




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (7): 30-43.DOI: 10.13304/j.nykjdb.2024.0621
胡懿1( ), 公杰2(
), 公杰2( ), 赵玮2, 程蓉2, 柳忠玉1(
), 赵玮2, 程蓉2, 柳忠玉1( ), 高世庆2(
), 高世庆2( ), 杨亚珍1(
), 杨亚珍1( )
)
收稿日期:2024-08-02
接受日期:2024-09-18
出版日期:2025-07-15
发布日期:2025-07-11
通讯作者:
柳忠玉,高世庆,杨亚珍
作者简介:胡懿 E-mail:2813092866@qq.com基金资助:
Yi HU1( ), Jie GONG2(
), Jie GONG2( ), Wei ZHAO2, Rong CHENG2, Zhongyu LIU1(
), Wei ZHAO2, Rong CHENG2, Zhongyu LIU1( ), Shiqing GAO2(
), Shiqing GAO2( ), Yazhen YANG1(
), Yazhen YANG1( )
)
Received:2024-08-02
Accepted:2024-09-18
Online:2025-07-15
Published:2025-07-11
Contact:
Zhongyu LIU,Shiqing GAO,Yazhen YANG
摘要:
植物光敏色素(phytochrome, PHY)不仅是光感受器接收光信号,还能够作为温度传感器响应环境温度的变化。为鉴定小麦PHY基因,采用生物信息学方法在小麦全基因组中进行鉴定,并对其理化性质、系统进化、基因结构、蛋白保守结构域、启动子顺式作用元件和基因表达特性等进行综合分析。结果表明,在小麦全基因组中共鉴定出9个TaPHY基因,随机分布在小麦6条染色体上;亚细胞定位预测这些蛋白均定位于细胞核内。系统进化分析将这9个TaPHY基因划分成3个亚类,同一亚类基因的结构和蛋白保守结构域相似度较高,推测其功能相似。顺式作用元件分析显示,TaPHY基因启动子区域存在多种光响应、激素响应顺式作用元件。此外,组织表达分析发现,不同TaPHY基因的表达丰度存在显著差异。结合耐热筛选和荧光定量检测发现,在热胁迫诱导下TaPHYA1和TaPHYC2的表达量先下调后上调,其他基因则呈现不同程度的下调表达趋势,推断TaPHY基因可能参与调控小麦耐热性。相关性分析表明,在耐热材料1-2-3中,TaPHYA3的表达与H2O2含量呈显著负相关;TaPHYC1的表达与超氧化物歧化酶(superoxide dismutase,SOD)活性呈极显著正相关;TaPHYC3的表达与SOD活性呈显著正相关。上述研究结果为深入解析TaPHY参与热胁迫响应机制提供了试验依据,也为小麦耐热品系筛选提供了重要功能基因资源。
中图分类号:
胡懿, 公杰, 赵玮, 程蓉, 柳忠玉, 高世庆, 杨亚珍. 小麦PHY基因家族鉴定及热胁迫下表达分析[J]. 中国农业科技导报, 2025, 27(7): 30-43.
Yi HU, Jie GONG, Wei ZHAO, Rong CHENG, Zhongyu LIU, Shiqing GAO, Yazhen YANG. Identification of PHY Gene Family in Wheat (Triticum aestivum L.)and Their Expression Analysis Under Heat Stress[J]. Journal of Agricultural Science and Technology, 2025, 27(7): 30-43.
| 基因 Gene | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaPHYA1 | CTCTCCTGTTGGAAGTTCTGTC | CCCTGGTGCTTGATCCTAAG |
| TaPHYA2 | GACAAAGTATGGCGTCTGGG | GATCTTAGCCACTGCCATCC |
| TaPHYA3 | AGCTGCTATGCAATACTGTGG | CTTGCGCGGACATCACAAA |
| TaPHYB1 | TCTTCAGATTTGCCTGTCCTG | CTCCGCTCTGACTCTCTGAT |
| TaPHYB2 | TTCAGATTTGCCTGTCCTGG | CTGATGTACTGCACCTCACC |
| TaPHYB3 | GAAGGCATTGGACTAAGCGT | AGCTCCGTTCCCTACTTTCT |
| TaPHYC1 | CGAGCATGGTGAGGTCATTG | CACAGGACTTGCAGCACAAT |
| TaPHYC2 | ATGTGATATGCTCCTCCGGG | GGTAACACAATGCTGCACCA |
| TaPHYC3 | GGTAGATCTCGTGGAGGGTG | TGCCCTGTCAAATCTTGTGC |
| 18S rRNA | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
表1 qRT-PCR引物
Table 1 qRT-PCR primers
| 基因 Gene | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaPHYA1 | CTCTCCTGTTGGAAGTTCTGTC | CCCTGGTGCTTGATCCTAAG |
| TaPHYA2 | GACAAAGTATGGCGTCTGGG | GATCTTAGCCACTGCCATCC |
| TaPHYA3 | AGCTGCTATGCAATACTGTGG | CTTGCGCGGACATCACAAA |
| TaPHYB1 | TCTTCAGATTTGCCTGTCCTG | CTCCGCTCTGACTCTCTGAT |
| TaPHYB2 | TTCAGATTTGCCTGTCCTGG | CTGATGTACTGCACCTCACC |
| TaPHYB3 | GAAGGCATTGGACTAAGCGT | AGCTCCGTTCCCTACTTTCT |
| TaPHYC1 | CGAGCATGGTGAGGTCATTG | CACAGGACTTGCAGCACAAT |
| TaPHYC2 | ATGTGATATGCTCCTCCGGG | GGTAACACAATGCTGCACCA |
| TaPHYC3 | GGTAGATCTCGTGGAGGGTG | TGCCCTGTCAAATCTTGTGC |
| 18S rRNA | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
指标 Index | 基因名称 Gene name | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TaPHYA1 | TaPHYA2 | TaPHYA3 | TaPHYB1 | TaPHYB2 | TaPHYB3 | TaPHYC1 | TaPHYC2 | TaPHYC3 | |
基因ID号 Gene ID | TraesCS4A02G262900 | TraesCS4B02G052000 | TraesCS4D02G052200 | TraesCS4A02G122500 | TraesCS4B02G182400 | TraesCS4D02G183400 | TraesCS5A02G391300 | TraesCS5B02G396200 | TraesCS5D02G401000 |
| 染色体位置 Chromosome location | 4A:575 025 345~575 033 010(+) | 4B:40 780 124~40 786 295(-) | 4D:28 456 337~28 461 004(-) | 4A:151 945 624~151 953 947(+) | 4B:399 939 743~399 948 265(+) | 4D:320 592 499~320 600 764(-) | 5A:586 594 493~586 599 825(+) | 5B: 573 216 965~ 573 221 131(+) | 5D: 466 220 578~466 225 769(+) |
| 亚细胞定位 Subcellular localization | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus |
氨基酸数目 Number of amino acids/aa | 1 130 | 885 | 1 134 | 1 166 | 1 166 | 1 164 | 1 164 | 1 152 | 1 130 |
分子量 Molecular weight/Da | 124 881.26 | 97 690.63 | 125 617.12 | 127 679.74 | 127 695.74 | 127 593.73 | 128 636.61 | 126 939.97 | 126 015.48 |
| 理论等电点 Theoretical pI | 5.52 | 5.37 | 5.51 | 5.71 | 5.71 | 5.75 | 5.79 | 5.69 | 5.79 |
负电荷残基数 (Asp+Glu ) Number of negatively charged residues (Asp+Glu ) | 146 | 115 | 148 | 137 | 137 | 137 | 139 | 143 | 138 |
正电荷残基数 (Arg+Lys) Number of positively charged residues (Arg+Lys) | 113 | 86 | 114 | 113 | 113 | 114 | 114 | 115 | 113 |
总原子数 Total number of atoms | 17 552 | 13 707 | 17 633 | 17 898 | 17 899 | 17 894 | 18 095 | 17 802 | 17 710 |
分子式 Formula | |||||||||
不稳定指数 Instability index | 52.41 | 52.47 | 51.94 | 48.98 | 48.98 | 48.89 | 49.63 | 49.39 | 48.81 |
脂溶指数 Aliphatic index | 94.67 | 92.33 | 93.31 | 88.94 | 88.85 | 89.09 | 94.74 | 90.76 | 93.32 |
| 亲水性 GRAVY | -0.094 | -0.101 | -0.113 | -0.127 | -0.130 | -0.127 | -0.122 | -0.209 | -0.147 |
表2 小麦PHY基因家族蛋白理化性质及亚细胞定位预测
Table 2 Prediction of physicochemical properties and subcellular localization of wheat PHY gene family proteins
指标 Index | 基因名称 Gene name | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TaPHYA1 | TaPHYA2 | TaPHYA3 | TaPHYB1 | TaPHYB2 | TaPHYB3 | TaPHYC1 | TaPHYC2 | TaPHYC3 | |
基因ID号 Gene ID | TraesCS4A02G262900 | TraesCS4B02G052000 | TraesCS4D02G052200 | TraesCS4A02G122500 | TraesCS4B02G182400 | TraesCS4D02G183400 | TraesCS5A02G391300 | TraesCS5B02G396200 | TraesCS5D02G401000 |
| 染色体位置 Chromosome location | 4A:575 025 345~575 033 010(+) | 4B:40 780 124~40 786 295(-) | 4D:28 456 337~28 461 004(-) | 4A:151 945 624~151 953 947(+) | 4B:399 939 743~399 948 265(+) | 4D:320 592 499~320 600 764(-) | 5A:586 594 493~586 599 825(+) | 5B: 573 216 965~ 573 221 131(+) | 5D: 466 220 578~466 225 769(+) |
| 亚细胞定位 Subcellular localization | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus |
氨基酸数目 Number of amino acids/aa | 1 130 | 885 | 1 134 | 1 166 | 1 166 | 1 164 | 1 164 | 1 152 | 1 130 |
分子量 Molecular weight/Da | 124 881.26 | 97 690.63 | 125 617.12 | 127 679.74 | 127 695.74 | 127 593.73 | 128 636.61 | 126 939.97 | 126 015.48 |
| 理论等电点 Theoretical pI | 5.52 | 5.37 | 5.51 | 5.71 | 5.71 | 5.75 | 5.79 | 5.69 | 5.79 |
负电荷残基数 (Asp+Glu ) Number of negatively charged residues (Asp+Glu ) | 146 | 115 | 148 | 137 | 137 | 137 | 139 | 143 | 138 |
正电荷残基数 (Arg+Lys) Number of positively charged residues (Arg+Lys) | 113 | 86 | 114 | 113 | 113 | 114 | 114 | 115 | 113 |
总原子数 Total number of atoms | 17 552 | 13 707 | 17 633 | 17 898 | 17 899 | 17 894 | 18 095 | 17 802 | 17 710 |
分子式 Formula | |||||||||
不稳定指数 Instability index | 52.41 | 52.47 | 51.94 | 48.98 | 48.98 | 48.89 | 49.63 | 49.39 | 48.81 |
脂溶指数 Aliphatic index | 94.67 | 92.33 | 93.31 | 88.94 | 88.85 | 89.09 | 94.74 | 90.76 | 93.32 |
| 亲水性 GRAVY | -0.094 | -0.101 | -0.113 | -0.127 | -0.130 | -0.127 | -0.122 | -0.209 | -0.147 |
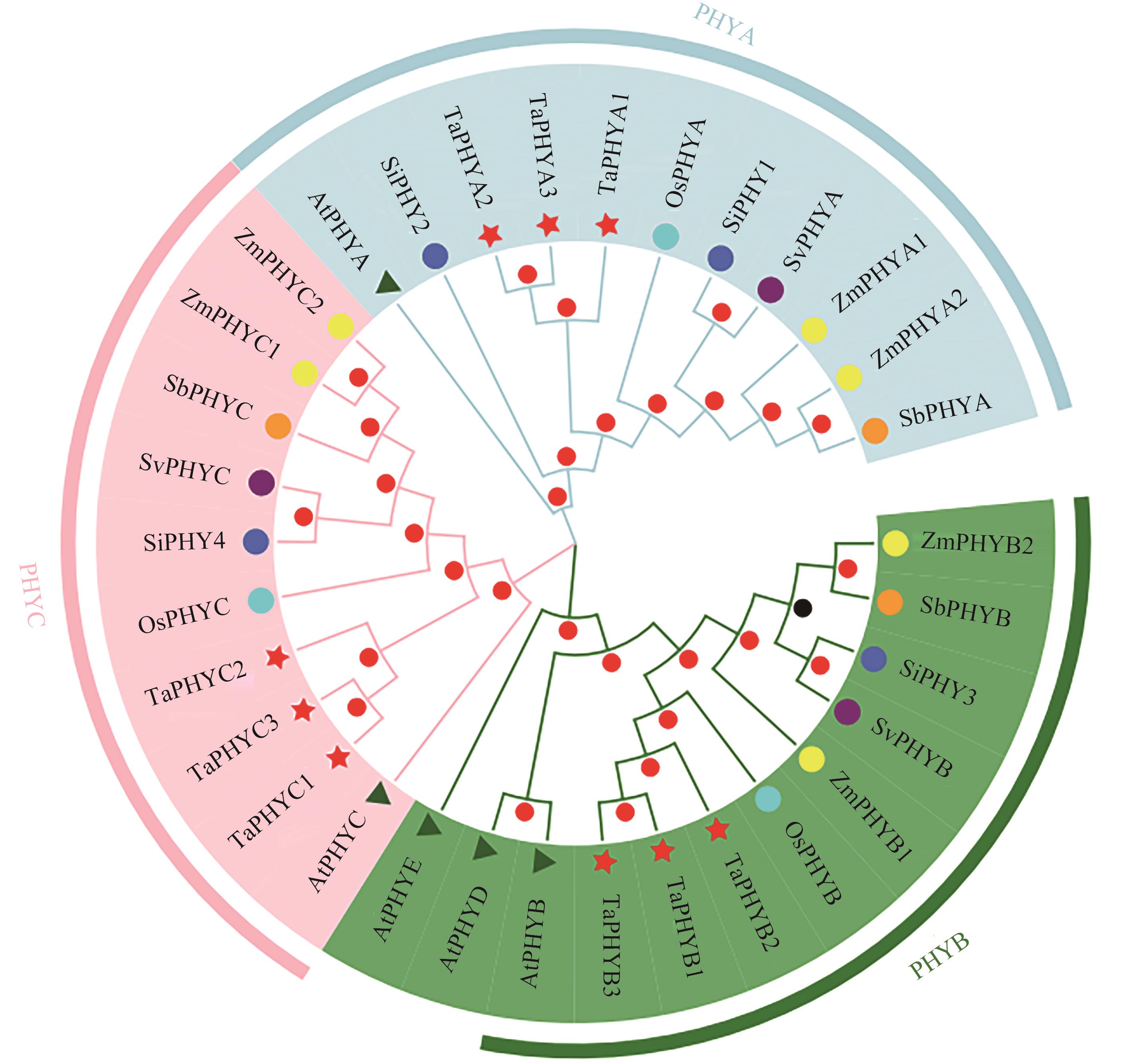
图2 TaPHY蛋白质系统进化分析注:At—拟南芥; Os—水稻; Ta—小麦; Zm—玉米; Si—谷子; Sb—高粱; Sv—狗尾草。
Fig. 2 Phylogeny analysis of TaPHY proteinsNote:At—Arabidopsis thaliana; Os—Oryza sativa L.; Ta—Triticum aestivum L.; Zm—Zea mays L.; Si —Setaria italica L.; Sb—Sorghum bicolor L.; Sv—Setaria viridis L.
蛋白名称 Protein name | α螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β转角 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| TaPHYA1 | 48.05 | 14.16 | 5.49 | 32.30 |
| TaPHYA2 | 45.54 | 14.24 | 5.65 | 34.58 |
| TaPHYA3 | 47.80 | 14.11 | 5.20 | 32.89 |
| TaPHYB1 | 46.91 | 13.89 | 5.40 | 33.79 |
| TaPHYB2 | 46.57 | 14.24 | 4.89 | 34.31 |
| TaPHYB3 | 47.16 | 14.60 | 5.07 | 33.16 |
| TaPHYC1 | 46.13 | 16.07 | 5.24 | 32.56 |
| TaPHYC2 | 45.49 | 13.54 | 5.56 | 35.42 |
| TaPHYC3 | 47.50 | 14.22 | 5.88 | 32.40 |
表 3 小麦 TaPHY 蛋白的二级结构及其占比
Table 3 Secondary structure of TaPHY proteins ant its percentage
蛋白名称 Protein name | α螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β转角 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| TaPHYA1 | 48.05 | 14.16 | 5.49 | 32.30 |
| TaPHYA2 | 45.54 | 14.24 | 5.65 | 34.58 |
| TaPHYA3 | 47.80 | 14.11 | 5.20 | 32.89 |
| TaPHYB1 | 46.91 | 13.89 | 5.40 | 33.79 |
| TaPHYB2 | 46.57 | 14.24 | 4.89 | 34.31 |
| TaPHYB3 | 47.16 | 14.60 | 5.07 | 33.16 |
| TaPHYC1 | 46.13 | 16.07 | 5.24 | 32.56 |
| TaPHYC2 | 45.49 | 13.54 | 5.56 | 35.42 |
| TaPHYC3 | 47.50 | 14.22 | 5.88 | 32.40 |
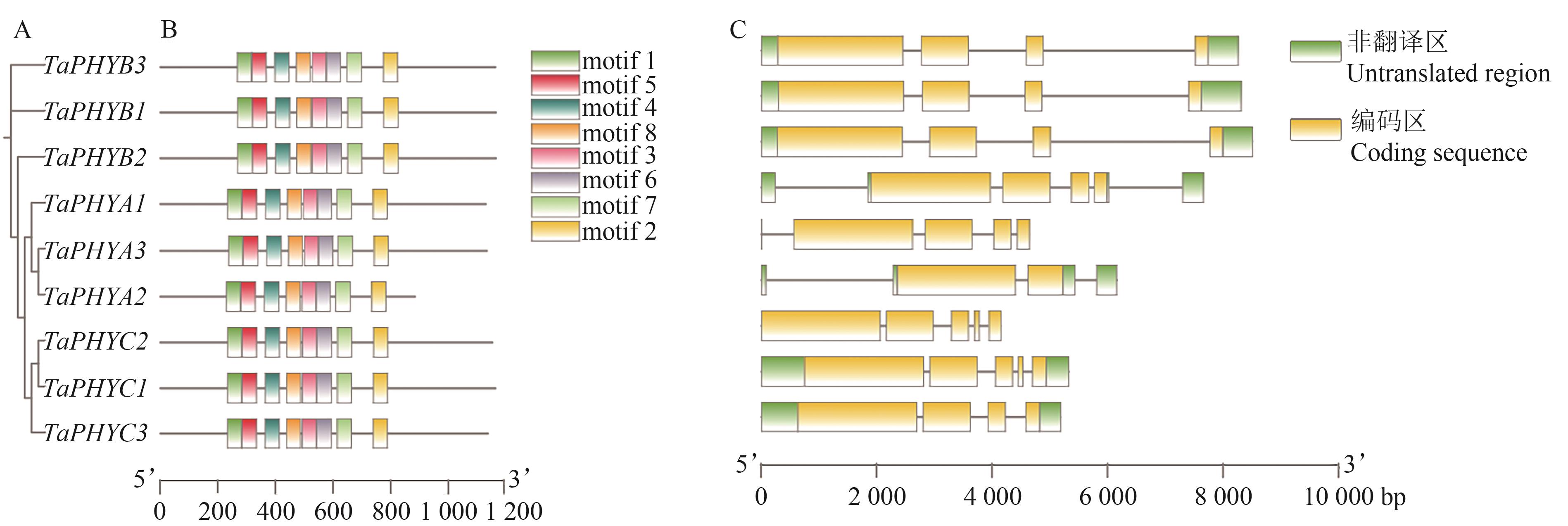
图4 小麦TaPHY基因结构和保守基序A:系统进化;B:保守结构域;C:基因结构
Fig. 4 Gene structure and conserved motifs of wheat TaPHY genesA: Phylogeny; B: Conserved domain; C: Gene structure

图6 TaPHY表达数据库热图分析A~C:基于WheatOmics 1.0网站的转录组数据,Z10—1叶期,Z13—3叶期,Z23—分蘖早期,Z30—起身期,Z32—拔节早期,Z39—拔节晚期,Z65—开花中期,Z71—开花后2 d,Z75—开花后10 d,Z85—开花后30 d;D~F:基于Wheat Expression Browser数据库,S1—幼苗期的根,S2—幼苗期的茎和叶,V1—营养生长期的茎和叶,V2—营养生长期的根,V3—营养生长期的穗,R1—生殖生长期的根和叶,R2—生殖生长期的根,R3—生殖生长期的穗,R4—生殖生长期的籽粒
Fig. 6 TaPHY expression database heat map analysisA~C: Based on transcriptome data of WheatOmics 1.0, Z10—1 leaf stage, Z13—3 leaves stage, Z23—Early tillering stage, Z30—Standing stage, Z32—Early jointing stage, Z39—Late jointing stage, Z65—Middle flowering stage, Z71—2 d after flowering, Z75—10 d after flowering, Z85—30 d after flowering; D~F: Based on wheat expression browser database, S1—Roots in seedling stage, S2—Stems and leaves in seedling stage, V1—Stems and leaves in vegetative stage, V2—Roots in vegetative stage, V3—Panicles in vegetative stage, R1—Roots and leaves in reproductive growth stage, R2—Roots in reproductive growth stage, R3—Panicles in reproductive growth stage, R4—Grain in reproductive growth stage
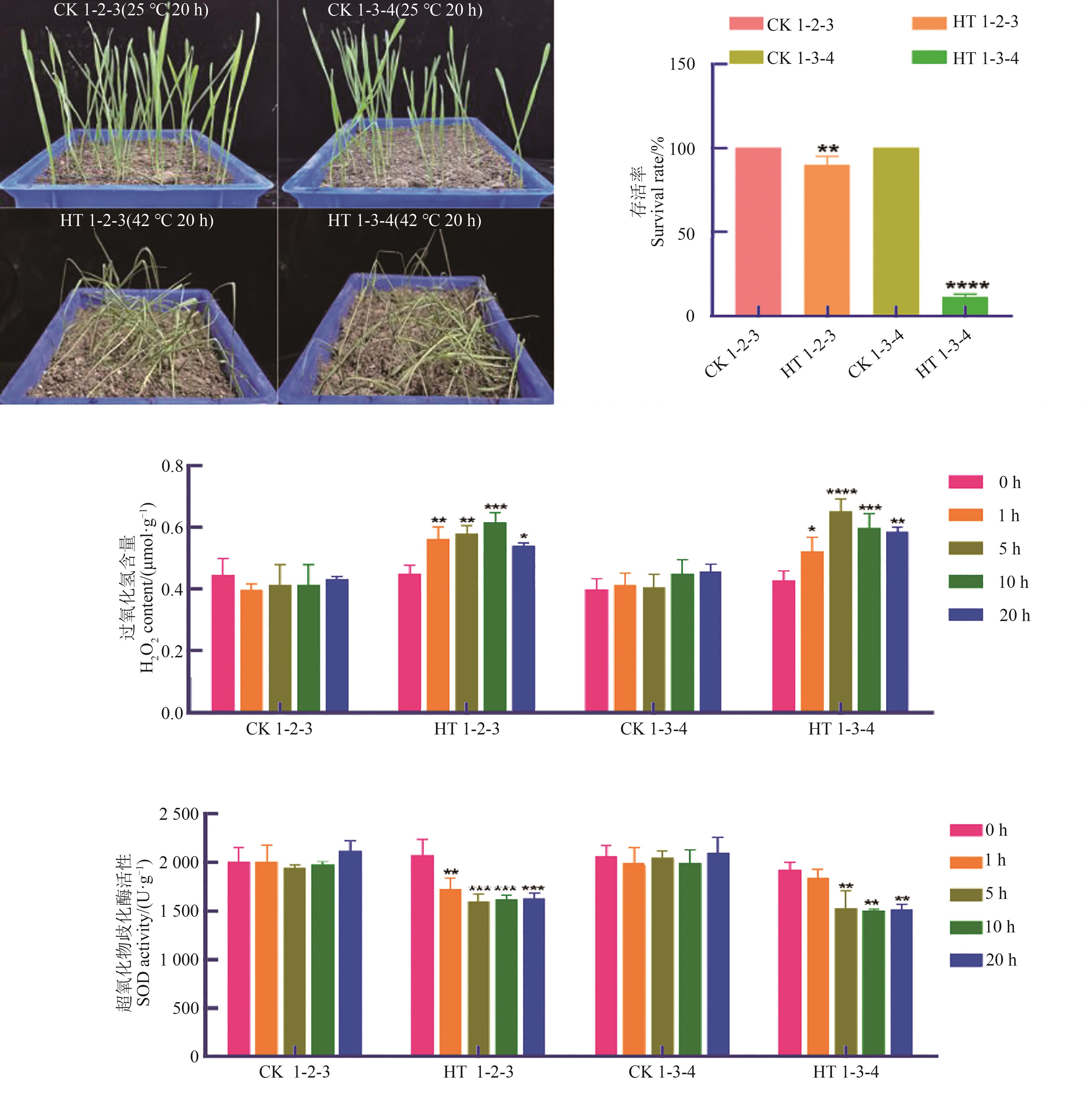
图7 1-2-3和1-3-4热胁迫处理后的存活率和生理指标注:*、**、***和****分别表示在P<0.05、P<0.01、P<0.001和P<0.000 1水平差异显著。
Fig. 7 Survival rate and physiological indexes of 1-2-3 and 1-3-4 under heat stressNote:*,**, *** and **** indicate significant at P<0.05,P<0.01, P<0.001 and P<0.000 1 levels, respectively.

图8 TaPHY基因不同处理下的相对表达量注:不同小写字母表示不同时间在P<0.05水平差异显著。
Fig. 8 Relative expression of TaPHY genes under the heat stress conditionNote:Different lowercase letters indicate significant differences between different times at P<0.05 level.
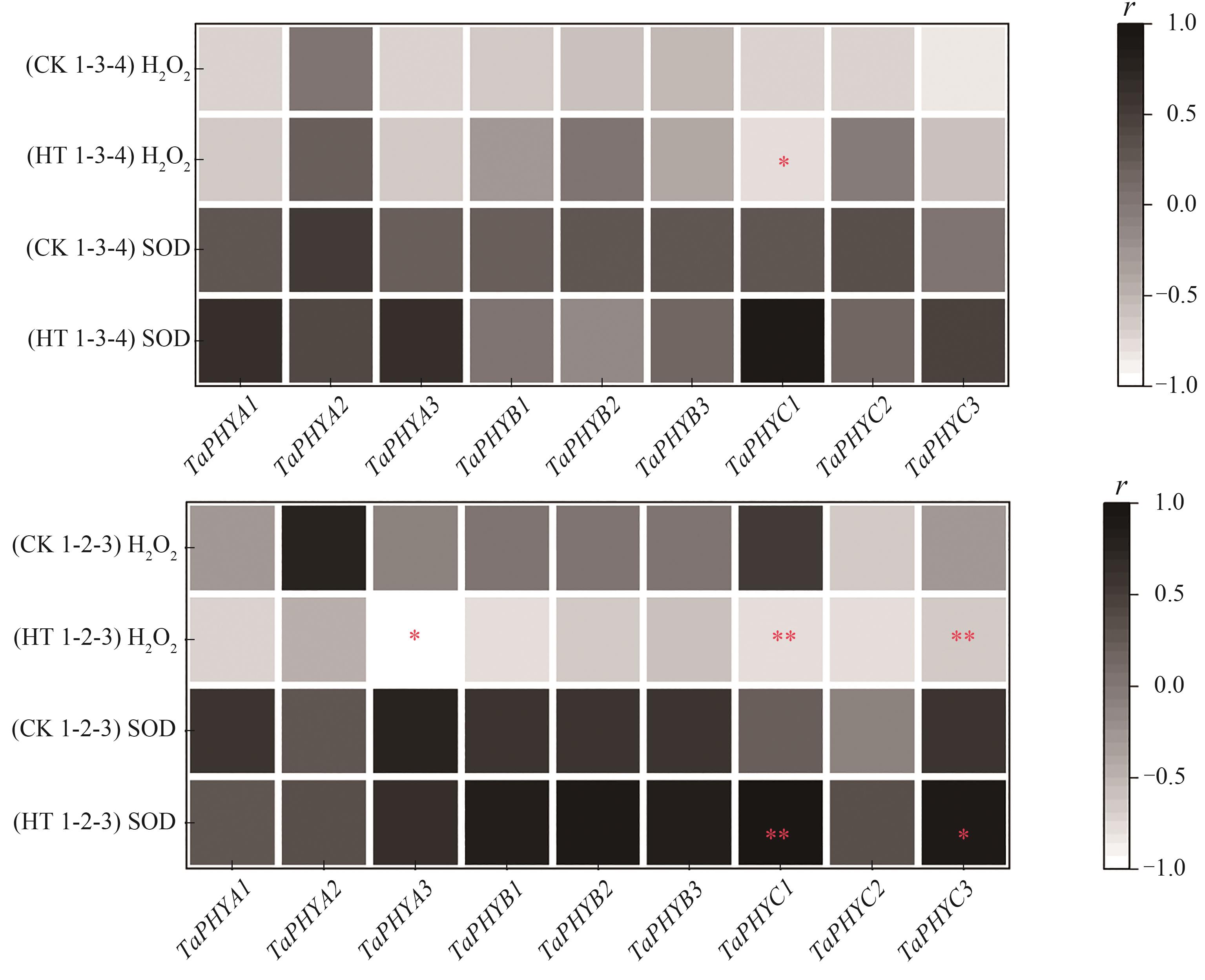
图9 H2O2含量、SOD活性与TaPHYs基因表达的相关性分析注:*和**分别表示在P<0.05和P<0.01水平相关显著。
Fig. 9 Correlation analysis of H2O2 content, SOD activity and relative expression of TaPHYsNote:* and ** indicate significant correlations at P<0.05 and P<0.01 levels, respectively.
| [1] | 王富刚, 张静, 张雄, 等. 光敏色素与植物的光形态建成 [J]. 基因组学与应用生物学, 2017, 36(8):3167-3171. |
| WANG F G, ZHANG J, ZHANG X, et al.. Phytochromes and plant photomorphogenesis [J]. Genomics Appl. Biol., 2017, 36(8):3167-3171. | |
| [2] | 王静,王艇.高等植物光敏色素的分子结构、生理功能和进化特征[J].植物学通报,2007,24(5):649-658. |
| WANG J, WANG T. Molecular structure,physiological function and evolution of phytochrome in higher plants [J]. Chin. Bull.Bot., 2007,24(5):649-658. | |
| [3] | SONG B, ZHAO H, DONG K,et al..Phytochrome A inhibits shade avoidance responses under strong shade through repressing the brassinosteroid pathway in Arabidopsis [J]. Plant J., 2020,104(6):1520-1534. |
| [4] | KIPPES N, VANGESSEL C, HAMILTON J, et al.. Effect of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods [J/OL]. BMC Plant Biol., 2020,20(1):197 [2024-07-10].. |
| [5] | XU P, LIAN H, XU F,et al..Phytochrome B and AGB1 coordinately regulate photomorphogenesis by antagonistically modulating PIF3 stability in Arabidopsis [J]. Mol. Plant, 2019, 12(2):229-247. |
| [6] | YANG L, JIANG Z, JING Y,et al..PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis [J].J.Integr.Plant Biol.,2020,62(9):1372-1384. |
| [7] | LIU S, YANG L, LI J, et al.. FHY3 interacts with phytochrome B and regulates seed dormancy and germination [J]. Plant Physiol., 2021,187(1):289-302. |
| [8] | 张芳,张晓枫,王进征,等.植物光敏色素PHYA、PHYB研究进展[J].生物学通报,2011,46(1):11-14. |
| ZHANG F, ZHANG X F, WANG J Z, et al.. Research advances on the phytochromes A and B [J]. Bull. Biol., 2011,46(1):11-14. | |
| [9] | SONG J, LIU Q, HU B, et al.. Photoreceptor PhyB involved in Arabidopsis temperature perception and heat-tolerance formation [J/OL]. Int. J. Mol. Sci., 2017,18(6):E1194 [2024-07-10]. . |
| [10] | JIANG B C, SHI Y T, PENG Y, et al.. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by sstabilizing the phyB thermosensor in Arabidopsis [J]. Mol. Plant, 2020, 13(6):894-906. |
| [11] | LEGRIS M, KLOSE C, BURGIE E S, et al.. Phytochrome B integrates light and temperature signals in Arabidopsis [J].Science, 2016,354(6314):897-900. |
| [12] | JUNG J H, DOMIJAN M, KLOSE C, et al.. Phytochromes function as thermosensors in Arabidopsis [J]. Science, 2016,354(6314):886-889. |
| [13] | HE Y, LI Y, CUI L, et al.. Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in rice [J/OL].Front. Plant Sci., 2016,7:1963 [2024-07-10]. . |
| [14] | GAO Y, JIANG W, DAI Y,et al..A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice [J]. Plant Mol. Biol., 2015,87(4/5):413-428. |
| [15] | 李敏, 苏慧, 李阳阳, 等. 黄淮海麦区小麦耐热性分析及其鉴定指标的筛选 [J]. 中国农业科学, 2021, 54 (16): 3381-3393. |
| LI M, SU H, LI Y Y, et al.. Analysis of heat tolerance of wheat with different genotypes and screening of identification indexes in Huang-Huai-Hai region [J]. Sci. Agric. Sin., 2021, 54 (16): 3381-3393. | |
| [16] | REI S, MICAEL P, VIEIRA C, et al..Fold change in regucalcin expression after chill-coma recovery (ChCR) obtained by qRT-PCR using the 2-ΔΔCT method [J]. Methods, 2001, 25 (4):402-408. |
| [17] | 王璐璐. 玉米光敏色素基因的克隆与苗期光暗条件下表达分析 [D]. 沈阳:沈阳农业大学, 2017. |
| WANG L L. The cloning of phytochrome gene in maize and the expression analysis in the light and dark conditionsat the seeding stage [D]. Shenyang: Shenyang Agricultural University, 2017. | |
| [18] | AUKERMAN M J, HIRSCHFELD M, WESTER L,et al..A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing [J]. Plant Cell,1997,9(8):1317-1326. |
| [19] | 李建平,杨建平,宋梅芳,等.拟南芥光敏色素D(AtphyD)在不同光质下的表达特性分析[J].新疆农业科学,2014,51(2):305-310. |
| LI J P, YANG J P, SONG M F,et al..Differential patterns of expression of the Arabidopsis phytochrome D under different light conditions [J]. Xinjiang Agric. Sci., 2014,51(2):305-310. | |
| [20] | 陈由, 王亚琴. 水稻光敏色素 A基因克隆及其在光形态建成中的作用 [J]. 华南师范大学学报(自然科学版), 2014, 46(1):71-76. |
| CHEN Y, WANG Y Q. Cloning and Function Analysis of rice Phytochrome A gene in photomorphogenesis [J]. J. South China Norm. Univ. (Nat. Sci.), 2014, 46(1):71-76. | |
| [21] | 丁武思,陈士瞻,刘磊,等.2个玉米光敏色素C基因的克隆及功能验证[J].河南农业科学,2021,50(1):16-26. |
| DING W S, CHEN S Z, LIU L,et al..Cloning and function verification of two phytochrome C genes in Zea mays [J]. J.Henan Agric. Sci., 2021,50(1):16-26. | |
| [22] | SÁNCHEZ-LAMAS M, LORENZO C D, CERDÁN P D.Bottom-up assembly of the phytochrome network [J/OL]. PLoS Genet., 2016,12(11):e1006413 [2024-07-10]. . |
| [23] | CHEN A, LI C X, HU W, et al.. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod [J]. Proc. Natl. Acad. Sci. USA, 2014,111(28):10037-10044. |
| [24] | PEARCE S, KIPPES N, CHEN A, et al.. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways [J/OL]. BMC Plant Biol., 2016,16(1):141 [2024-07-10]. . |
| [25] | 李丹阳, 张晓花, 刘灵芝, 等. 光敏色素互作因子OsPIL15基因对水稻磷、氮吸收利用的影响[J].河南农业科学,2021,50(5):24-31. |
| LI D Y, ZHANG X H, LIU L Z, et al.. Effects of phytochrome interacting factor OsPIL15 gene on absorption and utilization of phosphorus and nitrogen in rice [J]. J. Henan Agric. Sci., 2021, 50(5):24-31. | |
| [26] | SUN J J, LU J, BAI M J, et al.. Phytochrome-interacting factors interact with transcription factor CONSTANS to suppress flowering in rose [J].Plant Physiol.,2021,186(2):1186-1201. |
| [27] | QIU Y J, LI M N, KIM R J A,et al..Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA [J/OL].Nat.Commun., 2019,10(1):140 [2024-07-10]. . |
| [28] | BURKO Y, WILLIGE B C, SELUZICKI A, et al.. PIF7 is a master regulator of thermomorphogenesis in shade [J/OL].Nat. Commun.,2022,13(1):4942 [2024-07-10]. . |
| [1] | 朱强, 车宗贤, 崔恒, 张久东, 包兴国. 绿肥替代氮肥对麦田温室气体的影响[J]. 中国农业科技导报, 2025, 27(7): 182-189. |
| [2] | 呼斯乐, 包玉龙, 图布新巴雅尔null, 陶际峰, 郭恩亮. 基于无人机高光谱和集成学习的春小麦叶绿素含量反演[J]. 中国农业科技导报, 2025, 27(6): 93-103. |
| [3] | 史硕, 冯宇, 李亮, 孟瑞, 章延泽, 杨秀荣. 印度梨形孢介导小麦抗纹枯病的转录组分析及关键基因筛选[J]. 中国农业科技导报, 2025, 27(5): 133-145. |
| [4] | 马蓓, 公杰, 杜银柯, 甘雨薇, 程蓉, 朱波, 易丽霞, 马锦绣, 高世庆. 小麦花粉孔发育相关TaINP1基因鉴定及表达分析[J]. 中国农业科技导报, 2025, 27(4): 22-35. |
| [5] | 薛振宇, 张康康, 张元元, 闫强强, 姚立蓉, 张宏, 孟亚雄, 司二静, 李葆春, 马小乐, 王化俊, 汪军成. 优质抗旱小麦种质的筛选及功能基因检测[J]. 中国农业科技导报, 2025, 27(1): 35-49. |
| [6] | 孙宪印, 牟秋焕, 米勇, 吕广德, 亓晓蕾, 孙盈盈, 尹逊栋, 王瑞霞, 吴科, 钱兆国, 赵岩, 高明刚. 基于GT双标图对小麦新品系的分类评价[J]. 中国农业科技导报, 2024, 26(7): 14-24. |
| [7] | 鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21. |
| [8] | 刘萌, 林辰壹, 吴瑞, 曹爽, 梁志豪, 张若楠. 香菇菌丝热胁迫响应及耐热综合评价[J]. 中国农业科技导报, 2024, 26(5): 90-100. |
| [9] | 赵刚, 王淑英, 李尚中, 张建军, 党翼, 王磊, 李兴茂, 程万莉, 周刚, 倪胜利, 樊廷录. 黄土旱塬区近40年降水对冬小麦耗水和产量的影响[J]. 中国农业科技导报, 2024, 26(3): 164-173. |
| [10] | 张宏, 李卫国, 张晓东, 卢必慧, 张琤琤, 李伟, 马廷淮. 基于HJ-1星和GF-1号影像融合特征提取冬小麦种植面积[J]. 中国农业科技导报, 2024, 26(2): 109-119. |
| [11] | 张景云, 关峰, 石博, 万新建. 小麦根系分泌物对苦瓜幼苗生长及土壤生物学环境的影响[J]. 中国农业科技导报, 2024, 26(2): 181-190. |
| [12] | 李双, 王爱英, 焦浈, 池青, 孙昊, 焦涛. 盐胁迫下不同抗性小麦幼苗生理生化特性及转录组分析[J]. 中国农业科技导报, 2024, 26(2): 20-32. |
| [13] | 刘海霞, 张寅辉, 庄蕾, 郭梦娇, 赵李, 吴美娟, 侯健, 李甜, 刘红霞, 张学勇, 郝晨阳. 基于关联分析挖掘小麦SDS沉降值相关候选基因及KASP标记开发[J]. 中国农业科技导报, 2024, 26(12): 18-29. |
| [14] | 胡墨明, 孟琰珺, 苗淑媛, 胡西宁, 许晴, 马红珍, 王鹏飞, 刘国芹, 康国章. 小麦淀粉合成关键基因TaAGPL1功能标记的开发[J]. 中国农业科技导报, 2024, 26(11): 43-55. |
| [15] | 田鹏, 宋洁, 李霞, 王裕智, 武棒棒. 不同粒色小麦籽粒中叶酸及衍生物含量分析[J]. 中国农业科技导报, 2024, 26(11): 56-65. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号