




















Journal of Agricultural Science and Technology ›› 2023, Vol. 25 ›› Issue (7): 29-42.DOI: 10.13304/j.nykjdb.2022.0089
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Yaqian SUN1( ), Shiliang CHEN2, Jiahao CHU1, Xihuan LI1(
), Shiliang CHEN2, Jiahao CHU1, Xihuan LI1( ), Caiying ZHANG1(
), Caiying ZHANG1( )
)
Received:2022-02-07
Accepted:2022-04-12
Online:2023-07-15
Published:2023-08-25
Contact:
Xihuan LI,Caiying ZHANG
孙亚倩1( ), 陈士亮2, 褚佳豪1, 李喜焕1(
), 陈士亮2, 褚佳豪1, 李喜焕1( ), 张彩英1(
), 张彩英1( )
)
通讯作者:
李喜焕,张彩英
作者简介:孙亚倩 E-mail: 1602032390@qq.com
基金资助:CLC Number:
Yaqian SUN, Shiliang CHEN, Jiahao CHU, Xihuan LI, Caiying ZHANG. Mining of QTLs and Candidate Genes for Pod and Seed Traits via Combining BSA-seq and Linkage Mapping in Soybean[J]. Journal of Agricultural Science and Technology, 2023, 25(7): 29-42.
孙亚倩, 陈士亮, 褚佳豪, 李喜焕, 张彩英. 基于BSA-seq结合连锁分析发掘大豆荚粒性状QTLs及候选基因[J]. 中国农业科技导报, 2023, 25(7): 29-42.
项目 Item | 环境 Environment | 荚长 PL/cm | 荚宽 PW/cm | 荚重 PWT/g | 粒长 SL/cm | 粒宽 SW/cm | 粒重 SWT/g |
|---|---|---|---|---|---|---|---|
| LP | E1 | 4.68 | 1.21 | 1.81 | 1.41 | 0.86 | 0.51 |
| E2 | 5.22 | 1.20 | 2.25 | 1.42 | 0.86 | 0.64 | |
| E3 | 4.82 | 1.21 | 1.88 | 1.33 | 0.83 | 0.55 | |
| E4 | 5.13 | 1.21 | 2.07 | 1.38 | 0.85 | 0.57 | |
| 均值Mean | 4.96 | 1.21 | 2.00 | 1.39 | 0.85 | 0.57 | |
| SP | E1 | 4.06 | 1.10 | 1.49 | 1.30 | 0.79 | 0.45 |
| E2 | 4.52 | 1.09 | 1.74 | 1.31 | 0.81 | 0.54 | |
| E3 | 4.22 | 1.07 | 1.53 | 1.22 | 0.76 | 0.48 | |
| E4 | 4.35 | 1.07 | 1.58 | 1.24 | 0.78 | 0.47 | |
| 均值Mean | 4.29 | 1.08 | 1.59 | 1.27 | 0.79 | 0.49 | |
显著性 Significance (P-value) | E1 | 4.3e-07 | 5.5e-07 | 4.2e-06 | 3.5e-05 | 6.0e-07 | 8.7e-04 |
| E2 | 1.1e-12 | 1.3e-08 | 3.7e-09 | 6.4e-07 | 2.7e-07 | 2.1e-06 | |
| E3 | 3.8e-10 | 7.1e-10 | 1.5e-05 | 2.6e-07 | 4.2e-07 | 1.3e-03 | |
| E4 | 7.7e-11 | 9.5e-09 | 2.8e-08 | 2.2e-07 | 4.2e-06 | 5.1e-06 | |
| 多环境All environments | 5.6e-03 | 3.0e-06 | 1.0e-02 | 5.8e-03 | 2.6e-03 | 4.2e-02 |
Table 1 Genetic variation of extreme RIL lines on pod and seed traits for BSA-seq under different environments
项目 Item | 环境 Environment | 荚长 PL/cm | 荚宽 PW/cm | 荚重 PWT/g | 粒长 SL/cm | 粒宽 SW/cm | 粒重 SWT/g |
|---|---|---|---|---|---|---|---|
| LP | E1 | 4.68 | 1.21 | 1.81 | 1.41 | 0.86 | 0.51 |
| E2 | 5.22 | 1.20 | 2.25 | 1.42 | 0.86 | 0.64 | |
| E3 | 4.82 | 1.21 | 1.88 | 1.33 | 0.83 | 0.55 | |
| E4 | 5.13 | 1.21 | 2.07 | 1.38 | 0.85 | 0.57 | |
| 均值Mean | 4.96 | 1.21 | 2.00 | 1.39 | 0.85 | 0.57 | |
| SP | E1 | 4.06 | 1.10 | 1.49 | 1.30 | 0.79 | 0.45 |
| E2 | 4.52 | 1.09 | 1.74 | 1.31 | 0.81 | 0.54 | |
| E3 | 4.22 | 1.07 | 1.53 | 1.22 | 0.76 | 0.48 | |
| E4 | 4.35 | 1.07 | 1.58 | 1.24 | 0.78 | 0.47 | |
| 均值Mean | 4.29 | 1.08 | 1.59 | 1.27 | 0.79 | 0.49 | |
显著性 Significance (P-value) | E1 | 4.3e-07 | 5.5e-07 | 4.2e-06 | 3.5e-05 | 6.0e-07 | 8.7e-04 |
| E2 | 1.1e-12 | 1.3e-08 | 3.7e-09 | 6.4e-07 | 2.7e-07 | 2.1e-06 | |
| E3 | 3.8e-10 | 7.1e-10 | 1.5e-05 | 2.6e-07 | 4.2e-07 | 1.3e-03 | |
| E4 | 7.7e-11 | 9.5e-09 | 2.8e-08 | 2.2e-07 | 4.2e-06 | 5.1e-06 | |
| 多环境All environments | 5.6e-03 | 3.0e-06 | 1.0e-02 | 5.8e-03 | 2.6e-03 | 4.2e-02 |
| 样品Sample | 过滤后数据 Clean data/bp | Q20/% | Q30/% | GC/% | 总读段 Total reads | 读段比对率Mapped reads/% | 覆盖度 Coverage rate/% | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 1× | 4× | 10× | 20× | |||||||
| KN7 | 30 185 672 700 | 97.29 | 92.63 | 36.42 | 200 007 218 | 98.08 | 98.20 | 97.28 | 94.97 | 79.68 |
| C813 | 30 445 756 200 | 97.28 | 92.59 | 36.54 | 201 728 350 | 98.92 | 97.67 | 96.63 | 94.35 | 80.95 |
| SP | 31 026 257 700 | 97.42 | 92.91 | 36.32 | 205 658 536 | 98.98 | 98.54 | 97.79 | 95.76 | 82.33 |
| LP | 29 921 497 800 | 97.23 | 92.50 | 36.58 | 198 179 602 | 98.97 | 98.67 | 97.63 | 94.93 | 79.99 |
Table 2 Quality analysis of BSA-seq of two pools and soybean RIL parents
| 样品Sample | 过滤后数据 Clean data/bp | Q20/% | Q30/% | GC/% | 总读段 Total reads | 读段比对率Mapped reads/% | 覆盖度 Coverage rate/% | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 1× | 4× | 10× | 20× | |||||||
| KN7 | 30 185 672 700 | 97.29 | 92.63 | 36.42 | 200 007 218 | 98.08 | 98.20 | 97.28 | 94.97 | 79.68 |
| C813 | 30 445 756 200 | 97.28 | 92.59 | 36.54 | 201 728 350 | 98.92 | 97.67 | 96.63 | 94.35 | 80.95 |
| SP | 31 026 257 700 | 97.42 | 92.91 | 36.32 | 205 658 536 | 98.98 | 98.54 | 97.79 | 95.76 | 82.33 |
| LP | 29 921 497 800 | 97.23 | 92.50 | 36.58 | 198 179 602 | 98.97 | 98.67 | 97.63 | 94.93 | 79.99 |
| 项目 Item | C813 | KN7 | LP | SP | |
|---|---|---|---|---|---|
SNP数量 SNP number | 1 619 731 | 1 953 331 | 2 412 499 | 2 397 371 | |
SNP所在基因组位置 SNP genome position | 下游 Downstream | 82 313 | 97 262 | 123 627 | 122 700 |
| 外显子区 Exonic | 61 800 | 72 657 | 91 369 | 89 863 | |
| 基因间Intergenic | 1 171 840 | 1 421 376 | 1 736 589 | 1 728 224 | |
| 内含子区Intronic | 163 852 | 195 463 | 249 905 | 246 732 | |
| 可变剪切位点 Splicing | 456 | 543 | 697 | 685 | |
| 上游Upstream | 96 035 | 114 708 | 143 985 | 144 040 | |
| 3’-UTR | 21 461 | 25 807 | 33 101 | 32 432 | |
| 5’-UTR | 14 584 | 16 642 | 21 941 | 21 530 | |
| 外显子&可变剪切Exonic & splicing | 25 | 23 | 36 | 35 | |
| 上游&下游Upstream & downstream | 7 342 | 8 811 | 11 195 | 11 072 | |
| 5’-UTR & 3’-UTR | 23 | 39 | 54 | 58 | |
| SNP变异类型SNP mutation type | 非同义突变 Non-synonymous | 34 542 | 40 700 | 51 060 | 50 279 |
| 终止获得 Stop-gain | 726 | 894 | 1 089 | 1 077 | |
| 终止丢失 Stop-loss | 144 | 182 | 223 | 216 | |
| 同义突变Synonymous | 26 386 | 30 881 | 39 002 | 38 302 | |
| 未知 Unknown | 27 | 23 | 31 | 24 | |
Table 3 SNP number and quality analysis of BSA-seq of two pools and soybean RIL parents
| 项目 Item | C813 | KN7 | LP | SP | |
|---|---|---|---|---|---|
SNP数量 SNP number | 1 619 731 | 1 953 331 | 2 412 499 | 2 397 371 | |
SNP所在基因组位置 SNP genome position | 下游 Downstream | 82 313 | 97 262 | 123 627 | 122 700 |
| 外显子区 Exonic | 61 800 | 72 657 | 91 369 | 89 863 | |
| 基因间Intergenic | 1 171 840 | 1 421 376 | 1 736 589 | 1 728 224 | |
| 内含子区Intronic | 163 852 | 195 463 | 249 905 | 246 732 | |
| 可变剪切位点 Splicing | 456 | 543 | 697 | 685 | |
| 上游Upstream | 96 035 | 114 708 | 143 985 | 144 040 | |
| 3’-UTR | 21 461 | 25 807 | 33 101 | 32 432 | |
| 5’-UTR | 14 584 | 16 642 | 21 941 | 21 530 | |
| 外显子&可变剪切Exonic & splicing | 25 | 23 | 36 | 35 | |
| 上游&下游Upstream & downstream | 7 342 | 8 811 | 11 195 | 11 072 | |
| 5’-UTR & 3’-UTR | 23 | 39 | 54 | 58 | |
| SNP变异类型SNP mutation type | 非同义突变 Non-synonymous | 34 542 | 40 700 | 51 060 | 50 279 |
| 终止获得 Stop-gain | 726 | 894 | 1 089 | 1 077 | |
| 终止丢失 Stop-loss | 144 | 182 | 223 | 216 | |
| 同义突变Synonymous | 26 386 | 30 881 | 39 002 | 38 302 | |
| 未知 Unknown | 27 | 23 | 31 | 24 | |
| 项目Item | C813 | KN7 | LP | SP | |
|---|---|---|---|---|---|
Indel数量 Indel number | 304 227 | 346 870 | 432 243 | 434 173 | |
Indel所在基因组位置 Indel genome position | 下游 Downstream | 23 599 | 26 795 | 34 163 | 34 277 |
| 外显子区 Exonic | 3 823 | 4 331 | 5 338 | 5 270 | |
| 基因间Intergenic | 191 138 | 218 488 | 268 913 | 270 622 | |
| 内含子区Intronic | 41 422 | 47 052 | 60 068 | 59 808 | |
| 可变剪切位点 Splicing | 182 | 207 | 244 | 246 | |
| 上游Upstream | 30 262 | 34 121 | 43 159 | 43 784 | |
| 3’-UTR | 6 081 | 7 174 | 9 062 | 8 946 | |
| 5’-UTR | 5 163 | 5 894 | 7 571 | 7 475 | |
| 外显子&可变剪切Exonic & splicing | 3 | 2 | 3 | 3 | |
| 上游&下游Upstream & downstream | 2 549 | 2 798 | 3 716 | 3 735 | |
| 5’-UTR & 3’-UTR | 5 | 8 | 6 | 7 | |
Indel变异类型 Indel mutation type | 移码缺失Frameshift deletion | 1 128 | 1 247 | 1 534 | 1 512 |
| 移码插入Frameshift insertion | 987 | 1 134 | 1 314 | 1 294 | |
| 非移码缺失Non-frameshift deletion | 878 | 1 007 | 1 299 | 1 282 | |
| 非移码插入Non-frameshift insertion | 749 | 851 | 1 082 | 1 067 | |
| 终止获得 Stop-gain | 69 | 72 | 92 | 95 | |
| 终止丢失 Stop-loss | 10 | 17 | 15 | 18 | |
| 未知Unknown | 5 | 5 | 5 | 5 | |
Table 4 Indel number and quality analysis of BSA-seq of two pools and RIL parents
| 项目Item | C813 | KN7 | LP | SP | |
|---|---|---|---|---|---|
Indel数量 Indel number | 304 227 | 346 870 | 432 243 | 434 173 | |
Indel所在基因组位置 Indel genome position | 下游 Downstream | 23 599 | 26 795 | 34 163 | 34 277 |
| 外显子区 Exonic | 3 823 | 4 331 | 5 338 | 5 270 | |
| 基因间Intergenic | 191 138 | 218 488 | 268 913 | 270 622 | |
| 内含子区Intronic | 41 422 | 47 052 | 60 068 | 59 808 | |
| 可变剪切位点 Splicing | 182 | 207 | 244 | 246 | |
| 上游Upstream | 30 262 | 34 121 | 43 159 | 43 784 | |
| 3’-UTR | 6 081 | 7 174 | 9 062 | 8 946 | |
| 5’-UTR | 5 163 | 5 894 | 7 571 | 7 475 | |
| 外显子&可变剪切Exonic & splicing | 3 | 2 | 3 | 3 | |
| 上游&下游Upstream & downstream | 2 549 | 2 798 | 3 716 | 3 735 | |
| 5’-UTR & 3’-UTR | 5 | 8 | 6 | 7 | |
Indel变异类型 Indel mutation type | 移码缺失Frameshift deletion | 1 128 | 1 247 | 1 534 | 1 512 |
| 移码插入Frameshift insertion | 987 | 1 134 | 1 314 | 1 294 | |
| 非移码缺失Non-frameshift deletion | 878 | 1 007 | 1 299 | 1 282 | |
| 非移码插入Non-frameshift insertion | 749 | 851 | 1 082 | 1 067 | |
| 终止获得 Stop-gain | 69 | 72 | 92 | 95 | |
| 终止丢失 Stop-loss | 10 | 17 | 15 | 18 | |
| 未知Unknown | 5 | 5 | 5 | 5 | |
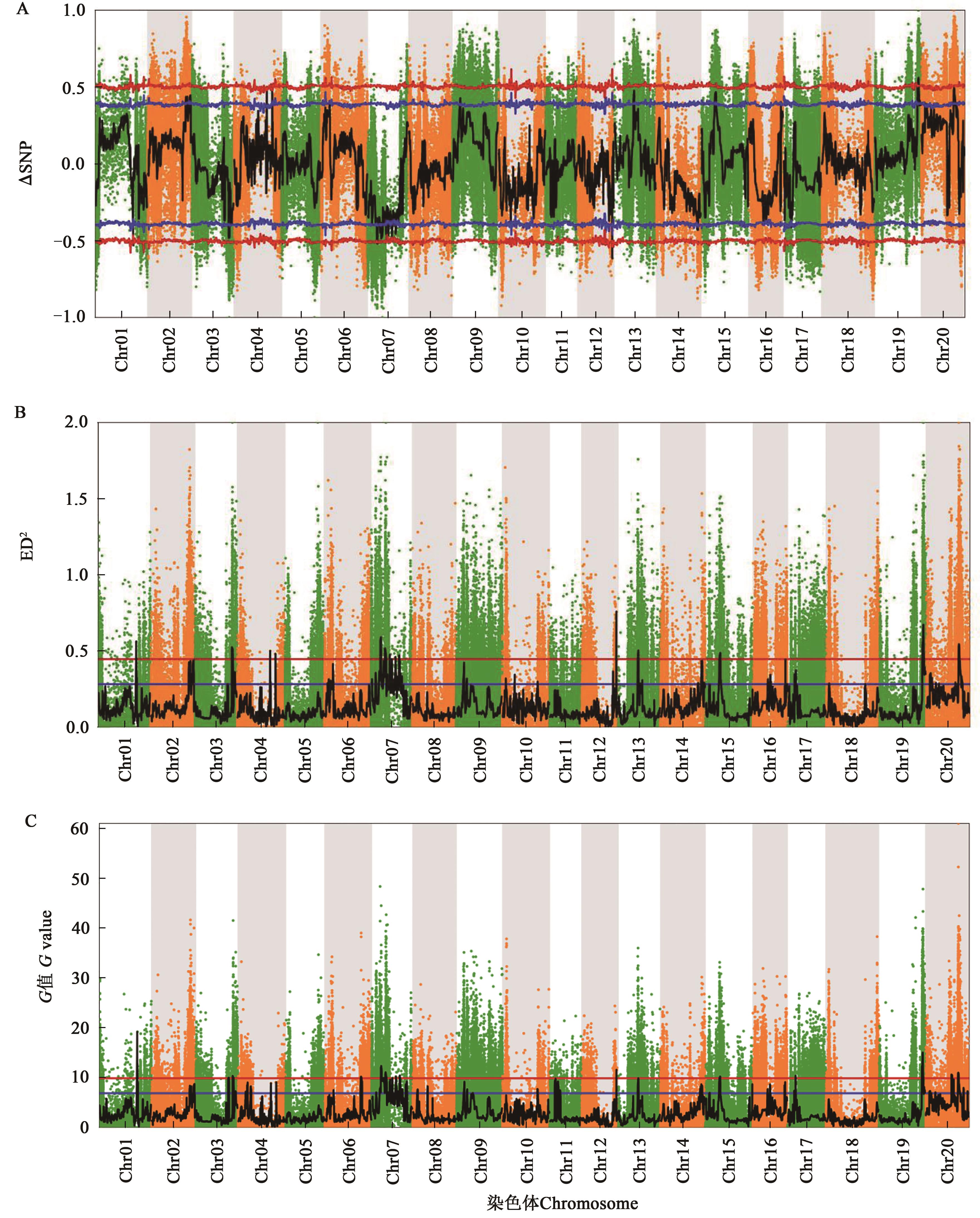
Fig. 1 Associated regions of soybean pod and seed traits via different methodsA: SNP-index method; B: ED method; C: G-statistic method; black lines indicate the fit curve; blue line indicate the confidence level 0.95 (P<0.05); red line indicate the confidence level 0.99 (P<0.01)
| 置信度Confidence level | 染色体 Chromosome | 起始位置 Start position /bp | 终止位置 End position /bp | 基因数量 Gene number |
|---|---|---|---|---|
| 0.95 | Chr01 | 40 690 001 | 41 790 000 | 19 |
| Chr02 | 41 180 001 | 44 100 000 | 246 | |
| Chr02 | 45 540 001 | 47 550 000 | 236 | |
| Chr03 | 39 550 001 | 41 670 000 | 228 | |
| Chr04 | 35 390 001 | 36 590 000 | 17 | |
| Chr04 | 41 290 001 | 42 390 000 | 30 | |
| Chr06 | 9 260 001 | 10 280 000 | 98 | |
| Chr07 | 8 310 001 | 11 590 000 | 173 | |
| Chr07 | 12 960 001 | 16 950 000 | 202 | |
| Chr07 | 19 560 001 | 20 870 000 | 20 | |
| Chr07 | 22 020 001 | 23 400 000 | 1 | |
| Chr07 | 25 340 001 | 27 930 000 | 12 | |
| Chr07 | 28 330 001 | 29 520 000 | 13 | |
| Chr07 | 29 640 001 | 32 490 000 | 25 | |
| Chr09 | 7 890 001 | 9 230 000 | 38 | |
| Chr12 | 37 500 001 | 38 500 000 | 82 | |
| Chr13 | 20 600 001 | 22 520 000 | 189 | |
| Chr14 | 44 180 001 | 46 170 000 | 136 | |
| Chr15 | 14 470 001 | 16 910 000 | 88 | |
| Chr16 | 15 650 001 | 16 710 000 | 7 | |
| Chr16 | 18 160 001 | 19 450 000 | 22 | |
| Chr16 | 34 770 001 | 35 850 000 | 49 | |
| Chr17 | 8 300 001 | 9 500 000 | 121 | |
| Chr19 | 46 210 001 | 50 580 000 | 495 | |
| Chr20 | 4 100 001 | 5 720 000 | 28 | |
| Chr20 | 10 380 001 | 11 500 000 | 6 | |
| Chr20 | 34 990 001 | 38 100 000 | 295 | |
| 0.99 | Chr01 | 40 790 001 | 41 790 000 | 15 |
| Chr07 | 9 770 001 | 11 210 000 | 57 | |
| Chr12 | 37 500 001 | 38 500 000 | 82 | |
| Chr19 | 46 970 001 | 48 610 000 | 182 |
Table 5 Genetic loci of soybean pod and seed traits via different methods
| 置信度Confidence level | 染色体 Chromosome | 起始位置 Start position /bp | 终止位置 End position /bp | 基因数量 Gene number |
|---|---|---|---|---|
| 0.95 | Chr01 | 40 690 001 | 41 790 000 | 19 |
| Chr02 | 41 180 001 | 44 100 000 | 246 | |
| Chr02 | 45 540 001 | 47 550 000 | 236 | |
| Chr03 | 39 550 001 | 41 670 000 | 228 | |
| Chr04 | 35 390 001 | 36 590 000 | 17 | |
| Chr04 | 41 290 001 | 42 390 000 | 30 | |
| Chr06 | 9 260 001 | 10 280 000 | 98 | |
| Chr07 | 8 310 001 | 11 590 000 | 173 | |
| Chr07 | 12 960 001 | 16 950 000 | 202 | |
| Chr07 | 19 560 001 | 20 870 000 | 20 | |
| Chr07 | 22 020 001 | 23 400 000 | 1 | |
| Chr07 | 25 340 001 | 27 930 000 | 12 | |
| Chr07 | 28 330 001 | 29 520 000 | 13 | |
| Chr07 | 29 640 001 | 32 490 000 | 25 | |
| Chr09 | 7 890 001 | 9 230 000 | 38 | |
| Chr12 | 37 500 001 | 38 500 000 | 82 | |
| Chr13 | 20 600 001 | 22 520 000 | 189 | |
| Chr14 | 44 180 001 | 46 170 000 | 136 | |
| Chr15 | 14 470 001 | 16 910 000 | 88 | |
| Chr16 | 15 650 001 | 16 710 000 | 7 | |
| Chr16 | 18 160 001 | 19 450 000 | 22 | |
| Chr16 | 34 770 001 | 35 850 000 | 49 | |
| Chr17 | 8 300 001 | 9 500 000 | 121 | |
| Chr19 | 46 210 001 | 50 580 000 | 495 | |
| Chr20 | 4 100 001 | 5 720 000 | 28 | |
| Chr20 | 10 380 001 | 11 500 000 | 6 | |
| Chr20 | 34 990 001 | 38 100 000 | 295 | |
| 0.99 | Chr01 | 40 790 001 | 41 790 000 | 15 |
| Chr07 | 9 770 001 | 11 210 000 | 57 | |
| Chr12 | 37 500 001 | 38 500 000 | 82 | |
| Chr19 | 46 970 001 | 48 610 000 | 182 |
染色体 Chromosome | 候选基因 Candidate gene | 非同义突变数量 Non-synonymous number | 基因注释 Gene annotation |
|---|---|---|---|
| Chr07 | Glyma.07G106700 | 2 | β-半乳糖苷酶β-galactosidase |
| Chr07 | Glyma.07G108700 | 1 | CCR4-NOT转录复合物亚基CCR4-NOT transcription complex subunit |
| Chr07 | Glyma.07G109500 | 1 | Squamosa启动子结合蛋白Squamosa promoter-binding-like protein |
| Chr19 | Glyma.19G219500 | 1 | 硼转运体Boron transporter |
| Chr19 | Glyma.19G219600 | 1 | NIM1-INTERACTING蛋白NIM1-INTERACTING protein |
| Chr19 | Glyma.19G219700 | 1 | 多聚半乳糖醛酸酶Polygalacturonase |
| Chr19 | Glyma.19G220000 | 4 | 锌指蛋白Zinc finger protein |
| Chr19 | Glyma.19G220100 | 13 | 含WD重复蛋白WD repeat-containing protein |
| Chr19 | Glyma.19G221700 | 5 | WRKY转录因子WRKY transcription factor |
| Chr19 | Glyma.19G222200 | 2 | MYB转录因子MYB transcription factor |
| Chr19 | Glyma.19G222300 | 2 | BnaC04g52040D |
| Chr19 | Glyma.19G222400 | 1 | 果胶乙酰酯酶Pectin acetylesterase |
| Chr19 | Glyma.19G222600 | 1 | 肽基脯氨酰顺反异构酶Peptidyl-prolyl cis-trans isomerase |
| Chr19 | Glyma.19G223000 | 2 | 1,4-β-甘露糖苷酶1,4-β-mannosidase |
| Chr19 | Glyma.19G223900 | 1 | BnaA06g39450D |
| Chr19 | Glyma.19G224000 | 8 | 翻译起始因子IF-2类蛋白Translation initiation factor IF-2-like |
| Chr19 | Glyma.19G224100 | 2 | 通用转录因子IIF亚基General transcription factor IIF subunit |
| Chr19 | Glyma.19G224400 | 3 | NRT1/ PTR家族蛋白NRT1/ PTR family protein |
| Chr19 | Glyma.19G224700 | 2 | 类PIF3转录因子Transcription factor PIF3-like |
| Chr19 | Glyma.19G226000 | 1 | 丝氨酸/苏氨酸蛋白激酶Serine/threonine-protein kinase |
| Chr19 | Glyma.19G227800 | 2 | 半乳醇合酶Galactinol synthase |
| Chr19 | Glyma.19G228600 | 1 | 肽链释放因子Peptide chain release factor |
| Chr19 | Glyma.19G229400 | 2 | 钙调素结合蛋白Calmodulin-binding protein |
Table 6 Candidate genes in the consistent regions with non-synonymous mutations in gene exons
染色体 Chromosome | 候选基因 Candidate gene | 非同义突变数量 Non-synonymous number | 基因注释 Gene annotation |
|---|---|---|---|
| Chr07 | Glyma.07G106700 | 2 | β-半乳糖苷酶β-galactosidase |
| Chr07 | Glyma.07G108700 | 1 | CCR4-NOT转录复合物亚基CCR4-NOT transcription complex subunit |
| Chr07 | Glyma.07G109500 | 1 | Squamosa启动子结合蛋白Squamosa promoter-binding-like protein |
| Chr19 | Glyma.19G219500 | 1 | 硼转运体Boron transporter |
| Chr19 | Glyma.19G219600 | 1 | NIM1-INTERACTING蛋白NIM1-INTERACTING protein |
| Chr19 | Glyma.19G219700 | 1 | 多聚半乳糖醛酸酶Polygalacturonase |
| Chr19 | Glyma.19G220000 | 4 | 锌指蛋白Zinc finger protein |
| Chr19 | Glyma.19G220100 | 13 | 含WD重复蛋白WD repeat-containing protein |
| Chr19 | Glyma.19G221700 | 5 | WRKY转录因子WRKY transcription factor |
| Chr19 | Glyma.19G222200 | 2 | MYB转录因子MYB transcription factor |
| Chr19 | Glyma.19G222300 | 2 | BnaC04g52040D |
| Chr19 | Glyma.19G222400 | 1 | 果胶乙酰酯酶Pectin acetylesterase |
| Chr19 | Glyma.19G222600 | 1 | 肽基脯氨酰顺反异构酶Peptidyl-prolyl cis-trans isomerase |
| Chr19 | Glyma.19G223000 | 2 | 1,4-β-甘露糖苷酶1,4-β-mannosidase |
| Chr19 | Glyma.19G223900 | 1 | BnaA06g39450D |
| Chr19 | Glyma.19G224000 | 8 | 翻译起始因子IF-2类蛋白Translation initiation factor IF-2-like |
| Chr19 | Glyma.19G224100 | 2 | 通用转录因子IIF亚基General transcription factor IIF subunit |
| Chr19 | Glyma.19G224400 | 3 | NRT1/ PTR家族蛋白NRT1/ PTR family protein |
| Chr19 | Glyma.19G224700 | 2 | 类PIF3转录因子Transcription factor PIF3-like |
| Chr19 | Glyma.19G226000 | 1 | 丝氨酸/苏氨酸蛋白激酶Serine/threonine-protein kinase |
| Chr19 | Glyma.19G227800 | 2 | 半乳醇合酶Galactinol synthase |
| Chr19 | Glyma.19G228600 | 1 | 肽链释放因子Peptide chain release factor |
| Chr19 | Glyma.19G229400 | 2 | 钙调素结合蛋白Calmodulin-binding protein |
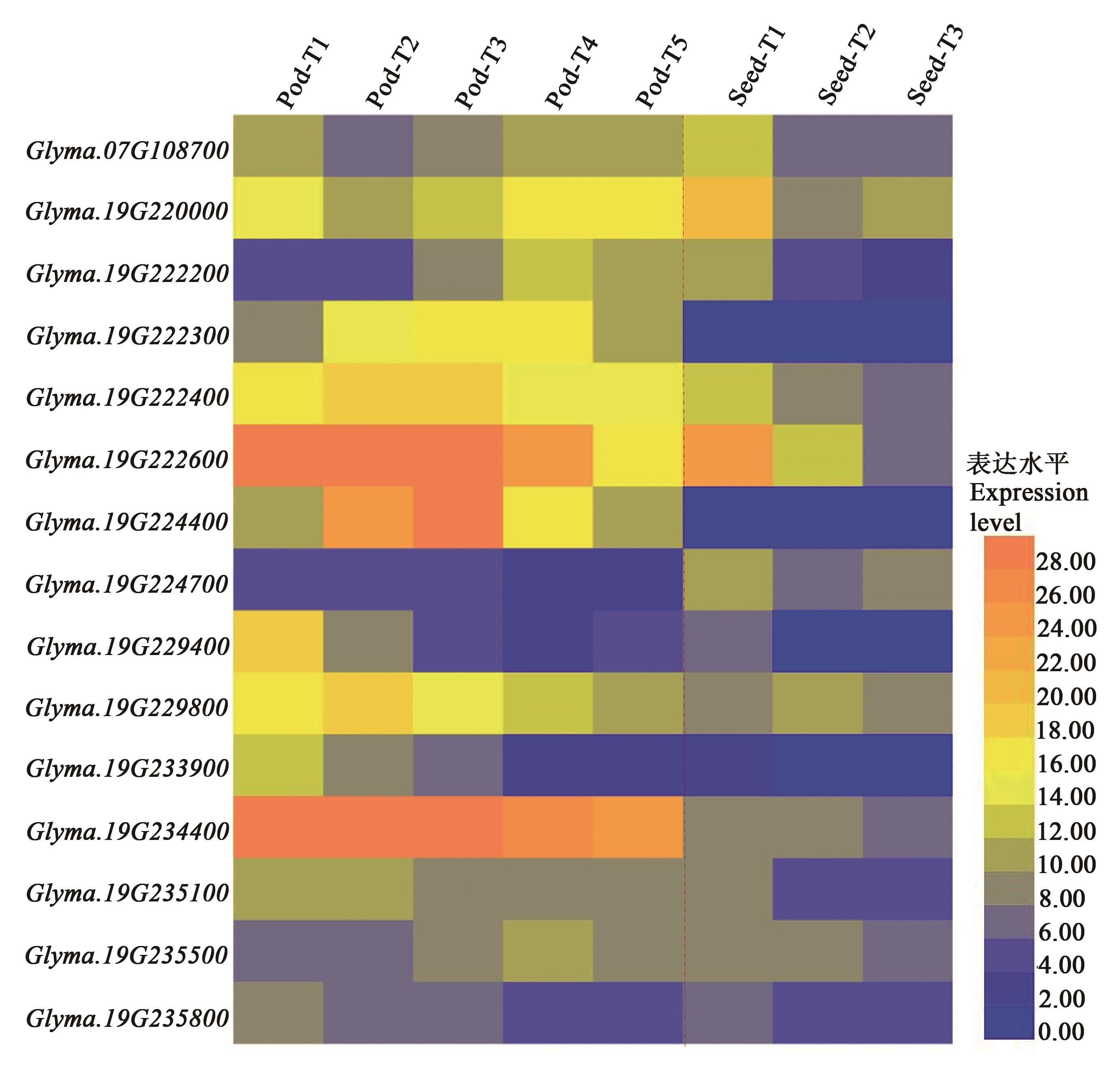
Fig. 2 Expressions of candidate genes related to soybean pod and seed traits in C813Note:Pod-T1~Pod-T5 indicate 5 developmental stages of pod, Seed-T1~Seed-T3 indicate 3 developmental stages of seed.
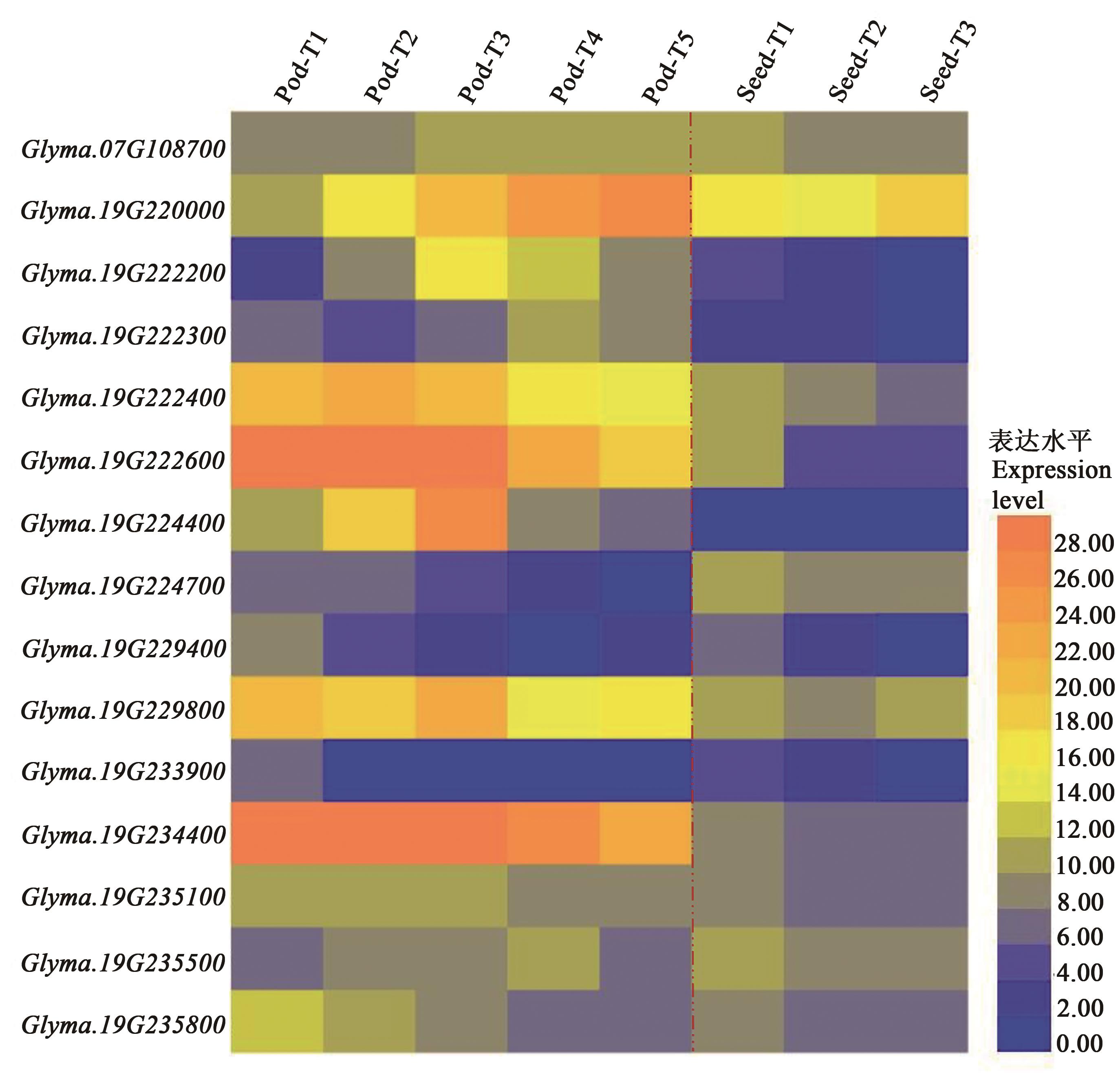
Fig. 3 Expressions of candidate genes for soybean pod and seed traits in KN7Note:Pod-T1~Pod-T5 indicate 5 developmental stages of pod, Seed-T1~Seed-T3 indicate 3 developmental stages of seed.
染色体 Chromosome | 候选基因 Candidate gene | 非同义突变数量 Non-synonymous number | 基因注释 Gene annotation |
|---|---|---|---|
| Chr19 | Glyma.19G229500 | 2 | 类SAR DEFICIENT蛋白SAR DEFICIENT like |
| Chr19 | Glyma.19G229800 | 1 | 类importin蛋白亚基Importin subunit alpha like |
| Chr19 | Glyma.19G230100 | 1 | 驱动蛋白Kinesin |
| Chr19 | Glyma.19G231000 | 3 | 类F-box蛋白F-box protein like |
| Chr19 | Glyma.19G231200 | 5 | 类F-box/富LRR蛋白F-box/LRR-repeat protein like |
| Chr19 | Glyma.19G231400 | 3 | 果胶酯酶Pectinesterase like |
| Chr19 | Glyma.19G231500 | 1 | 果胶酯酶抑制因子Pectinesterase inhibitor |
| Chr19 | Glyma.19G233900 | 1 | 呼吸氧化酶同源蛋白Respiratory burst oxidase homolog protein |
| Chr19 | Glyma.19G234300 | 1 | Tudor/PWWP/MBT家族蛋白Tudor/PWWP/MBT superfamily protein |
| Chr19 | Glyma.19G234400 | 3 | 酰基辅酶A结合域蛋白Acyl-CoA-binding domain-containing protein |
| Chr19 | Glyma.19G235100 | 1 | Xaa-pro氨基肽酶Xaa-pro aminopeptidase |
| Chr19 | Glyma.19G235300 | 1 | 微管结合蛋白Microtubule-binding protein |
| Chr19 | Glyma.19G235400 | 2 | 驱动蛋白Kinesin |
| Chr19 | Glyma.19G235500 | 1 | β-甘露糖苷酶β-mannosidase |
| Chr19 | Glyma.19G235800 | 2 | 类RRP15蛋白RRP15-like protein |
| Chr19 | Glyma.19G236700 | 1 | CAR1转录因子CAR1 transcription factor |
| Chr19 | Glyma.19G236900 | 1 | bHLH转录因子bHLH transcription factor |
Table 6 Candidate genes in the consistent regions with non-synonymous mutations in gene exons
染色体 Chromosome | 候选基因 Candidate gene | 非同义突变数量 Non-synonymous number | 基因注释 Gene annotation |
|---|---|---|---|
| Chr19 | Glyma.19G229500 | 2 | 类SAR DEFICIENT蛋白SAR DEFICIENT like |
| Chr19 | Glyma.19G229800 | 1 | 类importin蛋白亚基Importin subunit alpha like |
| Chr19 | Glyma.19G230100 | 1 | 驱动蛋白Kinesin |
| Chr19 | Glyma.19G231000 | 3 | 类F-box蛋白F-box protein like |
| Chr19 | Glyma.19G231200 | 5 | 类F-box/富LRR蛋白F-box/LRR-repeat protein like |
| Chr19 | Glyma.19G231400 | 3 | 果胶酯酶Pectinesterase like |
| Chr19 | Glyma.19G231500 | 1 | 果胶酯酶抑制因子Pectinesterase inhibitor |
| Chr19 | Glyma.19G233900 | 1 | 呼吸氧化酶同源蛋白Respiratory burst oxidase homolog protein |
| Chr19 | Glyma.19G234300 | 1 | Tudor/PWWP/MBT家族蛋白Tudor/PWWP/MBT superfamily protein |
| Chr19 | Glyma.19G234400 | 3 | 酰基辅酶A结合域蛋白Acyl-CoA-binding domain-containing protein |
| Chr19 | Glyma.19G235100 | 1 | Xaa-pro氨基肽酶Xaa-pro aminopeptidase |
| Chr19 | Glyma.19G235300 | 1 | 微管结合蛋白Microtubule-binding protein |
| Chr19 | Glyma.19G235400 | 2 | 驱动蛋白Kinesin |
| Chr19 | Glyma.19G235500 | 1 | β-甘露糖苷酶β-mannosidase |
| Chr19 | Glyma.19G235800 | 2 | 类RRP15蛋白RRP15-like protein |
| Chr19 | Glyma.19G236700 | 1 | CAR1转录因子CAR1 transcription factor |
| Chr19 | Glyma.19G236900 | 1 | bHLH转录因子bHLH transcription factor |
| 1 | WANG S D, LIU S L, WANG J, et al.. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication [J]. Nat. Sci. Rev., 2020, 7: 1776-1786. |
| 2 | CHEN S L, SUN Y Q, SHAO Z Q, et al.. Stable pleiotropic loci and candidate genes for fresh pod- and seed-related characteristics across multiple environments in soybean (Glycine max) [J]. Plant Breeding, 2021, 140: 630-642. |
| 3 | NIU Y, XU Y, LIU X F, et al.. Association mapping for seed size and shape traits in soybean cultivars [J]. Mol. Breeding, 2013, 31: 785-794. |
| 4 | XIE F T, NIU Y, ZHANG J, et al.. Fine mapping of quantitative trait loci for seed size traits in soybean [J]. Mol. Breeding, 2014, 34: 2165-2178. |
| 5 | DARGAHI H, TANYA P, SRINIVES P. Detection of quantitative trait loci for seed size traits in soybean (Glycine max L.) [J]. Kasetsart J., 2015, 49: 832-843. |
| 6 | YANG H Y, WANG W B, HE Q Y, et al.. Chromosome segment detection for seed size and shape traits using an improved population of wild soybean chromosome segment substitution lines [J]. Physiol. Mol. Biol. Plants, 2017, 23(4): 877-889. |
| 7 | TENG W L, SUI M N, LI W, et al.. Identification of quantitative trait loci underlying seed shape in soybean across multiple environments [J]. J. Agric. Sci., 2018, 156: 3-12. |
| 8 | CUI B F, CHEN L, YANG Y Q, et al.. Genetic analysis and map-based delimitation of a major locus qSS3 for seed size in soybean [J]. Plant Breeding, 2020, 139: 1145-1157. |
| 9 | HINA A, CAO Y C, SONG S Y, et al.. High-resolution mapping in two RIL populations refines major “QTL Hotspot” regions for seed size and shape in soybean (Glycine max L.) [J]. Int. J. Mol. Sci., 2020, 21(3): 1040-1073. |
| 10 | KUMAWAT G, XU D H. A major and stable quantitative trait locus qSS2 for seed size and shape traits in a soybean RIL population [J/OL]. Front. Genet., 2021, 12: 646102 [2022-05-05]. . |
| 11 | AGUADO E, GARC’IA A, IGLESIAS-MOYA J, et al.. Mapping a partial Andromonoecy locus in Citrullus lanatus using BSA-Seq and GWAS approaches [J/OL]. Front. Plant Sci., 2020, 11: 1243 [2022-05-05]. . |
| 12 | YANG T T, AMANULLAH S, PAN J H, et al.. Identification of putative genetic regions for watermelon rind hardness and related traits by BSA-seq and QTL mapping [J/OL]. Euphytica, 2021, 217: 19 [2022-05-05]. . |
| 13 | 张之昊,王俊,刘章雄,等.基于BSA-Seq技术挖掘大豆中黄622的多小叶基因[J].作物学报, 2020, 46(12): 1839-1849. |
| ZHANG Z H, WANG J, LIU Z X, et al.. Mapping of an incomplete dominant gene controlling multifoliolate leaf by BSA-Seq in soybean (Glycine max L.) [J]. Acta Agron. Sin., 2020, 46(12): 1839-1849. | |
| 14 | 曾维英,赖振光,孙祖东,等.基于BSA-Seq和RNA-Seq方法鉴定大豆抗豆卷叶螟候选基因[J]. 作物学报, 2021, 47(8): 1460-1471. |
| ZENG W Y, LAI Z G, SUN Z D, et al.. Identification of the candidate genes of soybean resistance to bean pyralid (Lamprosema indicata Fabricius) by BSA-Seq and RNA-Seq [J]. Acta Agron. Sin., 2021, 47(8): 1460-1471. | |
| 15 | 刁卫楠,袁平丽,龚成胜,等.西瓜果肉柠檬黄色的遗传分析和基因定位[J]. 中国农业科学, 2021, 54(18): 3945-3958. |
| DIAO W N, YUAN P L, GONG C S, et al.. Genetic analysis and gene mapping of canary yellow in watermelon flesh [J]. Sci. Agric. Sin., 2021, 54(18): 3945-3958. | |
| 16 | ZHAO H Y, ZHENG Y B, BAI F, et al.. Bulked segregation analysis coupled with whole-genome sequencing (BSA-Seq) and identification of a novel locus, qGL 3.5, that regulates grain length [J/OL]. Theor. Appl. Genet., 2020, 21(6): 2162 [2022-05-05]. . |
| 17 | ZHANG B, QI F X, HU G, et al.. BSA-seq-based identification of a major additive plant height QTL with an effect equivalent to that of semi-dwarf 1 in a large rice F2 population [J]. Crop J., 2021,9(6): 1428-1437. |
| 18 | CHEN S L, SUN Y Q, SHAO Z Q, et al.. Genetic loci and responsible genes for pod and seed traits under diverse environments via linkage mapping analysis in soybean [Glycine max (L.) Merr.] [J]. Genet. Resour. Crop Evol., 2021,69:1089-1105. |
| 19 | FANG Y X, ZHANG X Q, ZHANG X, et al.. A high-density genetic linkage map of SLAFs and QTL analysis of grain size and weight in barley (Hordeum vulgare L.) [J]. Front. Plant Sci., 2020, 11: 620922 [2022-05-05]. . |
| 20 | LEI L, WANG L F, WANG S M, et al.. Marker-trait association analysis of seed traits in accessions of common bean (Phaseolus vulgaris L.) in China [J/OL]. Front. Genet., 2020, 11: 698 [2022-05-05]. . |
| 21 | ZOU C, WANG P X, XU Y B. Bulked sample analysis in genetics, genomics and crop improvement [J]. Plant Biotechnol. J., 2016, 14: 1-15. |
| 22 | 郭微.利用QTL定位和BSA-seq分析鉴定碱胁迫下水稻产量相关性状的候选基因[D].哈尔滨:东北农业大学, 2019. |
| GUO W. QTL mapping and BSA-seq identify candidate genes for rice yield-related traits under alkali stress [D]. Harbin: Northeast Agricultural University, 2019. | |
| 23 | 李君.玉米抗圆斑病QTL分析及候选基因发掘[D].延吉:延边大学, 2021. |
| LI J. QTL mapping and candidate gene discovery of resistance to spot disease in maize population [D]. Yanji: Yanbian University, 2021. | |
| 24 | VOGEL G, LAPLANT K E, MAZOUREK M, et al.. A combined BSA-Seq and linkage mapping approach identifies genomic regions associated with Phytophthora root and crown rot resistance in squash [J]. Theor. Appl. Genet., 2021, 134: 1015-1031. |
| 25 | ZHAO X, LI W J, ZHAO X Y, et al.. Genome-wide association mapping and candidate gene analysis for seed shape in soybean (Glycine max) [J]. Crop Pasture Sci., 2019, 70(8): 684-693. |
| 26 | 王明晓,黄怀玲,赵传志,等.豆科植物种子大小性状相关基因及其研究进展[J].基因组学与应用生物学, 2021 [2022-05-05]. . |
| WANG M X, HUANG H L, ZHAO C Z, et al.. Progress on genes related to seed size traits in legumes [J]. Genomics Appl. Biol., 2021 [2022-05-05]. . | |
| 27 | LU D D, WANG T, PERSSON S, et al.. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development [J]. Nat. Commun., 2014, 5: 3767. |
| 28 | WU P P, PENG M S, LI Z G, et al.. DRMY1, a myb-like protein, regulates cell expansion and seed production in Arabidopsis thaliana [J]. Plant Cell Physiol., 2019, 60(2): 285-302. |
| 29 | 魏一鸣.玉米籽粒发育调控基因ZmNPF 7.9和ZmAPP1的克隆与功能分析[D].泰安:山东农业大学, 2021. |
| WEI Y M. Cloning and functional analysis of ZmNPF 7.9 and ZmAPP1 regulating maize kernel development [D]. Tai’an: Shandong Agricultural University, 2021. |
| [1] | Jian HU, Gang CHE, Lin WAN, Huiru ZHOU, Guang LI. Study of Soybean Row Line Extraction Method Under High Light Conditions [J]. Journal of Agricultural Science and Technology, 2023, 25(5): 106-111. |
| [2] | Zhanwu YANG, Hui DU, Xinzhu XING, Wenlong LI, Youbin KONG, Xihuan LI, Caiying ZHANG. Functional Analysis of Cytochrome P450 Family GmCYP78A71 in Soybean Nodulation [J]. Journal of Agricultural Science and Technology, 2023, 25(1): 50-57. |
| [3] | Zhenxiang TIAN, Wei DING, Zhuo CHENG, Hangyu DAI. Isolation of Endophytic Bacteria in Soybean and Its Action Effect [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 47-57. |
| [4] | Kuiyuan CHEN, Hui LIU, Wei DING. Effect of Glyphosate on Soil Nutrient and the Functional Enzyme Activities in Soybean Fields [J]. Journal of Agricultural Science and Technology, 2022, 24(5): 180-188. |
| [5] | Ning QIN, Junru LI, Rui TIAN, Zhenqi SHAO, Xihuan LI, Caiying ZHANG. Mining of Genetic Loci and Screening of Candidate Genes for Seed Tocopherol Content in Soybean [J]. Journal of Agricultural Science and Technology, 2022, 24(3): 48-56. |
| [6] | Huifeng KE, Zhengwen SUN, Guoning WANG, Chengsheng MENG, Liqiang WU. Mining and Screening of Associated Markers and Candidate Genes Related to Seed Size and Shape in Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 76-86. |
| [7] | Kaiqiang WANG, Xue YANG, Changfeng LI, Xiao DUAN, Qing PENG, Yu QIAO, Bo SHI. Optimization of Glyceollins Synthesis Condition Induced by Xylooligosaccharides Based on Response Surface Methodology [J]. Journal of Agricultural Science and Technology, 2022, 24(10): 208-217. |
| [8] | Xinzhu XING, Zhanwu YANG, Youbin KONG, Wenlong LI, Hui DU, Xihuan LI, Caiying ZHANG. Functional Analysis of Carotenoid Cleavage Dioxygenases GmCCD8 in Soybean Nodulation [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 46-53. |
| [9] | FAN Wenhua, DAI Jianjun, ZHANG Shushan, SUN Lingwei, WU Caifeng, ZHANG Defu. Effect of Proanthocyanidins Addition in Soybean Lecithin-based Semen Extender on Goat Semen Freezing [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 78-86. |
| [10] | CHEN Shiliang, SUN Yaqian, SHAO Zhenqi, LI Wenlong, KONG Youbin, DU Hui, LI Xihuan, ZHANG Caiying. Analysis of Epistasis QTL and Its Interaction Effects on Controlling Fresh Pod and Seed Related Traits in Soybean [J]. Journal of Agricultural Science and Technology, 2021, 23(8): 25-36. |
| [11] | ZHANG Yongfang, GAO Zhihui, SHI Pengqing, HAN Zhiping*. Adaptability Analysis of Different Soybean Varieties Based on Agronomic and Quality Traits [J]. Journal of Agricultural Science and Technology, 2020, 22(8): 25-32. |
| [12] |
LI Zhi, WANG Hongfu*, WANG Yuyun, YANG Jing, YU Bingxing, HUANG Shanshan.
Impact of Millet and Soybean Intercropping on Their Photosynthetic Characteristics and Yield
[J]. Journal of Agricultural Science and Technology, 2020, 22(6): 168-175.
|
| [13] | JIN Linxue1, TANG Hongyan1*, WU Rongsheng1, WANG Huizhen1, LIU Linchun2. Risk Analysis and Regionalization of Soybean Drought Disasters in Inner Mongolia [J]. Journal of Agricultural Science and Technology, 2020, 22(1): 106-115. |
| [14] | BAI Yuzhe1, MA Yucong1, MENG Yize1, LIAN Ruijiao1, WANG Ying1, LI Xihuan1*, ZHANG Caiying2*. Epistatic QTLs Analysis of Protein and Oil Contents in Soybean (Glycine Max L. Merr.) Seeds [J]. Journal of Agricultural Science and Technology, 2019, 21(6): 36-42. |
| [15] | ZHANG Limei, MA Xin, HAN Baoji, SHI Lei*. Present and Residual Effectiveness of Different Boron Fertilizers on the Crop Yield in Soybean-Rapeseed Rotation [J]. Journal of Agricultural Science and Technology, 2019, 21(10): 133-139. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号