimna loreyi based on antennal transcriptome analysis [J/OL]. J. Insect Sci., 2022, 25( 3): 101934 [ 2023-03-25]. . |
| 25 | LIU P J, ZHANG X F, MENG R J, et al.. Identification of chemosensory genes from the antennal transcriptome of Semiothisa cinerearia [J/OL]. PLoS One, 2020, 15( 8):e 0237134 [ 2023-03-25]. . |
| 26 | PELOSI P, IOVINELLA I, FELICIOLI A, et al.. Soluble proteins of chemical communication: an overview across arthropods [J/OL]. Front. Physiol., 2014, 5: 320 [ 2023-03-25]. . |
| 27 | PELOSI P, IOVINELLA I, ZHU J, et al.. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects [J]. Biol. Rev. Cambridge Philos. Soc., 2018, 93: 184- 200. |
| 28 | ZHANG Y, FENG K, MEI R L, et al.. Analysis of the antennal transcriptome and identification of tissue-specific expression of olfactory-related genes in Micromelalopha troglodyta (Lepidoptera: Notodontidae) [J/OL]. J. Insect Sci., 2022, 22( 5): 8 [ 2023-03-25]. . |
| 29 | HA T S, SMITH D P. Odorant and pheromone receptors in insects [J/OL]. Front. Cell. Neurosci., 2009, 3: 10 [ 2023-03-25]. . |
| 30 | WICHER D, MIAZZI F. Functional properties of insect olfactory receptors: ionotropic receptors and odorant receptors [J]. J. Cell Tissue Res., 2021, 383( 1SI): 7- 19. |
| 31 | HU P, TAO J, CUI M M, et al.. Antennal transcriptome analysis and expression profiles of odorant binding proteins in Eogystia hippophaecolus (Lepidoptera: Cossidae) [J/OL]. BMC Genomics, 2016, 17: 651 [ 2023-03-25]. . |
| 32 | NIE H, XU S P, XIE C Q, et al.. Comparative transcriptome analysis of Apis mellifera antennae of workers performing different tasks [J]. Mol. Genet. Genomics, 2018, 293: 237- 248. |
| 33 | LIU Y P, DU L X, ZHU Y, et al.. Identification and sex-biased profiles of candidate olfactory genes in the antennal transcriptome of the parasitoid wasp Cotesia vestalis [J/OL]. CBP, Part D: Genomics Proteomics, 2020, 34: 100657 [ 2023-03-25]. . |
| 34 | CLAUDIANOS C, LIM J, YOUNG M, et al.. Odor memories regulate olfactory receptor expression in the sensory periphery [J]. Eur. J. Neurosci., 2014, 39: 1642- 1654. |
| 35 | WANG S N, PENG Y, LU Z Y, et al.. Identification and expression analysis of putative chemosensory receptor genes in microplitis mediator by antennal transcriptome Screening [J]. Int. J. Biol. Sci., 2015, 11( 7): 737- 751. |
| 36 | XIA Q Y, ZHOU Z Y, LU C, et al.. A draft sequence for the genome of the domesticated silkworm ( Bombyx mori) [J]. Science, 2004, 306( 5703): 1937- 1940. |
| 37 | ZHAO H T, GAO P F, ZHANG G X, et al.. Expression and localization analysis of the odorant receptor gene Orco in drones antennae of Apis cerana cerana [J]. Sci. Agric. Sin., 2015, 48( 4): 796- 803. |
| 38 | SAXENA K N, GOYAL S. Host-plant relations of the citrus butterfly Papilio demoleus L.: Orientational and ovipositional responses [J]. Entomol. Exp. Appl., 1978, 24( 1): 1- 10. |
| 39 | RYTZ R, CROSET V, BENTON R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond Insect [J]. Insect Biochem. Mol. Biol., 2013, 43( 9): 888- 897. |
| 40 | HU J, WANG X Y, TAN L S, et al.. Identification of chemosensory genes, including candidate pheromone receptors, in Phauda flammans (Walker) (Lepidoptera: Phaudidae) through transcriptomic analyses [J/OL]. Front. Physiol., 2022, 13: 907694 [ 2023-03-25]. . |
| 41 | POUDEL S, KIM Y, KIM Y T, et al.. Gustatory receptors required for sensing umbelliferone in Drosophila melanogaster [J]. Insect Biochem. Mol. Biol., 2015, 66: 110- 118. |
| 42 | SCOTT K, BRADY R J, CRAVCHIK A, et al.. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila [J]. Cell, 2001, 104: 661- 673. |
| 43 | CHANG X Q, NIE X P, ZHANG Z, et al.. De novo analysis of the oriental armyworm Mythimna separata antennal transcriptome and expression patterns of odorant-binding proteins [J]. CBP, Part D: Genomics Proteomics, 2017, 22: 120- 130. |
| [1] | Yiwei LU, Xueyan XIA, Yu ZHAO, Jihan CUI, Meng LIU, Meihong HUANG, Cheng CHU, Jianjun LIU, Shunguo LI. Transcriptome Profiling and Gene Mining of Millet Response to Potassium Deficiency Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 30-44. |
| [2] | Jiarui XU, Yiru WANG, Shaogeng ZHAO, Kun LI, Jun ZHENG. Functional Study and Transcriptome Analysis of Corn Gene ZmCCoAOMT1 Involved in Lignin Synthesis Pathway [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 30-43. |
| [3] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [4] | Xiangwu LI, Ziyang LIU, Yujun XU, Jianbo ZHU, Yanmin WU. Explore of Molecular Mechanism on Fungal Elicitors Regulating Shikonin Synthesis [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 78-88. |
| [5] | Xiaoran WANG, Xiaoyu LI, Hui SUN, Haidong YU, Yongchun SHI. Transcriptome Analysis of Tobacco Leaves Under Boron Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 53-64. |
| [6] | Jingying JIA, Yahui LI, Bingzhe FU, Yun MA, Xiaoyan CAI. Analysis on miRs Expression Profiles of Alfalfa and Screening of Trans-border Potential miRs [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 43-53. |
| [7] | Yunsheng WANG, Yincui CHEN, Zai CHENG, Jin ZHANG, Chuanbo ZHANG. Effects of Overexpression of veA Gene on Secondary Metabolism of Eurotium cristatus [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 77-86. |
| [8] | Yuqing ZHOU, Yongfei YANG, Changwei GE, Qian SHEN, Siping ZHANG, Shaodong LIU, Huijuan MA, Jing CHEN, Ruihua LIU, Shicong LI, Xinhua ZHAO, Cundong LI, Chaoyou PANG. Identification of Cold-related Co-expression Modules in Cotton Cotyledon by WGCNA [J]. Journal of Agricultural Science and Technology, 2022, 24(4): 52-62. |
| [9] | LI Shuxin, ZHANG Hao, ZHENG Housheng, ZHENG Peihe, PANG Shifeng, XU Shiquan. Transcriptome Analysis of Phenotypic Differences Between Ermaya and Changbo of Forest Cultivated Panax ginseng [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 56-68. |
| [10] | LIU Yuan, ZHANG Xiuyan, XU Miaoyun, ZHENG Hongyan, ZOU Junjie, ZHANG Lan, WANG Lei. Global Small RNA Transcriptome Profiling of Rice Under Drought Stress [J]. Journal of Agricultural Science and Technology, 2021, 23(6): 23-32. |
| [11] | ZHANG Wenyun1, ZHANG Jiancheng2, YAO Jingzhen2*. Comparative Transcriptome Analysis of Wheat Leaf in Response to Low Nitrogen Stress#br# [J]. Journal of Agricultural Science and Technology, 2020, 22(11): 26-34. |
| [12] | CHEN Cheng1, LIU Xiaofei2, LI Qiang3, WANG Jian1, FU Rongtao1, ZHANG Hong1, LU Daihua1*. Bioinformatic Analysis of Simple Sequence Repeat (SSR) Loci in Ganoderma oregonense Transcriptome [J]. Journal of Agricultural Science and Technology, 2018, 20(7): 48-55. |
| [13] | WANG Lin1, YANG Lijun2, ZHAO Jiajia2, HUANG Haitang2, XU Zicheng1*. Application of Omics Analysis Technology in Tobacco Research [J]. Journal of Agricultural Science and Technology, 2018, 20(7): 56-62. |
| [14] | HUANG Juan, DENG Jiao, CHEN Qingfu*. Transcriptome Analysis of Fagopyrum Root and Identification of Genes Involved in Flavonoid Biosynthesis [J]. Journal of Agricultural Science and Technology, 2017, 19(2): 9-19. |
| [15] | YUE Chun-jiang1§, CHEN Chuan-chuan1§, GUO Feng-xian1, LI Hua1, SUN Hong-bo1, PEI Dan-ning1, MA Xiao-qing1, CHEN Fu-xin1, YANG Huo-li1, LI Qin1, LIU Yue1,2*. Data Mining of Simple Sequence Repeats in Transcriptome Sequences of Mongolia Medicinal Plant Artemisia frigida Willd [J]. Journal of Agricultural Science and Technology, 2016, 18(6): 31-43. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号