




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (3): 71-82.DOI: 10.13304/j.nykjdb.2023.0591
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Jinxian LIU1,2( ), Lijuan WANG2(
), Lijuan WANG2( ), Jie LIU2, Xianyu FU1,2, Guangheng WU3
), Jie LIU2, Xianyu FU1,2, Guangheng WU3
Received:2023-08-08
Accepted:2023-10-18
Online:2025-03-15
Published:2025-03-14
Contact:
Jinxian LIU
刘金仙1,2( ), 王丽娟2(
), 王丽娟2( ), 刘杰2, 傅仙玉1,2, 武广珩3
), 刘杰2, 傅仙玉1,2, 武广珩3
通讯作者:
刘金仙
作者简介:刘金仙 E-mail:liujinxian@163.com基金资助:CLC Number:
Jinxian LIU, Lijuan WANG, Jie LIU, Xianyu FU, Guangheng WU. Identification and Expression Analysis of Calmodulin-binding Transcription Activator (CAMTA) Family Genes in Tea Plants[J]. Journal of Agricultural Science and Technology, 2025, 27(3): 71-82.
刘金仙, 王丽娟, 刘杰, 傅仙玉, 武广珩. 茶树钙调蛋白结合转录激活因子(CAMTA)家族基因鉴定与表达分析[J]. 中国农业科技导报, 2025, 27(3): 71-82.
基因 Gene | 引物名称 Primer name | 引物序列 Primer sequence (5’-3’) |
|---|---|---|
| CsCAMAT10 | RT-CsCAMAT10-F | GCCATAGGAGCCTTAGCATA |
| RT-CsCAMAT10-R | CCAGGGCAAACTCAACAACT | |
| CsCAMAT9 | RT-CsCAMAT9-F | TGATCCTTTGGTGCAGAACG |
| RT-CsCAMAT9-R | CCAATCCAGCAACCATGTGA | |
| CsCAMAT8 | RT-CsCAMAT8-F | AGCATAATCTGGTCCGTTGG |
| RT-CsCAMAT8-R | CGTTCTTCTGTTTGCTTTCG | |
| CsCAMAT7 | RT-CsCAMAT7-F | GCCATAACTGGAGGAAGAAG |
| RT-CsCAMAT7-R | ATAGCTTCGCCTCTGGAAAT | |
| CsCAMAT6 | RT-CsCAMAT6-F | GCTGCTCGCATTCATCAAGT |
| RT-CsCAMAT6-R | GAATCCACGCAAACCACTCC | |
| CsCAMAT5 | RT-CsCAMAT5-F | GTCCGCAGATGTTACTGGCTAC |
| RT-CsCAMAT5-R | GCACTCAACTTGCTGGGATC | |
| CsCAMAT4 | RT-CsCAMAT4-F | AGGATGGCAAGACCGTCAAA |
| RT-CsCAMAT4-R | GTTCAAGGGCAACCGCACCA | |
| CsCAMAT3 | RT-CsCAMAT3-F | GTTCTCCCGTCACTCCTGTC |
| RT-CsCAMAT3-R | CCCTTCACGCTGATGATGTA | |
| CsCAMAT2 | RT-CsCAMAT2-F | TATGTTGTCTGGCTCCTCCC |
| RT-CsCAMAT2-R | CGGCTGATTACATCCTCCTC | |
| CsCAMAT1 | RT-CsCAMAT1-F | GGAAATTGTGAAATGGGCAGTC |
| RT-CsCAMAT1-R | TCCTCATCGTCATCGGCATC | |
| CsGAPDH | RT-CsGAPDH-F | TTGGCATCGTTGAGGGTCT |
| RT-CsGAPDH-R | CAGTGGGAACACGGAAAGC |
Table 1 qRT-PCR primers of CsCAMTAs
基因 Gene | 引物名称 Primer name | 引物序列 Primer sequence (5’-3’) |
|---|---|---|
| CsCAMAT10 | RT-CsCAMAT10-F | GCCATAGGAGCCTTAGCATA |
| RT-CsCAMAT10-R | CCAGGGCAAACTCAACAACT | |
| CsCAMAT9 | RT-CsCAMAT9-F | TGATCCTTTGGTGCAGAACG |
| RT-CsCAMAT9-R | CCAATCCAGCAACCATGTGA | |
| CsCAMAT8 | RT-CsCAMAT8-F | AGCATAATCTGGTCCGTTGG |
| RT-CsCAMAT8-R | CGTTCTTCTGTTTGCTTTCG | |
| CsCAMAT7 | RT-CsCAMAT7-F | GCCATAACTGGAGGAAGAAG |
| RT-CsCAMAT7-R | ATAGCTTCGCCTCTGGAAAT | |
| CsCAMAT6 | RT-CsCAMAT6-F | GCTGCTCGCATTCATCAAGT |
| RT-CsCAMAT6-R | GAATCCACGCAAACCACTCC | |
| CsCAMAT5 | RT-CsCAMAT5-F | GTCCGCAGATGTTACTGGCTAC |
| RT-CsCAMAT5-R | GCACTCAACTTGCTGGGATC | |
| CsCAMAT4 | RT-CsCAMAT4-F | AGGATGGCAAGACCGTCAAA |
| RT-CsCAMAT4-R | GTTCAAGGGCAACCGCACCA | |
| CsCAMAT3 | RT-CsCAMAT3-F | GTTCTCCCGTCACTCCTGTC |
| RT-CsCAMAT3-R | CCCTTCACGCTGATGATGTA | |
| CsCAMAT2 | RT-CsCAMAT2-F | TATGTTGTCTGGCTCCTCCC |
| RT-CsCAMAT2-R | CGGCTGATTACATCCTCCTC | |
| CsCAMAT1 | RT-CsCAMAT1-F | GGAAATTGTGAAATGGGCAGTC |
| RT-CsCAMAT1-R | TCCTCATCGTCATCGGCATC | |
| CsGAPDH | RT-CsGAPDH-F | TTGGCATCGTTGAGGGTCT |
| RT-CsGAPDH-R | CAGTGGGAACACGGAAAGC |
基因名称 Gene name | 基因ID Gene ID | 氨基酸 Amino acid/aa | 分子量 Molecular weight/kD | 等电点 pI | 亚细胞定位Subcellular prediction | 总平均疏水性Grand average hydropathicity | 信号肽预测 Singal petides |
|---|---|---|---|---|---|---|---|
| CsCAMTA1 | TEA005634 | 124 | 14.29 | 9.811 | 细胞核Nucleus | -0.388 | 无No |
| CsCAMTA2 | TEA006574 | 984 | 109.83 | 6.153 | 细胞核Nucleus | -0.518 | 无No |
| CsCAMTA3 | TEA011186 | 953 | 107.62 | 6.828 | 细胞核Nucleus | -0.475 | 无No |
| CsCAMTA4 | TEA011489 | 155 | 17.63 | 9.515 | 细胞核Nucleus | -0.484 | 无No |
| CsCAMTA5 | TEA011490 | 864 | 97.26 | 6.028 | 细胞核Nucleus | -0.401 | 无No |
| CsCAMTA6 | TEA012477 | 1 419 | 157.99 | 6.669 | 细胞核Nucleus | -0.346 | 无No |
| CsCAMTA7 | TEA015813 | 139 | 16.33 | 8.722 | 细胞核Nucleus | -0.637 | 无No |
| CsCAMTA8 | TEA015818 | 970 | 108.43 | 4.976 | 细胞核Nucleus | -0.587 | 无No |
| CsCAMTA9 | TEA025813 | 1 028 | 114.80 | 6.326 | 细胞核Nucleus | -0.512 | 无No |
| CsCAMTA10 | TEA028584 | 325 | 36.55 | 9.820 | 细胞核Nucleus | 0.006 | 无No |
Table 2 Physicochemical properties of CsCAMTAs genes
基因名称 Gene name | 基因ID Gene ID | 氨基酸 Amino acid/aa | 分子量 Molecular weight/kD | 等电点 pI | 亚细胞定位Subcellular prediction | 总平均疏水性Grand average hydropathicity | 信号肽预测 Singal petides |
|---|---|---|---|---|---|---|---|
| CsCAMTA1 | TEA005634 | 124 | 14.29 | 9.811 | 细胞核Nucleus | -0.388 | 无No |
| CsCAMTA2 | TEA006574 | 984 | 109.83 | 6.153 | 细胞核Nucleus | -0.518 | 无No |
| CsCAMTA3 | TEA011186 | 953 | 107.62 | 6.828 | 细胞核Nucleus | -0.475 | 无No |
| CsCAMTA4 | TEA011489 | 155 | 17.63 | 9.515 | 细胞核Nucleus | -0.484 | 无No |
| CsCAMTA5 | TEA011490 | 864 | 97.26 | 6.028 | 细胞核Nucleus | -0.401 | 无No |
| CsCAMTA6 | TEA012477 | 1 419 | 157.99 | 6.669 | 细胞核Nucleus | -0.346 | 无No |
| CsCAMTA7 | TEA015813 | 139 | 16.33 | 8.722 | 细胞核Nucleus | -0.637 | 无No |
| CsCAMTA8 | TEA015818 | 970 | 108.43 | 4.976 | 细胞核Nucleus | -0.587 | 无No |
| CsCAMTA9 | TEA025813 | 1 028 | 114.80 | 6.326 | 细胞核Nucleus | -0.512 | 无No |
| CsCAMTA10 | TEA028584 | 325 | 36.55 | 9.820 | 细胞核Nucleus | 0.006 | 无No |
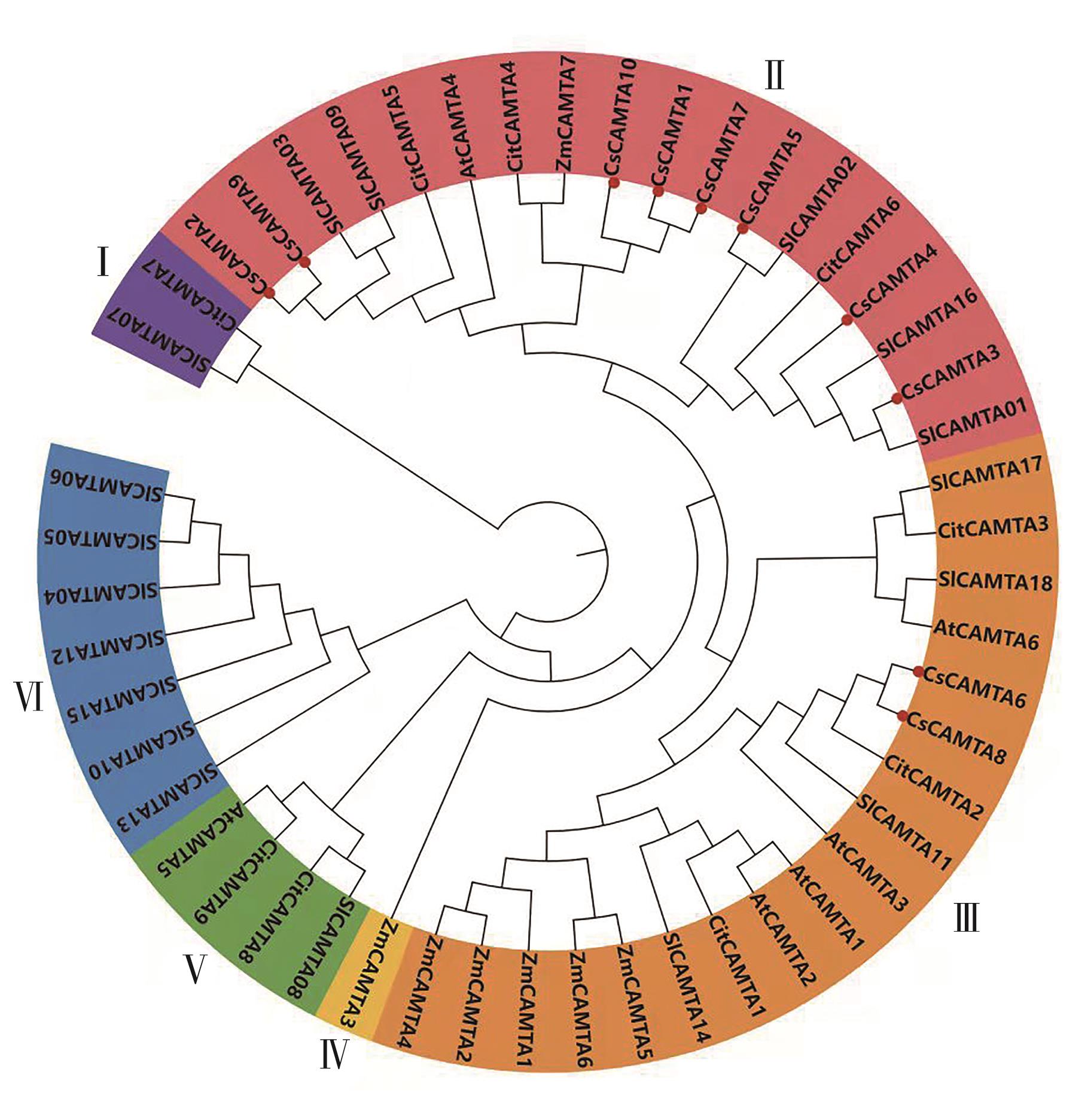
Fig. 6 Phylogenetic tree of CAMTA gene from different originNote:Cs—Camellia sinensis; At—Arabidopsis thaliana; Sl—Solanum lycopersicum; Zm—Zea mays; Cit—Citrus L.
| 1 | KUDLA J, BATISTIC O, HASHIMOTO K. Calcium signals: the lead currency of plant information processing [J]. Plant Cell, 2010, 22(3):541-563 |
| 2 | DODD A N, KUDLA J, SANDERS D. The language of calcium signaling [J]. Annu. Rev. Plant Biol., 2010, 61 (1):593-620. |
| 3 | KIM Y, GILMOUR S J, CHAO L, et al.. Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes [J]. Mol. Plant, 2019, 13(1):157-168. |
| 4 | LORENZO O. bZIP edgetic mutations: at the frontier of plant metabolism, development and stress trade-off [J]. J. Exp. Bot., 2019, 70(20):5517-5520. |
| 5 | PEI J, FLIEDER D B, PATCHEFSKY A, et al.. Detecting MYB and MYBL1 fusion genes in tracheobronchial adenoid cystic carcinoma by targeted RNA-sequencing [J]. Mod. Pathol., 2019, 32(10):1416-1420. |
| 6 | ZHU D, HOU L X, XIAO P L, et al.. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress [J]. Plant Sci., 2019, 280:132-142. |
| 7 | YANG Y, YOO C G, ROTTMANN W, et al.. PdWND 3A, NAC domain-containing proteinawood-associated, affects lignin biosynthesis and composition in populus [J/OL]. BMC Plant Biol., 2019, 19:486 [2023-07-05]. . |
| 8 | BOUCHE N, SCHARLAT A, SNEDDEN W, et al.. A novel family of calmodulin-binding transcription activators in multicellular organisms [J].J. Biol. Chem., 2002, 277 (24):21851-21861. |
| 9 | CHOI M S, KIM M C, YOO J H, et al.. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.) [J]. J. Biol. Chem., 2005, 280(49):40820-40831. |
| 10 | POOVAIAH B W, DU L Q, WANG H Z, et al.. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions [J]. Plant Physiol., 2013, 163 (2):531-542. |
| 11 | 李卉梓. SlGLRs 和SlCaMs 在调控番茄低温抗性中的机制研究[D].杭州:浙江大学, 2019. |
| LI H Z. Roles and mechanisms of SlGLRs and SlCaMs regulated cold tolerance in tomato [D]. Hangzhou: Zhejiang University, 2019. | |
| 12 | MEER L, MUMTAZ S, LABBO A M, et al.. Genome-wide identification and expression analysis of calmodulin-binding transcription activator genes in banana under drought stress [J]. Sci. Hortic., 2019, 244:10-14. |
| 13 | NOMAN M, JAMEEL A, QIANG W D, et al.. Overexpression of GmCAMTA12 enhanced drought tolerance in Arabidopsis and soybean [J/OL]. Int. J. Mol. Sci., 2019, 20(19):4849 [2023-07-05]. . |
| 14 | PRASAD K V S K, ABDEL-HAMEED A A E, XING D H, et al.. Global gene expression analysis using RNA-seq uncovered a new role for SR1/CAMTA3 transcription factor in salt stress [J/OL]. Sci. Rep., 2016, 6:27021 [2023-07-05]. . |
| 15 | 褚桂花.番茄SR/CAMTA 转录因子家族基因SlSR4的克隆与功能研究[D].重庆:重庆大学,2015. |
| CHU G H. Cloning and functional research of tomato SR/CAMTA family gene SlSR4 [D]. Chongqing: Chongqing University, 2015. | |
| 16 | DOHERTY C J, VAN BUSKIRK H A, MYERS S J, et al.. Roles for Arabidopsis CAMTA transcription factors in cold regulated gene expression and freezing tolerance [J]. Plant Cell, 2009, 21(3):972-984. |
| 17 | 王晴晴.油菜CAMTA1参与冷胁迫应答的功能研究[D].武汉:华中师范大学,2013. |
| WANG Q Q. Role of Brassica napus CAMTA1 protein in response to cold stress [D]. Wuhan: Huazhong Normal University, 2013. | |
| 18 | KIM Y S, AN C, PARK S, et al.. CAMTA-mediated regulation of salicylic acid immunity pathway genes in Arabidopsis exposed to low temperature and pathogen infection [J]. Plant Cell, 2017, 29 (10):2465-2477. |
| 19 | PANDEY N, RANJAN A, PANT P, et al.. CAMTA1 regulates drought responses in Arabidopsis thaliana [J/OL]. BMC Genomics, 2013, 14: 216 [2023-07-05]. . |
| 20 | DU L Q, ALI G S, SIMONS K A, et al.. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity [J]. Nature, 2009, 457(7233):1154-1158. |
| 21 | YANG T, POOVAIAH B W. An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death [J]. J. Biol. Chem., 2000, 275(49):38467-38473. |
| 22 | GALON Y, ALONI R, NACHMIAS D, et al.. Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis [J]. Planta, 2010, 232(1):165-178. |
| 23 | GU J M, WANG C Y, WANG F, et al.. Roles of CAMTA/SR in plant growth and development and stress response [J]. Acta Hortic. Sin., 2021, 48(4):613-631. |
| 24 | BOUCHE N, SCHARLAT A, SNEDDEN W, et al.. A novel family of calmodulin-binding transcription activators in multicellular organisms [J]. J. Biol. Chem., 2002, 277(24):21851-21861. |
| 25 | WANG G P, ZENG H Q, HU X Y, et al.. Identification and expression analyses of calmodulin-binding transcription activator genes in soybean [J]. Plant Soil, 2015, 386:205-221. |
| 26 | YUE R Q, LU C X, SUN T, et al.. Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses [J/OL]. Front. Plant Sci., 2015, 6:576 [2023-07-05]. . |
| 27 | 张静.柑橘CAMTA基因家族的鉴定与功能的初步研究[D].重庆:西南大学, 2019. |
| ZHANG J. Identification and functional analysis of citrus CAMTA genes [D]. Chongqing: Southwest University, 2019. | |
| 28 | 林显祖,肖小虎,阳江华,等.巴西橡胶树CAMTA 转录因子全基因组鉴定与表达分析[J].热带作物学报,2021,42(10): 2859-2868. |
| LIN X Z, XIAO X H, YANG J H, et al.. Genome-wide identification and expression analysis of the CAMTA family in rubber tree (Hevea brasiliensis) [J]. Chin. J. Tropic. Crops, 2021, 42(10):2859-2868. | |
| 29 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method [J]. Methods, 2001, 25(4):402-408. |
| 30 | DEL CAMPO E M, CASANO L M, BARREO E. Evolutionary implications of intron-exon distribution and the properties and sequences of the RPL10A gene in eukaryotes [J]. Mol. Phylogenet. Evol., 2013, 66(3):857-867. |
| 31 | 王宝强,朱晓林,魏小红,等.番茄钙调蛋白结合转录因子CAMTA家族基因的鉴定及其低温胁迫下表达模式的分析[J].农业生物技术学报,2021,29(5):871-884. |
| WANG B Q, ZHU X L, WEI X H, et al.. Identification of calmodulin-binding transcription factor CAMTA gene family and its expression analysis under low-temperature stress in tomato (Solanum lycopersicum) [J]. J. Agric. Biotechnol., 2021, 29(5):871-884. | |
| 32 | LI B, HE S, ZHENG Y Q, et al.. Genome-wide identifcation and expression analysis of the calmodulin-binding transcription activator (CAMTA) family genes in tea plant [J/OL]. BMC Genomics, 2022, 23:667 [2023-07-05]. . |
| 33 | ZHOU Q Y, ZHAO M W, XING F, et al.. Identification and expression analysis of CAMTA Genes in tea plant reveal their complex regulatory role in stress responses [J/OL]. Front. Plant Sci., 2022, 13:910768 [2023-07-05]. . |
| 34 | 吴娴.小麦CAMTA基因家族鉴定及TaCAMTA1b-B .1功能验证[D].荆州:长江大学,2022. |
| WU X. Genome-wide identification of CAMTA gene families in wheat (Triticum asetivum L.) and functional analysis of TaCAMTA1b-B.1 [D]. Jingzhou:Yangtze University, 2022. | |
| 35 | SHANGGUAN L F, WANG X M, LENG X P, et al.. Identification and bioinformatic analysis of signal responsive/calmodulin-binding transcription activators gene models in Vitis vinifera [J]. Mol. Biol. Rep., 2014, 41(5):2937-2949. |
| 36 | MEER L, MUMTAZ S, LABBO A M, et al.. Genome-wide identification and expression analysis of calmodulin-binding transcription activator genes in banana under drought stress [J]. Sci. Hortic., 2019, 244:10-14. |
| 37 | BOUCHE N, SCHARLAT A, SNEDDEN W, et al.. A novel family of calmodulin-binding transcription activators in multicellular organisms [J]. J. Biol. Chem., 2002, 277(24):21851-21861. |
| 38 | CHOI M S, KIM M C, YOO J H, et al.. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.) [J]. J. Biol. Chem., 2005, 280(49):40820-40831. |
| 39 | 刘志鑫,孙宇,叶子,等.杧果钙调蛋白转录激活因子基因家族的鉴定及分析[J].果树学报,2021,38(8):1252-1263. |
| LIU Z X, SUN Y, YE Z, et al.. Identification and analysis of calmodulin-binding transcription activator gene family in mango [J]. J. Fruit Sci., 2021, 38(8):1252-1263. | |
| 40 | RAHMAN H, XU Y P, ZHANG X R, et al.. Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to pamp triggered immunity and resistance to Sclerotinia sclerotiorum [J/OL]. Front. Plant. Sci., 2016, 7:581 [2023-07-05]. . |
| 41 | KAKAR K U, NAWAZ Z, CUI Z Q, et al.. Evolutionary and expression analysis of CAMTA gene family in Nicotiana tabacum yielded insights into their origin, expansion and stress responses [J/OL]. Sci. Rep., 2018, 8(1):10322 [2023-07-05]. . |
| 42 | YANG F, DONG F S, HU F H,et al.. Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (CAMTA) gene family in wheat(Triticum aestivum L.) [J/OL]. BMC Genetics, 2020, 21:105 [2023-07-05]. . |
| 43 | WEI M, XU X M, LI C H. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress [J/OL]. Sci. Rep., 2017, 7(1): 17910 [2023-07-05]. . |
| 44 | CHANG Y L, BAI Y J, WEI Y X, et al.. CAMTA3 negatively regulates disease resistance through modulating immune response and extensive transcriptional reprogramming in cassava [J]. Tree Physiol., 2020, 40(11):1520-1533. |
| 45 | RAHMAN H, YANG J, XU Y P, et al.. Phylogeny of plant CAMTAs and role of AtCAMTAs in nonhost resistance to Xanthomonas oryzae pv. Oryzae [J/OL]. Front. Plant Sci., 2016, 7:177 [2023-07-05]. . |
| 46 | SCHWEIGHOFER A, HIRT H, MESKIENE I. Plant PP2C phos-phatases: emerging functions in stress signaling [J].Trends Plant Sci., 2004, 9(5):236-243. |
| 47 | 刘文宇,杜何为,黄敏.玉米CAMTA基因家族的全基因组分析[J].分子植物育种,2021,19(11):3499-3505. |
| LIU W Y, DU H W, HUANG M. Genome-wide analysis of the CAMTA gene family in maize (Zea mays L.) [J]. Mol. Plant Breeding, 2021, 19(11):3499-3505. | |
| 48 | YANG T B, PENG H, WHITAKER B D, et al.. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening [J/OL]. BMC Plant Biol., 2012, 12:19 [2023-07-05]. . |
| 49 | YANG Y J, SUN T, XU L Q, et al.. Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments [J/OL]. Front. Plant Sci., 2015, 6:459 [2023-07-05]. . |
| 50 | IQBAL Z, IQBAL M S, SINGH S P, et al.. Ca2+/calmodulin complex triggers CAMTA transcriptional machinery under stress in plants: signaling cascade and molecular regulation [J/OL]. Front. Plant Sci., 2020, 11:598327 [2023-07-05]. . |
| 51 | REDDY A S N, ALI G S, CELESNIK H, et al.. Coping with stresses:roles of calcium- and calcium/calmodulin-regulated gene expression[J]. Plant Cell, 2011, 23(6):2010-2032. |
| 52 | DU L Q, ALI G S, SIMONS K A, et al.. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity [J]. Nature, 2009, 457(7233):1154-1158. |
| [1] | Xianguo LI, Qi DAI, Zepeng WANG, Zhaolong CHEN, Huizhuan YAN, Ning LI. Identification and Phylogenetic Analysis of Tomato CCCH-like Zinc Finger Protein Family [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 80-95. |
| [2] | Xiaorong GUO, Ying LIU, Jiazhen FAN, Tao HUANG, Rong ZHOU. Bioinformatics Analysis of Porcine CREBRF Gene and Its Expression Pattern [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 44-53. |
| [3] | Fulin ZHANG, Rui XI, Yuxiang LIU, Zhaolong CHEN, Qinghui YU, Ning LI. Genome-wide Identification and Expression Analysis of Tomato BURP Structural Domain Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 51-62. |
| [4] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [5] | Bo LIU, Wangtian WANG, Li MA, Junyan WU, Yuanyuan PU, Lijun LIU, Yan FANG, Wancang SUN, Yan ZHANG, Ruimin LIU, Xiucun ZENG. Identification and Characterization of IPT Gene Family in Brassica rapa L. [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 56-66. |
| [6] | Zhenwei ZHANG, Xiangshu DONG, Jing YANG, Xuejun LI, Meijun QI, Kuaile JIANG, Yonglin YANG, Butian WANG, Xuedong SHI, Junchao QIU, Zhihua CHEN, Yu GE. Genome-wide Identification and Expression Analysis of Key Chlorophyll Synthesis Related Gene CaPOR in Coffea arabica [J]. Journal of Agricultural Science and Technology, 2024, 26(10): 83-97. |
| [7] | Yurong DENG, Lian HAN, Jinlong WANG, Xinghan WEI, Xudong WANG, Ying ZHAO, Xiaohong WEI, Chaozhou LI. Identification of SOD Family Genes in Chenopodium quinoa and Their Response to Mixed Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 28-39. |
| [8] | Jinfeng ZHAO, Aili YU, Yanfang LI, Yanwei DU, Gaohong WANG, Zhenhua WANG. Response Characteristics of SiCBL3 to Abiotic Stresses in Foxtail Millet [J]. Journal of Agricultural Science and Technology, 2022, 24(11): 68-75. |
| [9] | Qinqin WANG, Xiugui CHEN, Xuke LU, Shuai WANG, Yuexin ZHANG, Yapeng FAN, Quanjia CHEN, Wuwei YE. Bioinformatics Analysis and Functional Verification of GhPKE1 inUpland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 38-45. |
| [10] | LIU Zhengwen, WANG Xingfen, MENG Chengsheng, ZHANG Yan, SUN Zhengwen, WU Liqiang, MA Zhiying, ZHANG Guiyin. Genome-Wide Identification and Analysis of GH9 Gene Family in Gossypium barbadense L. [J]. Journal of Agricultural Science and Technology, 2021, 23(9): 30-45. |
| [11] | PU Quanming, YANG Peng, YONG Lei, DENG Yuchuan, HE Zihan, LIN Bangmin, SHI Songmei, XIANG Chengyong, FANG Fang. Studies on Pigment Content and Photosyntheic Characteristics of Purple-red Leaf Color Mutant in Radish [J]. Journal of Agricultural Science and Technology, 2021, 23(8): 45-54. |
| [12] | CUI Jianghui§, YANG Puyuan§, CHANG Jinhua*. Identification and Expression Analysis Under Abiotic Stress of GRF Gene Family in Sorghum [J]. Journal of Agricultural Science and Technology, 2021, 23(4): 37-46. |
| [13] | GUAN Sijing, WANG Nan, XU Rongrong, GE Tiantian, GAO Jing, YAN Yonggang, ZHANG Gang, CHEN Ying, ZHANG Mingying. [J]. Journal of Agricultural Science and Technology, 2021, 23(12): 66-75. |
| [14] |
LIU Peng1,WEI Jie1,YANG Yiqing1, ZHANG Na1,WEN Xiaolei1,2, FAN Xuefeng1,YANG Wenxiang1*,LIU Daqun1*.
A New Subtype of Calmodulinlike TaCML25/26 in Wheat Regulate Resistance to Leaf Rust
[J]. Journal of Agricultural Science and Technology, 2020, 22(4): 120-128.
|
| [15] | LI Xia1,2, FAN Xin2, LIANG Chengzhen2, WANG Yuan2, ZHANG Rui2, MENG Zhigang2*, LIU Xiaodong1*, SUN Guoqing2*. Functional Analysis of BS2 Gene in Tobacco [J]. Journal of Agricultural Science and Technology, 2019, 21(11): 43-50. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号