




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (3): 49-59.DOI: 10.13304/j.nykjdb.2024.0111
李娜( ), 张华, 邢馨竹, 邵振启, 杨占武, 李喜焕(
), 张华, 邢馨竹, 邵振启, 杨占武, 李喜焕( ), 张彩英
), 张彩英
收稿日期:2024-02-12
接受日期:2024-03-28
出版日期:2025-03-15
发布日期:2025-03-14
通讯作者:
李喜焕
作者简介:李娜 E-mail: 1784437078@qq.com;
基金资助:
Na LI( ), Hua ZHANG, Xinzhu XING, Zhenqi SHAO, Zhanwu YANG, Xihuan LI(
), Hua ZHANG, Xinzhu XING, Zhenqi SHAO, Zhanwu YANG, Xihuan LI( ), Caiying ZHANG
), Caiying ZHANG
Received:2024-02-12
Accepted:2024-03-28
Online:2025-03-15
Published:2025-03-14
Contact:
Xihuan LI
摘要:
扩展蛋白是一类通过松弛细胞壁释放胞内膨胀压力使植物细胞得以扩张的蛋白,在植物生长发育以及抵御各种逆境胁迫中发挥重要功能。然而,有关扩展蛋白在荚果发育中的作用尚不明确,鉴于此,克隆大豆扩展蛋白基因GmEXLA1,并研究其在荚果发育中的功能。结果发现,GmEXLA1在大豆荚粒中优势表达,开放阅读框长度780 bp,编码259个氨基酸,其中第1~19位氨基酸属于信号肽序列,具有扩展蛋白发挥其生物学功能所必需的2个保守结构域;烟草叶片亚细胞定位发现,编码蛋白GmEXLA1位于植物细胞壁;进一步分析发现,GmEXLA1超表达极显著增加转基因拟南芥的果荚长度、宽度和种子重量,其中种子重量的增加幅度为61.2%~80.6%;而GmEXLA1突变则显著降低大豆的荚长、粒长、粒宽和粒重等性状,其中粒长、粒宽和粒重的降低幅度分别为13.5%、7.0%和25.6%,说明GmEXLA1在植物荚果发育过程中发挥重要作用。另外,分析不同大豆品种资源GmEXLA1等位变异发现15个SNPs,其中位于上游的SNP(A/G)在野生大豆与栽培大豆中的分布频率(A85% vs A38%;G15% vs G62%)存在极显著差异。研究结果为通过生物育种手段改良大豆产量性状提供了重要的扩展蛋白基因。
中图分类号:
李娜, 张华, 邢馨竹, 邵振启, 杨占武, 李喜焕, 张彩英. 大豆扩展蛋白基因GmEXLA1在荚果发育中的功能鉴定[J]. 中国农业科技导报, 2025, 27(3): 49-59.
Na LI, Hua ZHANG, Xinzhu XING, Zhenqi SHAO, Zhanwu YANG, Xihuan LI, Caiying ZHANG. Function Analysis of Soybean Expansin Gene GmEXLA1 inPlantPod and Seed Development[J]. Journal of Agricultural Science and Technology, 2025, 27(3): 49-59.
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) | 引物用途 Primer usage |
|---|---|---|
| GmEXLA1-F | GCAGTCGACATGGCTTTCTTTATTCTCTCC | 扩增开放阅读框;检测转基因拟南芥;检测大豆突变体 Open reading frame amplification; detection of transgenic Arabidopsis and soybean mutant |
| GmEXLA1-R | TATGGATCCTGTCCCATCATCGCAGGG | |
| GmEXLA1-RT-F1 | TGGCCGTTCGAGTTGAAGAA | 转基因拟南芥实时定量分析 Real-time quantitative analysis of transgenic Arabidopsis |
| GmEXLA1-RT-R1 | ATACTGCCCCGTGGTTTCTG | |
| GmEXLA1-Mu-F | GCTGGATCCATGTGTGATCGCTGCT | gmexla1突变体目的基因原核表达 Prokaryotic expression of target gene in gmexla1 mutant |
| GmEXLA1-Mu-R | TTAGAGCTCTCATGTCCCATCATCGCAG | |
| GmEXLA1-RT-F2 | ACTGAAGCTTGGCATTGCTG | gmexla1突变体实时定量分析 Real-time quantitative analysis of gmexla1 mutant |
| GmEXLA1-RT-R2 | CATACTGCCCCGTGGTTTCT | |
| GmActin11-F | ATCTTGACTGAGCGTGGTTATTCC | |
| GmActin11-R | GCTGGTCCTGGCTGTCTCC |
表1 本研究所用引物序列
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) | 引物用途 Primer usage |
|---|---|---|
| GmEXLA1-F | GCAGTCGACATGGCTTTCTTTATTCTCTCC | 扩增开放阅读框;检测转基因拟南芥;检测大豆突变体 Open reading frame amplification; detection of transgenic Arabidopsis and soybean mutant |
| GmEXLA1-R | TATGGATCCTGTCCCATCATCGCAGGG | |
| GmEXLA1-RT-F1 | TGGCCGTTCGAGTTGAAGAA | 转基因拟南芥实时定量分析 Real-time quantitative analysis of transgenic Arabidopsis |
| GmEXLA1-RT-R1 | ATACTGCCCCGTGGTTTCTG | |
| GmEXLA1-Mu-F | GCTGGATCCATGTGTGATCGCTGCT | gmexla1突变体目的基因原核表达 Prokaryotic expression of target gene in gmexla1 mutant |
| GmEXLA1-Mu-R | TTAGAGCTCTCATGTCCCATCATCGCAG | |
| GmEXLA1-RT-F2 | ACTGAAGCTTGGCATTGCTG | gmexla1突变体实时定量分析 Real-time quantitative analysis of gmexla1 mutant |
| GmEXLA1-RT-R2 | CATACTGCCCCGTGGTTTCT | |
| GmActin11-F | ATCTTGACTGAGCGTGGTTATTCC | |
| GmActin11-R | GCTGGTCCTGGCTGTCTCC |
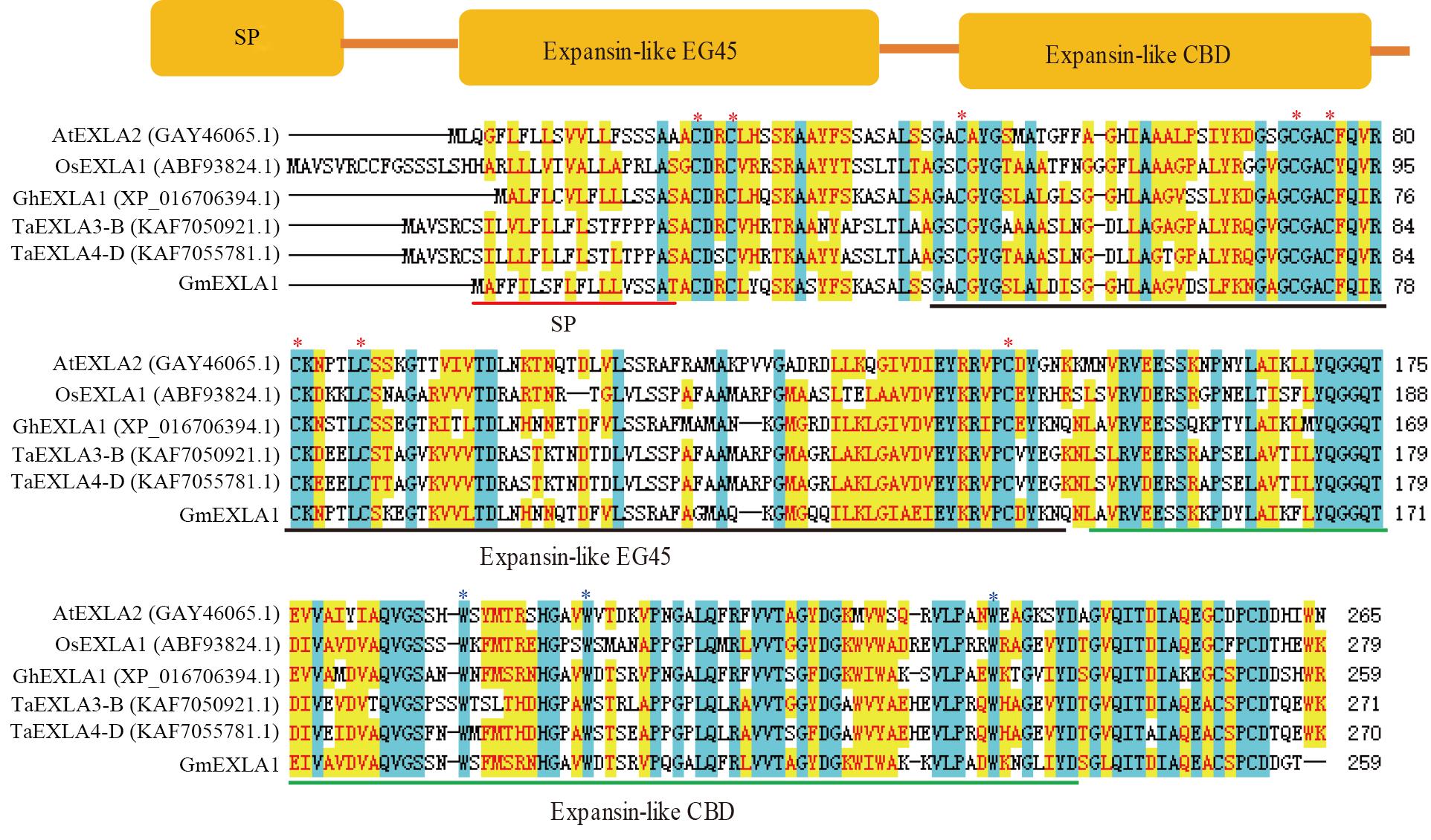
图1 大豆扩展蛋白GmEXLA1保守结构域分析注:SP—信号肽;Expansin-like EG45—催化区;Expansin-like CBD—结合区;*为保守半胱氨酸C和色氨酸W。
Fig. 1 Conserved domain analysis of GmEXLA1Note: SP—Signal peptide; Expansin-like EG45—Catalytic domain; Expansin-like CBD—Binding domain;* indicates conserved cysteine and tryptophan.
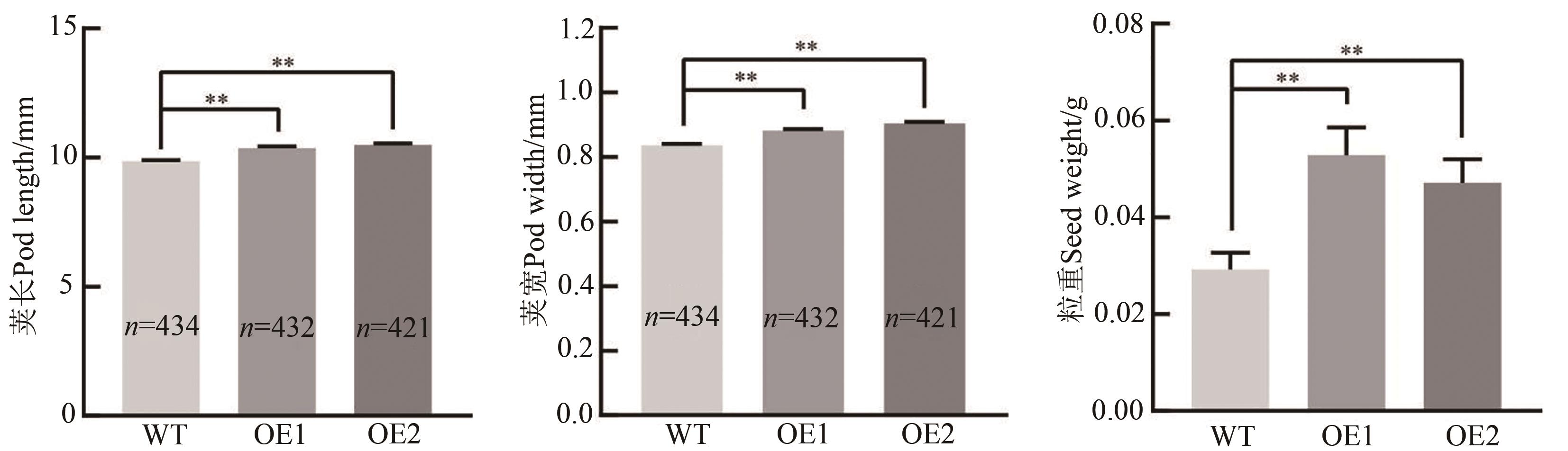
图4 超表达GmEXLA1拟南芥荚长、荚宽与种子重量分析注:WT—野生型;OE1~OE2—超表达转GmEXLA1拟南芥纯合株系;**表示P<0.01水平差异显著。
Fig. 4 Pod length, pod width and seed weight per plant of Arabidopsis over-expressed GmEXLA1Note: WT—Wild-type; OE1~OE2—Over-expression lines with GmEXLA1; ** indicates significant difference at P<0.01 level.

图5 大豆gmexla1突变体分子检测及其突变位置A:PCR检测,M—DNA marker DL2000,1—Williams 82,2—突变体,3—空白对照;B:PCR产物测序结果,黄色背景标出突变碱基C/T;C:突变体目的基因qRT-PCR检测;D:gmexla1及其突变基因的原核诱导表达蛋白检测,M—蛋白分子量标准,1—诱导前的Williams82的GmEXLA1,2—诱导后的Williams82的GmEXLA1,3—诱导前的突变体gmexla1,4—诱导后的突变体gmexla1;E:gmexla1突变位置
Fig. 5 Molecular identification of soybean gmexla1 mutant and its mutation positionA: PCR identification of soybean gmexla1 mutant, M—DNA marker DL2000, 1—Williams 82, 2—gmexla1 mutant, 3—Blank control; B: DNA sequencing result of PCR amplification, the mutant C/T was marked in yellow background; C: qRT-PCR identification of soybean gmexla1 mutant; D: Detection of induced protein in soybean gmexla1 mutant, M—Protein marker, 1—GmEXLA1 in Williams 82 before induced, 2—GmEXLA1 in Williams 82 after induced, 3—gmexla1 in mutant before induced, 4—gmexla1 in mutant after induced; E: gmexla1 mutation position
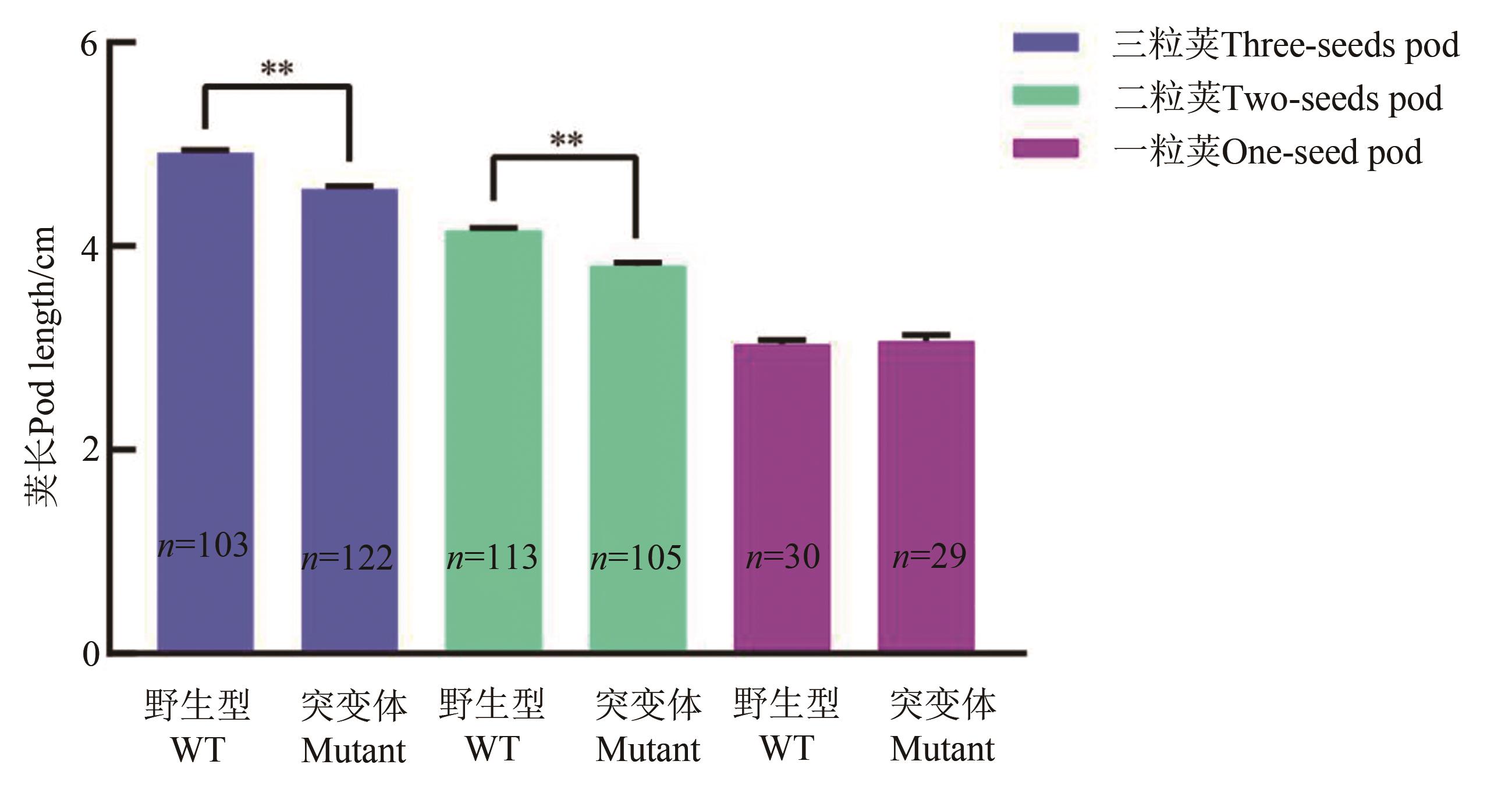
图6 大豆gmexla1突变体豆荚长度分析注:WT—Williams 82野生型;Mutant—gmexla1突变体;**表示P<0.01水平差异显著。
Fig. 6 Comparison of pod length between Williams82 and its gmexla1 mutantNote: WT—Williams 82 wild-type control; Mutant— gmexla1 mutant; ** indicates significant difference at P<0.01 level.
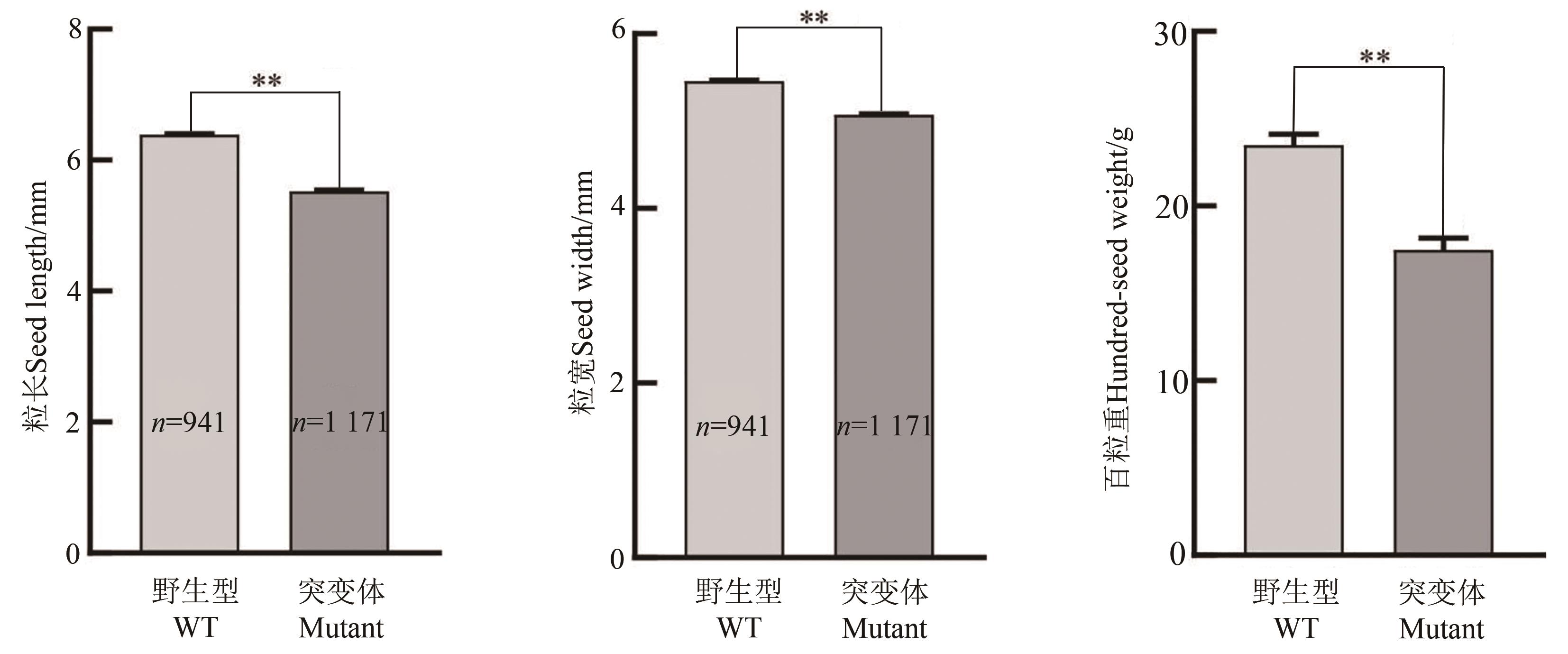
图7 大豆gmexla1突变体籽粒长度、宽度和百粒重分析注:**表示P<0.01水平差异显著。
Fig. 7 Comparison of seed-length, -width and hundred-seed weight between Williams82 and its gmexla1 mutantNote: ** indicates significant difference at P<0.01 level.
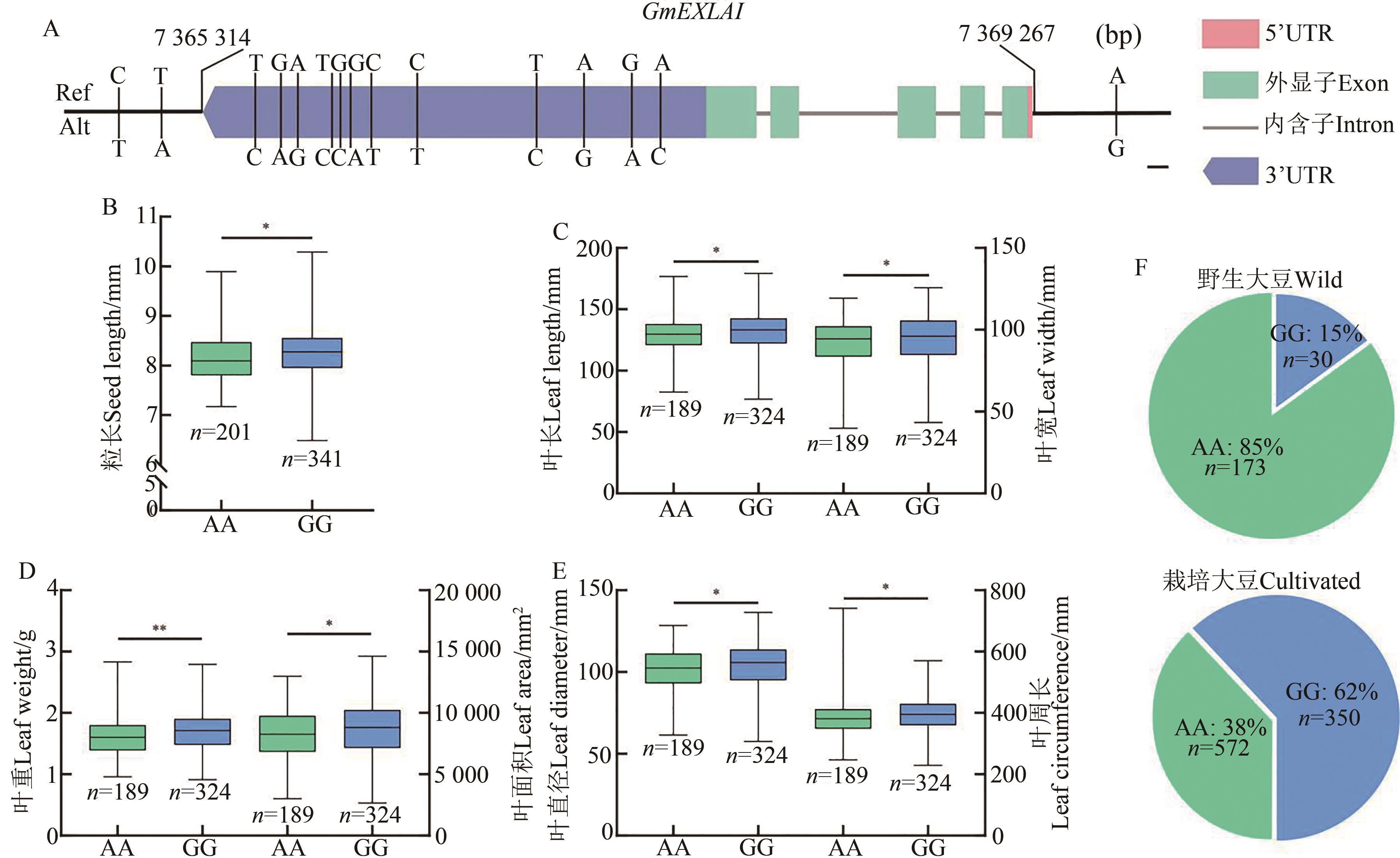
图8 GmEXLA1等位变异及其在野生和栽培大豆中的分布频率分析A:GmEXLA1等位变异分析,上游启动子SNP为A/G变异;B~E:AA型与GG型大豆相关性状差异分析,依据上游SNP(A/G)分型,*和**分别表示P<0.05和P<0.01水平差异显著;F:上游启动子SNP(A/G)在野生大豆与栽培大豆中的分布频率
Fig. 8 SNPs and distribution frequency of GmEXLA1 in wild and cultivated soybeansA: SNPs in GmEXLA1, the SNP in promoter region is A/G variation; B~E: AA- and GG-type related traits, based of on (upstream SNP (A/G)), * and ** indicate significant difference at P<0.05 and P<0.01 levels, respectively; F: Distribution frequency of upstream SNP (A/G) in wild and cultivated soybeans;
| 1 | SHEN Y T, LIU J, GENG H Y, et al.. De novo assembly of a Chinese soybean genome [J]. Sci. China Life Sci., 2018, 61: 871-884. |
| 2 | MCQUEEN-MASON S, DURACHKO D M, COSGROVE D J. Two endogenous proteins that induce cell wall extension in plants [J]. Plant Cell, 1992, 4(11): 1425-1433. |
| 3 | KOK B Ö, ALTUNOGLU Y C, ÖNCÜL A B, et al.. Expansin gene family database: a comprehensive bioinformatics resource for plant expansin multigene family [J/OL]. J. Bioinf. Comput. Biol., 2023, 21(3): 2350015 [2024-03-13].. |
| 4 | HAN Z S, LIU Y L, DENG X, et al.. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.) [J/OL]. BMC Genomics, 2019, 20: 101 [2024-03-13]. . |
| 5 | PARK S, LI F F, RENAUD J, et al.. NbEXPA1, an α-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection [J]. Plant J., 2017, 92: 846-861. |
| 6 | HEPLER N K, BOWMAN A, CAREY R E, et al.. Expansin gene loss is a common occurrence during adaptation to an aquatic environment [J]. Plant J., 2020, 101: 666-680. |
| 7 | 蔡兆琴,龙婷晞,肖冬,等.甘薯扩展蛋白IbEXPA2基因克隆和表达分析[J].分子植物育种,2023, 21(5): 1401-1407. |
| CAI Z Q, LONG T X, XIAO D, et al.. Cloning and expression analysis of IbEXPA2 from sweet potato [J]. Mol. Plant Breeding, 2023, 21(5): 1401-1407. | |
| 8 | LIU X P, DONG S Y, MIAO H, et al.. Genome-wide analysis of expansins and their role in fruit spine development in cucumber (Cucumis sativus L.) [J]. Hortic. Plant J., 2022, 8(6): 757-768. |
| 9 | BEREZHNEVA Z A, MUSIN K G, KULUEV B R. Root growth of transgenic tobacco plants with overexpression of expansin and xyloglucan endotransglycosylase genes under cadmium stress [J/OL]. Russ. J. Plant Physiol., 2022, 69: 96 [2024-03-10]. . |
| 10 | 朱益民.桂花OfSVP和扩张蛋白基因在开花过程中的作用[D].杭州:浙江农林大学,2019. |
| ZHU Y M. The role of OfSVP transcription factors and dilated proteins in Osmanthus fragrans flowering [D]. Hangzhou: Zhejiang A&F University, 2019. | |
| 11 | 邹寒艳.水稻OsLIR1维持叶绿体功能的机制及OsEXPB2参与水稻根系发育的功能研究[D].重庆:重庆大学,2015. |
| ZOU H Y. Study on mechanisms of OsLIR1 maintaining chloroplast function and roles of OsEXPB2 in root development in rice (Oryza sativa) [D]. Chongqing: Chongqing University, 2015. | |
| 12 | YAQOOB A, BASHIR S, RAO A Q, et al.. Transformation of α-EXPA1 gene leads to an improved fibre quality in Gossypium hirsutum [J]. Plant Breeding, 2020, 139: 1213-1220. |
| 13 | 赵湾湾,冯力,胡绍彬,等.番木瓜果实软化相关CpEXPA2基因的克隆与表达分析[J].果树学报,2018, 35(7):785-793. |
| ZHAO W W, FENG L, HU S B, et al.. Cloning and expression analysis of CpEXPA2 gene related to softening of papaya fruit [J]. J. Fruit Sci., 2018, 35(7): 785-793. | |
| 14 | 廖嘉明,包钰韬,陈媛,等.黄梁木NcEXPA8基因提高拟南芥种子萌发速度的研究[J].广西植物, 2021, 41(4): 654-661. |
| LIAO J M, BAO Y T, CHEN Y, et al.. Enhancement of Arabidopsis thaliana seed germination speed by NcEXPA8 gene of Neolamarckia cadamba [J]. Guihaia, 2021, 41(4): 654-661. | |
| 15 | LI X X, ZHAO J, TAN Z Y, et al.. GmEXPB2, a cell wall β-expansin gene, affects soybean nodulation through modifying root architecture and promoting nodule formation and development [J]. Plant Physiol., 2015, 169(4): 2640-2653. |
| 16 | SUN Q, LI Y F, GONG D M, et al.. A NAC-EXPANSIN module enhances maize kernel size by controlling nucellus elimination [J/OL]. Nat. Commun., 2022, 13: 5708 [2024-03-13]. . |
| 17 | CALDERINI D F, CASTILLO F M, ARENAS-M A, et al.. Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential [J]. New Phytol., 2021, 230: 629-640. |
| 18 | MAROWA P, DING A M, KONG Y Z. Expansins: roles in plant growth and potential applications in crop improvement [J]. Plant Cell Rep., 2016, 35: 949-965. |
| 19 | GUO W B, ZHAO J, LI X X, et al.. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses [J]. Plant J., 2011, 66: 541-552. |
| 20 | YANG Z J, ZHENG J K, ZHOU H W, et al.. The soybean β-expansin gene GmINS1 contributes to nodule development in response to phosphate starvation [J]. Physiol. Plant, 2021, 172(4): 2034-2047. |
| 21 | 孙丽.大豆膨胀素GmEXPA4基因转化拟南芥及功能验证[D].哈尔滨:东北农业大学,2013. |
| SUN L. The soybean expansion GmEXPA4 gene transformed Arabidopsis thaliana and functional verification [D]. Harbin: Northeast Agricultural University, 2013. | |
| 22 | LEE D, AHN J H, SONG S, et al.. Expression of an expansin gene is correlated with root elongation in soybean [J]. Plant Physiol., 2003, 131: 985-997. |
| 23 | KONG Y B, WANG B, DU H, et al.. GmEXLB1, a soybean expansin-like B gene, alters root architecture to improve phosphorus acquisition in Arabidopsis [J/OL]. Front. Plant Sci., 2019, 10: 808 [2024-03-13]. . |
| 24 | ZHANG M Z, ZHANG X Y, JIANG X Y, et al.. iSoybean: a database for the mutational fingerprints of soybean [J]. Plant Biotechnol. J., 2022, 20: 1435-1437. |
| 25 | SHAO Z Q, SHAO J B, HUO X B, et al.. Identifcation of closely associated SNPs and candidate genes with seed size and shape via deep re-sequencing GWAS in soybean [J]. Theor. Appl. Genet., 2022, 135: 2341-2351. |
| 26 | ZHANG H Y, JIANG H, HU Z B, et al.. Development of a versatile resource for post-genomic research through consolidating and characterizing 1500 diverse wild and cultivated soybean genomes [J/OL]. BMC Genomics, 2022, 23(1): 250 [2024-03-13]. . |
| 27 | CHEN Y H, REN Y Q, ZHANG G Q, et al.. Overexpression of the wheat expansin gene TaEXPA2 improves oxidative stress tolerance in transgenic Arabidopsis plants [J]. Plant Physiol. Biochem., 2018, 124: 190-198. |
| 28 | REN Y Q, CHEN Y H, AN J, et al.. Wheat expansin gene TaEXPA2 is involved in conferring plant tolerance to Cd toxicity [J]. Plant Sci., 2018, 270: 245-256. |
| 29 | 丁安明,陈志华,杨懿德,等. NtabEXPA12基因过表达对烟草叶片发育及抗逆性的影响[J].中国烟草科学,2021,42(4):58-66. |
| DING A M, CHEN Z H, YANG Y D, et al.. Overexpression of NtabEXPA12 affects leaf development and abiotic stress tolerance in tobacco [J]. Chin. Tobacco Sci., 2021,42(4): 58-66. | |
| 30 | KRISHNAMURTHY P, MUTHUSAMY M, KIM J A, et al.. Brassica rapa expansin-like B1 gene (BrEXLB1) regulate growth and development in transgenic Arabidopsis and elicits response to abiotic stresses [J]. J. Plant Biochem. Biot., 2019, 28(4): 437-446. |
| 31 | WANG M, LI W Z, FANG C, et al.. Parallel selection on a dormancy gene during domestication of crops from multiple families [J]. Nat. Genet., 2018, 50: 1435-1441. |
| 32 | YONG B, ZHU W W, WEI S M, et al.. Parallel selection of loss-of-function alleles of Pdh1 orthologous genes in warm-season legumes for pod indehiscence and plasticity is related to precipitation [J]. New Phytol., 2023, 240(2): 863-879. |
| [1] | 杨冰. 大豆疫霉菌的致病机制及寄主分子响应和防控方法研究进展[J]. 中国农业科技导报, 2025, 27(3): 133-142. |
| [2] | 顿国强, 吴星澎, 纪欣鑫, 张福利, 纪文义, 杨永振. 双摆盘式大豆小区排种器的仿真优化[J]. 中国农业科技导报, 2024, 26(6): 82-90. |
| [3] | 王文月, 姚志鹏, 于洋, 葛毅强. 我国大豆种业科技创新发展现状及对策建议[J]. 中国农业科技导报, 2024, 26(3): 1-6. |
| [4] | 杨皓森. 中国大豆生物育种产业化对贸易依存度的影响[J]. 中国农业科技导报, 2024, 26(11): 15-22. |
| [5] | 姚建民, 马俊奎, 王忠祥, 毕昕媛, 李瑞珍, 杨瑞平, 刘钊, 郭丰辉. 全生物降解渗水地膜在大豆-玉米带状复合种植中的应用效果研究[J]. 中国农业科技导报, 2023, 25(9): 178-185. |
| [6] | 田蕊, 张华, 黄玫红, 邵振启, 李喜焕, 张彩英. 大豆抗旱遗传位点及候选基因发掘[J]. 中国农业科技导报, 2023, 25(9): 69-82. |
| [7] | 张晨阳, 徐明岗, 王斐, 李然, 孙楠. 施用有机肥对我国大豆产量及土壤养分的影响[J]. 中国农业科技导报, 2023, 25(8): 148-156. |
| [8] | 柯博洋, 李文龙, 张彩英. 大豆SWEET 基因在荚粒发育过程中与逆境胁迫下的表达[J]. 中国农业科技导报, 2023, 25(8): 33-52. |
| [9] | 孙亚倩, 陈士亮, 褚佳豪, 李喜焕, 张彩英. 基于BSA-seq结合连锁分析发掘大豆荚粒性状QTLs及候选基因[J]. 中国农业科技导报, 2023, 25(7): 29-42. |
| [10] | 胡健, 车刚, 万霖, 周慧茹, 李光. 高光照条件下的大豆行线提取方法研究[J]. 中国农业科技导报, 2023, 25(5): 106-111. |
| [11] | 邵振启, 李文龙, 孔佑宾, 杜汇, 李占军, 李喜焕, 张彩英. 黄淮海大豆种质资源品质鉴定与优异种质筛选[J]. 中国农业科技导报, 2023, 25(11): 58-69. |
| [12] | 杨占武, 杜汇, 邢馨竹, 李文龙, 孔佑宾, 李喜焕, 张彩英. 大豆细胞色素P450家族GmCYP78A71固氮功能解析[J]. 中国农业科技导报, 2023, 25(1): 50-57. |
| [13] | 田振祥, 丁伟, 程茁, 戴航宇. 大豆内生细菌的分离及其作用效果研究[J]. 中国农业科技导报, 2022, 24(6): 47-57. |
| [14] | 陈奎元, 刘卉, 丁伟. 草甘膦对大豆田土壤养分及其功能酶活性的影响[J]. 中国农业科技导报, 2022, 24(5): 180-188. |
| [15] | 程名, 朱莹, 王晓楠, 罗平, 陈勇, 郝转芳, 席章营. 玉米ZmSNAC13等位变异对抗旱性的调控研究[J]. 中国农业科技导报, 2022, 24(5): 24-31. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号