




















中国农业科技导报 ›› 2024, Vol. 26 ›› Issue (9): 12-24.DOI: 10.13304/j.nykjdb.2024.0216
收稿日期:2024-03-20
接受日期:2024-05-21
出版日期:2024-09-15
发布日期:2024-09-13
通讯作者:
王文生,崔彦茹
作者简介:魏荣华 E-mail:weironghua1999@163.com
Ronghua WEI1( ), Ming YIN2, Wensheng WANG2(
), Ming YIN2, Wensheng WANG2( ), Yanru CUI1(
), Yanru CUI1( )
)
Received:2024-03-20
Accepted:2024-05-21
Online:2024-09-15
Published:2024-09-13
Contact:
Wensheng WANG,Yanru CUI
摘要:
抽穗期作为水稻重要的性状之一,不仅与水稻生育时期密切相关,还与水稻产量、品质和抗逆性密切相关,决定了水稻品种的种植地区和季节适应性,因此,定位水稻抽穗期相关基因在水稻生产中有着重要的作用。利用HQ20和金23B及其杂交构建的F4群体为试验材料,根据群体中水稻抽穗期划分3个等级:抽穗期早、抽穗期适中、大青棵(不抽穗),并构建早、中、青3个混池,采用BSA-seq(bulked-segregant analysis sequencing)方法挖掘水稻抽穗期基因。结果表明,BSA-seq结果与参考基因组比对率在94%以上,4×基因组覆盖率在80%以上;基于此,在置信度为0.95时,经SNP-index算法获得位于4条染色体(6、7、8和9号)的17个SNPs与Indels一致的关联区间,进一步分析区域内的候选基因,发现61个基因存在非同义突变。GO与KEGG富集分析表明,与水稻抽穗期密切相关的途径有UDP-糖基转移酶活性、细胞周期G2/M期转变的正向调控和植物激素信号转导等。结合3K水稻数据库中的单倍型分析与已公布的表达量数据,挖掘出3个抽穗期关键候选基因:LOC_Os07g22720、LOC_Os07g23740、LOC_Os08g07200。以上结果为开展水稻抽穗期性状遗传改良以及遗传基础解析奠定了基础。
中图分类号:
魏荣华, 尹明, 王文生, 崔彦茹. 基于BSA-seq发掘水稻抽穗期相关QTLs及候选基因[J]. 中国农业科技导报, 2024, 26(9): 12-24.
Ronghua WEI, Ming YIN, Wensheng WANG, Yanru CUI. Discovering of QTLs and Candidate Genes Related to Rice Heading Period Traits Based on BSA-seq[J]. Journal of Agricultural Science and Technology, 2024, 26(9): 12-24.
样品 Sample | 过滤后数据 Clean data/bp | Q30/% | GC/% | 总读段 Total reads | 读段比对率 Mapped reads/% | 覆盖度 Coverage rate/% | |
|---|---|---|---|---|---|---|---|
| 1× | 4× | ||||||
| HQ31 | 5 744 148 300 | 92.02 | 41.25 | 35 621 290 | 96.69 | 89.98 | 81.90 |
| OHQ20 | 5 343 193 500 | 91.24 | 42.09 | 38 294 322 | 96.87 | 90.43 | 82.90 |
| 适中Moderate | 19 863 114 000 | 91.69 | 44.01 | 12 1353 842 | 96.86 | 95.22 | 92.36 |
| 青棵Green | 18 203 076 300 | 91.94 | 45.05 | 132 420 760 | 96.70 | 95.83 | 92.77 |
| 早熟Early | 19 540 310 100 | 91.57 | 44.81 | 130 268 734 | 94.75 | 96.09 | 92.93 |
表1 3个混池及其亲本BSA-seq测序质量分析
Table 1 BSA-seq sequencing quality analysis of three mixed pools and their parents
样品 Sample | 过滤后数据 Clean data/bp | Q30/% | GC/% | 总读段 Total reads | 读段比对率 Mapped reads/% | 覆盖度 Coverage rate/% | |
|---|---|---|---|---|---|---|---|
| 1× | 4× | ||||||
| HQ31 | 5 744 148 300 | 92.02 | 41.25 | 35 621 290 | 96.69 | 89.98 | 81.90 |
| OHQ20 | 5 343 193 500 | 91.24 | 42.09 | 38 294 322 | 96.87 | 90.43 | 82.90 |
| 适中Moderate | 19 863 114 000 | 91.69 | 44.01 | 12 1353 842 | 96.86 | 95.22 | 92.36 |
| 青棵Green | 18 203 076 300 | 91.94 | 45.05 | 132 420 760 | 96.70 | 95.83 | 92.77 |
| 早熟Early | 19 540 310 100 | 91.57 | 44.81 | 130 268 734 | 94.75 | 96.09 | 92.93 |

图2 子代△SNPI和△IndelI在染色体上的分布A:△SNPI;B:△IndelI。图中的点代表SNP(Indel)位点,其横坐标为SNP(Indel)位点所在窗口的中间值,纵坐标为指数的差值,不同染色体间用不同颜色区分;图中黑色线是均值,蓝色线为95%阈值,紫色的线为99%阈值
Fig. 2 Distribution of offspring gradient difference △SNPIand △IndelI on chromosomesA: △SNPI; B: △InDelI. The point in the figure represents SNP (Indel) site. Its abscissa is the middle value of the window where the SNP site is located, and the ordinate is the difference in index. Different chromosomes are distinguished by different colors; the black line in the figure is the mean value, the blue line is 95% threshold, and the purple line is 99% threshold
染色体 Chromosome | 起始位置 Start position/bp | 终止位置 End position/bp | 基因数量 Gene number |
|---|---|---|---|
| Chr5 | 9 171 001 | 9 172 000 | 1 |
| Chr5 | 15 980 001 | 15 981 000 | 1 |
| Chr6 | 819 788 | 862 809 | 2 |
| Chr6 | 24 267 001 | 24 268 000 | 1 |
| Chr7 | 3 282 221 | 3 515 234 | 5 |
| Chr7 | 7 078 687 | 7 184 099 | 6 |
| Chr7 | 7 661 654 | 8 210 957 | 19 |
| Chr7 | 9 362 884 | 13 851 724 | 151 |
| Chr7 | 14 502 162 | 16 910 700 | 57 |
| Chr7 | 17 474 000 | 17 474 209 | 1 |
| Chr7 | 26 393 554 | 26 913 290 | 5 |
| Chr7 | 28 410 165 | 28 428 916 | 3 |
| Chr8 | 559 292 | 634 638 | 3 |
| Chr8 | 1 038 578 | 1 075 302 | 5 |
| Chr8 | 3 660 741 | 4 179 420 | 13 |
| Chr8 | 5 032 940 | 5 032 946 | 1 |
| Chr8 | 6 217 190 | 6 217 199 | 1 |
| Chr8 | 9 001 410 | 9 001 425 | 1 |
| Chr8 | 9 310 563 | 9 510 022 | 2 |
| Chr8 | 10 365 320 | 10 920 326 | 6 |
| Chr8 | 11 279 434 | 11 279 468 | 1 |
| Chr8 | 11 589 580 | 11 589 588 | 1 |
| Chr8 | 12 272 300 | 12 272 330 | 1 |
| Chr8 | 13 063 007 | 13106 062 | 2 |
| Chr8 | 13 692 000 | 13 692 049 | 1 |
| Chr8 | 14 083 249 | 14 330 070 | 4 |
| Chr8 | 14 732 433 | 14 913 038 | 2 |
| Chr8 | 15 216 765 | 15 796 339 | 6 |
| Chr8 | 16 630 600 | 16 630 619 | 1 |
| Chr8 | 17 148 084 | 17 153 579 | 2 |
| Chr8 | 18 006 500 | 18 006 529 | 1 |
| Chr8 | 18 345 182 | 18 452 701 | 2 |
| Chr8 | 18 803 600 | 18 803 609 | 1 |
| Chr8 | 19 730 003 | 20 312 233 | 16 |
| Chr9 | 19 462 800 | 19 462 880 | 1 |
| Chr9 | 20 098 700 | 20 098 749 | 1 |
| Chr10 | 260 200 | 260 233 | 1 |
| Chr11 | 6 523 296 | 6 613 238 | 3 |
| Chr11 | 19 092 100 | 19 092 108 | 1 |
| Chr12 | 3 918 600 | 3 918 631 | 1 |
| Chr12 | 10 337 833 | 10 337 834 | 1 |
表2 SNP关联区间
Table 2 SNP association interval
染色体 Chromosome | 起始位置 Start position/bp | 终止位置 End position/bp | 基因数量 Gene number |
|---|---|---|---|
| Chr5 | 9 171 001 | 9 172 000 | 1 |
| Chr5 | 15 980 001 | 15 981 000 | 1 |
| Chr6 | 819 788 | 862 809 | 2 |
| Chr6 | 24 267 001 | 24 268 000 | 1 |
| Chr7 | 3 282 221 | 3 515 234 | 5 |
| Chr7 | 7 078 687 | 7 184 099 | 6 |
| Chr7 | 7 661 654 | 8 210 957 | 19 |
| Chr7 | 9 362 884 | 13 851 724 | 151 |
| Chr7 | 14 502 162 | 16 910 700 | 57 |
| Chr7 | 17 474 000 | 17 474 209 | 1 |
| Chr7 | 26 393 554 | 26 913 290 | 5 |
| Chr7 | 28 410 165 | 28 428 916 | 3 |
| Chr8 | 559 292 | 634 638 | 3 |
| Chr8 | 1 038 578 | 1 075 302 | 5 |
| Chr8 | 3 660 741 | 4 179 420 | 13 |
| Chr8 | 5 032 940 | 5 032 946 | 1 |
| Chr8 | 6 217 190 | 6 217 199 | 1 |
| Chr8 | 9 001 410 | 9 001 425 | 1 |
| Chr8 | 9 310 563 | 9 510 022 | 2 |
| Chr8 | 10 365 320 | 10 920 326 | 6 |
| Chr8 | 11 279 434 | 11 279 468 | 1 |
| Chr8 | 11 589 580 | 11 589 588 | 1 |
| Chr8 | 12 272 300 | 12 272 330 | 1 |
| Chr8 | 13 063 007 | 13106 062 | 2 |
| Chr8 | 13 692 000 | 13 692 049 | 1 |
| Chr8 | 14 083 249 | 14 330 070 | 4 |
| Chr8 | 14 732 433 | 14 913 038 | 2 |
| Chr8 | 15 216 765 | 15 796 339 | 6 |
| Chr8 | 16 630 600 | 16 630 619 | 1 |
| Chr8 | 17 148 084 | 17 153 579 | 2 |
| Chr8 | 18 006 500 | 18 006 529 | 1 |
| Chr8 | 18 345 182 | 18 452 701 | 2 |
| Chr8 | 18 803 600 | 18 803 609 | 1 |
| Chr8 | 19 730 003 | 20 312 233 | 16 |
| Chr9 | 19 462 800 | 19 462 880 | 1 |
| Chr9 | 20 098 700 | 20 098 749 | 1 |
| Chr10 | 260 200 | 260 233 | 1 |
| Chr11 | 6 523 296 | 6 613 238 | 3 |
| Chr11 | 19 092 100 | 19 092 108 | 1 |
| Chr12 | 3 918 600 | 3 918 631 | 1 |
| Chr12 | 10 337 833 | 10 337 834 | 1 |
染色体 Chromosome | 起始位置 Start position/bp | 终止位置 End position/bp | 基因数量 Gene number |
|---|---|---|---|
| Chr5 | 7 966 753 | 8 080 035 | 2 |
| Chr6 | 867 000 | 867 072 | 2 |
| Chr6 | 24 267 600 | 24 267 692 | 1 |
| Chr7 | 5 651 500 | 5 651 596 | 1 |
| Chr7 | 7 145 117 | 7 206 410 | 3 |
| Chr7 | 7 928 606 | 8 179 003 | 5 |
| Chr7 | 9 564 466 | 10 124 576 | 4 |
| Chr7 | 10 499 474 | 11 646 584 | 12 |
| Chr7 | 12 529 769 | 13 851 742 | 29 |
| Chr7 | 14 503 097 | 15 243 927 | 10 |
| Chr7 | 15 876 095 | 16 832 885 | 12 |
| Chr7 | 17 399 500 | 17 399 542 | 1 |
| Chr8 | 559 300 | 559 352 | 1 |
| Chr8 | 1 040 057 | 1 050 460 | 3 |
| Chr8 | 3 948 972 | 4 283 090 | 4 |
| Chr8 | 5 623 000 | 5 623 045 | 1 |
| Chr8 | 10 447 300 | 10 447 314 | 1 |
| Chr8 | 14 970 700 | 14 970 730 | 1 |
| Chr8 | 15 598 900 | 15 598 941 | 1 |
| Chr8 | 18 006 500 | 18 006 549 | 1 |
| Chr8 | 18 452 681 | 18 452 693 | 1 |
| Chr8 | 19 739 048 | 20 197 724 | 11 |
| Chr9 | 19 759 500 | 19 759 596 | 2 |
| Chr9 | 20 098 700 | 20 098 775 | 1 |
| Chr11 | 5 919 041 | 6 002 538 | 2 |
| Chr11 | 18 350 200 | 18 350 292 | 1 |
| Chr12 | 10 137 200 | 10 137 217 | 1 |
表3 Indel关联区间
Table 3 Indel correlation interval
染色体 Chromosome | 起始位置 Start position/bp | 终止位置 End position/bp | 基因数量 Gene number |
|---|---|---|---|
| Chr5 | 7 966 753 | 8 080 035 | 2 |
| Chr6 | 867 000 | 867 072 | 2 |
| Chr6 | 24 267 600 | 24 267 692 | 1 |
| Chr7 | 5 651 500 | 5 651 596 | 1 |
| Chr7 | 7 145 117 | 7 206 410 | 3 |
| Chr7 | 7 928 606 | 8 179 003 | 5 |
| Chr7 | 9 564 466 | 10 124 576 | 4 |
| Chr7 | 10 499 474 | 11 646 584 | 12 |
| Chr7 | 12 529 769 | 13 851 742 | 29 |
| Chr7 | 14 503 097 | 15 243 927 | 10 |
| Chr7 | 15 876 095 | 16 832 885 | 12 |
| Chr7 | 17 399 500 | 17 399 542 | 1 |
| Chr8 | 559 300 | 559 352 | 1 |
| Chr8 | 1 040 057 | 1 050 460 | 3 |
| Chr8 | 3 948 972 | 4 283 090 | 4 |
| Chr8 | 5 623 000 | 5 623 045 | 1 |
| Chr8 | 10 447 300 | 10 447 314 | 1 |
| Chr8 | 14 970 700 | 14 970 730 | 1 |
| Chr8 | 15 598 900 | 15 598 941 | 1 |
| Chr8 | 18 006 500 | 18 006 549 | 1 |
| Chr8 | 18 452 681 | 18 452 693 | 1 |
| Chr8 | 19 739 048 | 20 197 724 | 11 |
| Chr9 | 19 759 500 | 19 759 596 | 2 |
| Chr9 | 20 098 700 | 20 098 775 | 1 |
| Chr11 | 5 919 041 | 6 002 538 | 2 |
| Chr11 | 18 350 200 | 18 350 292 | 1 |
| Chr12 | 10 137 200 | 10 137 217 | 1 |
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation |
|---|---|---|
| Chr6 | LOC_Os06g40704 | 基质膜相关蛋白 Stromal membrane-associated protein |
| Chr7 | LOC_Os07g12520 | 锌离子结合蛋白 Zinc ion binding protein |
| Chr7 | LOC_Os07g12530 | ABC1家族结构域蛋白 ABC1 family domain containing protein |
| Chr7 | LOC_Os07g14160 | 多聚半乳糖醛酸酶 Polygalacturonase |
| Chr7 | LOC_Os07g18560 | 含有OsFBLD6-F-box,LRR和FBD结构域的蛋白 OsFBLD6 - F-box, LRR and FBD domain containing protein |
| Chr7 | LOC_Os07g19070 | 前折叠蛋白 Prefoldin |
| Chr7 | LOC_Os07g19210 | 细胞色素P450 Cytochrome P450 |
| Chr7 | LOC_Os07g22710 | 钙依赖性蛋白激酶 CAMK_CAMK_like.32-CAMK includes calcium/calmodulin depedent protein kinases |
| Chr7 | LOC_Os07g22720 | 含氧酸脱氢酶酰基转移酶结构域蛋白 2-oxo acid dehydrogenases acyltransferase domain containing protein |
| Chr7 | LOC_Os07g22930 | 淀粉合酶 Starch synthase |
| Chr7 | LOC_Os07g23169 | 甲基转移酶 Methyltransferase |
| Chr7 | LOC_Os07g23470 | 索马甜 Thaumatin |
| Chr7 | LOC_Os07g23740 | 甾醇3-β-葡萄糖基转移酶 Sterol 3-beta-glucosyltransferase |
| Chr7 | LOC_Os07g23890 | 含有OsFBDUF36-F-box和DUF结构域的蛋白 OsFBDUF36-F-box and DUF domain containing protein |
| Chr7 | LOC_Os07g23900 | 含有OsFBX234-F-box结构域的蛋白 OsFBX234-F-box domain containing protein |
| Chr7 | LOC_Os07g23944 | 糖基水解酶31家族 Glycosyl hydrolase, family 31 |
| Chr7 | LOC_Os07g24350 | 含有同源盒结构域的蛋白 Homeobox domain containing protein |
| Chr7 | LOC_Os07g28110 | 细胞色素P450 Cytochrome P450 |
| Chr7 | LOC_Os07g28480 | 谷胱甘肽S-转移酶 Glutathione S-transferase |
| Chr8 | LOC_Os08g02520 | OsSAUR31-生长素响应SAUR基因家族成员 OsSAUR31-Auxin-responsive SAUR gene family member |
表4 SNP与Indel一致区间内的候选基因
Table 4 Candidate genes within the consistent interval between SNP and Indel
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation |
|---|---|---|
| Chr6 | LOC_Os06g40704 | 基质膜相关蛋白 Stromal membrane-associated protein |
| Chr7 | LOC_Os07g12520 | 锌离子结合蛋白 Zinc ion binding protein |
| Chr7 | LOC_Os07g12530 | ABC1家族结构域蛋白 ABC1 family domain containing protein |
| Chr7 | LOC_Os07g14160 | 多聚半乳糖醛酸酶 Polygalacturonase |
| Chr7 | LOC_Os07g18560 | 含有OsFBLD6-F-box,LRR和FBD结构域的蛋白 OsFBLD6 - F-box, LRR and FBD domain containing protein |
| Chr7 | LOC_Os07g19070 | 前折叠蛋白 Prefoldin |
| Chr7 | LOC_Os07g19210 | 细胞色素P450 Cytochrome P450 |
| Chr7 | LOC_Os07g22710 | 钙依赖性蛋白激酶 CAMK_CAMK_like.32-CAMK includes calcium/calmodulin depedent protein kinases |
| Chr7 | LOC_Os07g22720 | 含氧酸脱氢酶酰基转移酶结构域蛋白 2-oxo acid dehydrogenases acyltransferase domain containing protein |
| Chr7 | LOC_Os07g22930 | 淀粉合酶 Starch synthase |
| Chr7 | LOC_Os07g23169 | 甲基转移酶 Methyltransferase |
| Chr7 | LOC_Os07g23470 | 索马甜 Thaumatin |
| Chr7 | LOC_Os07g23740 | 甾醇3-β-葡萄糖基转移酶 Sterol 3-beta-glucosyltransferase |
| Chr7 | LOC_Os07g23890 | 含有OsFBDUF36-F-box和DUF结构域的蛋白 OsFBDUF36-F-box and DUF domain containing protein |
| Chr7 | LOC_Os07g23900 | 含有OsFBX234-F-box结构域的蛋白 OsFBX234-F-box domain containing protein |
| Chr7 | LOC_Os07g23944 | 糖基水解酶31家族 Glycosyl hydrolase, family 31 |
| Chr7 | LOC_Os07g24350 | 含有同源盒结构域的蛋白 Homeobox domain containing protein |
| Chr7 | LOC_Os07g28110 | 细胞色素P450 Cytochrome P450 |
| Chr7 | LOC_Os07g28480 | 谷胱甘肽S-转移酶 Glutathione S-transferase |
| Chr8 | LOC_Os08g02520 | OsSAUR31-生长素响应SAUR基因家族成员 OsSAUR31-Auxin-responsive SAUR gene family member |
染色体 Chromosome | 起始位置 Start position/bp | 终止位置 End position/bp | 基因数量 Gene number |
|---|---|---|---|
| Chr12 | 10 696 100 | 10 696 167 | 1 |
| Chr12 | 11 928 400 | 11 928 496 | 1 |
| Chr12 | 13 782 500 | 13 782 529 | 1 |
| Chr12 | 21 876 500 | 21 876 519 | 1 |
| Chr12 | 22 742 600 | 22 742 664 | 1 |
表2 SNP关联区间 (续表Continued)
Table 2 SNP association interval
染色体 Chromosome | 起始位置 Start position/bp | 终止位置 End position/bp | 基因数量 Gene number |
|---|---|---|---|
| Chr12 | 10 696 100 | 10 696 167 | 1 |
| Chr12 | 11 928 400 | 11 928 496 | 1 |
| Chr12 | 13 782 500 | 13 782 529 | 1 |
| Chr12 | 21 876 500 | 21 876 519 | 1 |
| Chr12 | 22 742 600 | 22 742 664 | 1 |
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation |
|---|---|---|
| Chr8 | LOC_Os08g02550 | 蛋白酶体亚基 Proteasome subunit |
| Chr8 | LOC_Os08g07060 | 叶绿体呼吸 Chlororespiratory reduction 6 |
| Chr8 | LOC_Os08g07080 | 萜烯合酶 Terpene synthase |
| Chr8 | LOC_Os08g07200 | UDP-葡萄糖醛酸基/UDP-葡萄糖基转移酶 UDP-glucoronosyl/UDP-glucosyl transferase |
| Chr8 | LOC_Os08g30020 | 膜蛋白 Membrane protein |
| Chr8 | LOC_Os08g31830 | 含有UPF0041结构域的蛋白 UPF0041 domain containing protein |
| Chr8 | LOC_Os08g32160 | 氧化还原酶,含2OG-Fell加氧酶结构域的蛋白 Oxidoreductase, 2OG-FeII oxygenase domain containing protein |
| Chr8 | LOC_Os08g32370 | 粘蛋白相关表面蛋白 Mucin-associated surface protein |
| Chr9 | LOC_Os09g34060 | 转录因子RF2a Transcription factor RF2a |
表4 SNP与Indel一致区间内的候选基因 (续表Continued)
Table 4 Candidate genes within the consistent interval between SNP and Indel
染色体 Chromosome | 候选基因 Candidate gene | 基因注释 Gene annotation |
|---|---|---|
| Chr8 | LOC_Os08g02550 | 蛋白酶体亚基 Proteasome subunit |
| Chr8 | LOC_Os08g07060 | 叶绿体呼吸 Chlororespiratory reduction 6 |
| Chr8 | LOC_Os08g07080 | 萜烯合酶 Terpene synthase |
| Chr8 | LOC_Os08g07200 | UDP-葡萄糖醛酸基/UDP-葡萄糖基转移酶 UDP-glucoronosyl/UDP-glucosyl transferase |
| Chr8 | LOC_Os08g30020 | 膜蛋白 Membrane protein |
| Chr8 | LOC_Os08g31830 | 含有UPF0041结构域的蛋白 UPF0041 domain containing protein |
| Chr8 | LOC_Os08g32160 | 氧化还原酶,含2OG-Fell加氧酶结构域的蛋白 Oxidoreductase, 2OG-FeII oxygenase domain containing protein |
| Chr8 | LOC_Os08g32370 | 粘蛋白相关表面蛋白 Mucin-associated surface protein |
| Chr9 | LOC_Os09g34060 | 转录因子RF2a Transcription factor RF2a |
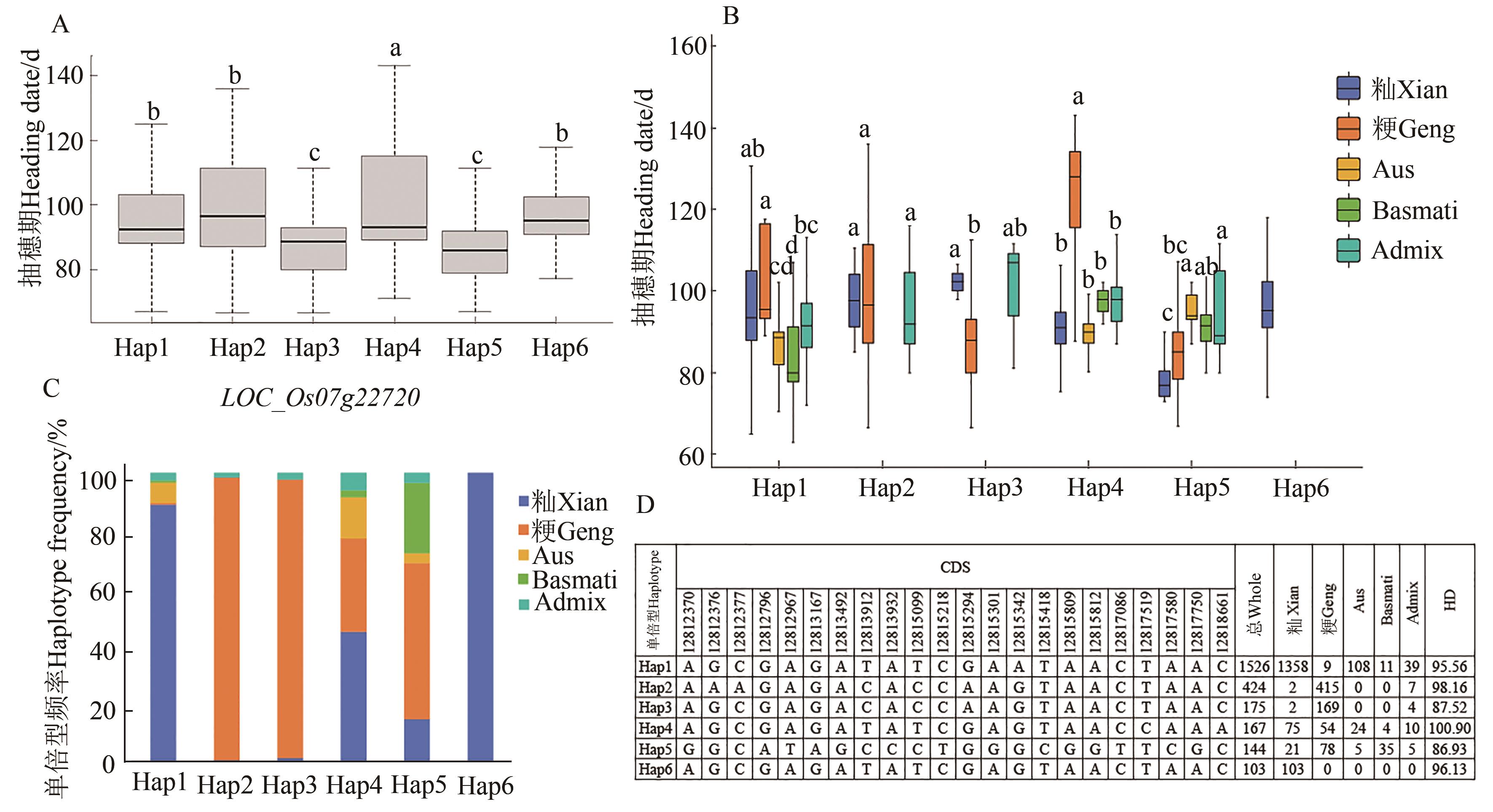
图6 LOC_Os07g22720单倍型分析A:LOC_Os07g22720单倍型的抽穗期分布;B:LOC_Os07g22720不同单倍型在亚群中的分布;C:LOC_Os07g22720不同单倍型在亚群中的频率;D:LOC_Os07g22720的CDS区的单倍型。不同小写字母表示单倍型之间在P<0.05水平差异显著
Fig. 6 LOC_Os07g22720 haplotype analysisA: Heading stage distribution of LOC_Os07g22720 haplotypes; B: Distribution of different haplotypes of LOC_Os07g22720 in subpopulations; C: Frequency of different haplotypes of LOC_Os07g22720 in subpopulations; D: Haplotypes of CDS region of LOC_Os07g22720. Different small letters indicate significant differences between haplotypes at P<0.05 level

图7 LOC_Os07g23740单倍型分析A:LOC_Os07g23740单倍型的抽穗期分布;B:LOC_Os07g23740不同单倍型在亚群中的分布;C:LOC_Os07g23740不同单倍型在亚群中的频率;D:LOC_Os07g23740的CDS区的单倍型。不同小写字母表示单倍型之间在P<0.05水平差异显著
Fig. 7 LOC_Os07g23740 haplotype analysisA: Heading stage distribution of LOC_Os07g23740 haplotypes; B: Distribution of different haplotypes of LOC_Os07g23740 in subpopulations; C: Frequency of different haplotypes of LOC_Os07g23740 in subpopulations; D: Haplotypes of CDS region of LOC_Os07g23740. Different small letters indicate significant differences between haplotypes at P<0.05 level
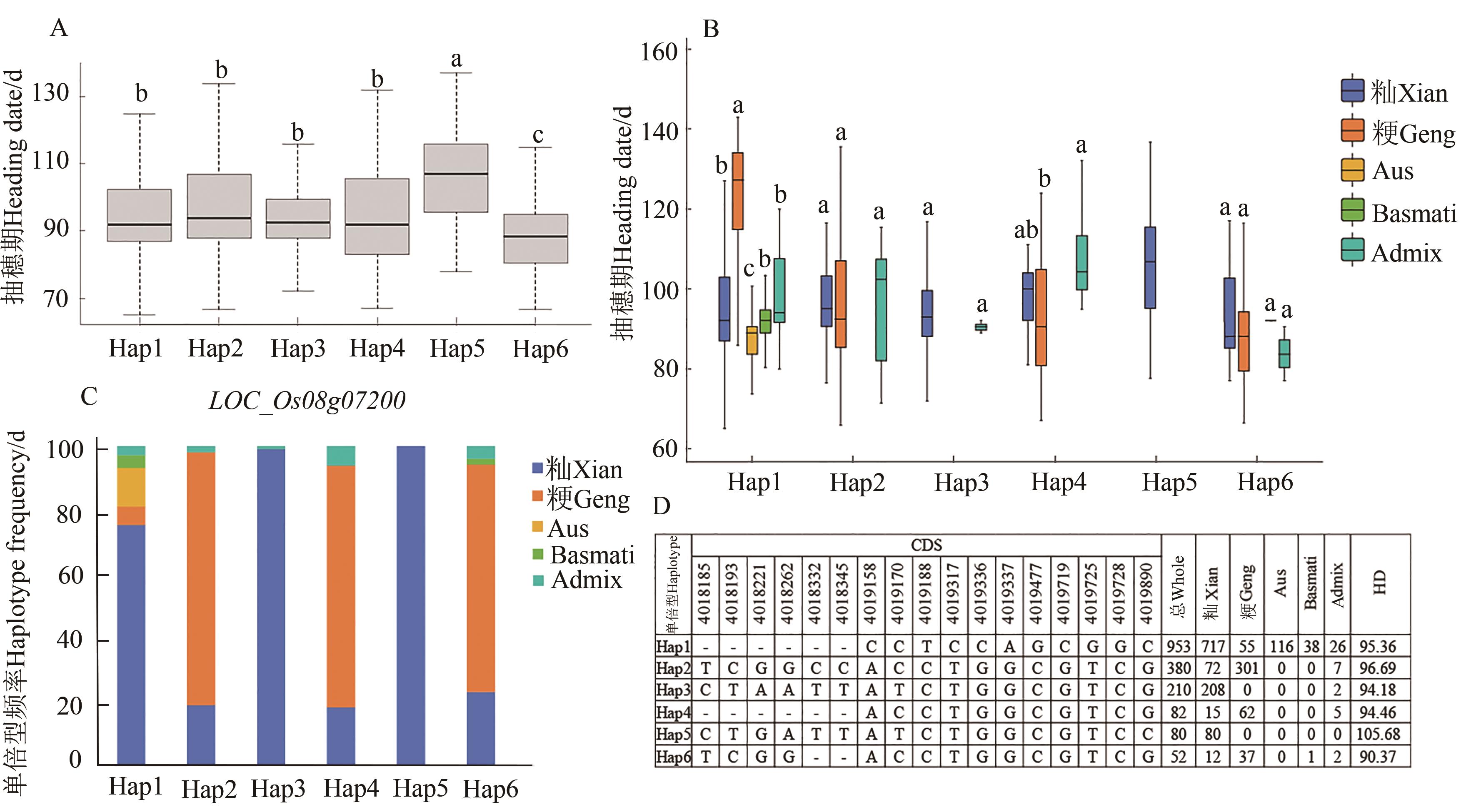
图8 LOC_Os08g07200单倍型显著性分析A:LOC_Os08g07200单倍型的抽穗期分布;B:LOC_Os0g07200不同单倍型在亚群中的分布;C:LOC_Os08g07200不同单倍型在亚群中的频率;D:LOC_Os08g07200的CDS区的单倍型。不同小写字母表示单倍型之间在P<0.05水平差异显著
Fig. 8 LOC_Os08g07200 haplotype significance analysisA: Heading stage distribution of LOC_Os08g07200 haplotypes; B: Distribution of different haplotypes of LOC_Os0g07200 in subpopulations; C: Frequency of different haplotypes of LOC_Os08g07200 in subpopulations; D: Haplotypes of CDS region of LOC_Os08g07200. Different small letters indicate significant differences between haplotypes at P<0.05 level
| 1 | 崔月.水稻抽穗期基因组合对产量相关性状的影响及其优化调控[D].沈阳:沈阳农业大学, 2020. |
| CUI Y. Optimization of rice productivity across a large latitudinal gradient using induce mutation in heading date [D]. Shenyang: Shenyang Agricultural University, 2020. | |
| 2 | 赵凌,梁文化,赵春芳,等.利用高密度Bin遗传图谱定位水稻抽穗期QTL[J].作物学报, 2023,49(1):119-128. |
| ZHAO L, LIANG W H, ZHAO C F, et al.. Mapping of QTLs for heading date of rice with high-density bin genetic map [J]. Acta Agron. Sin., 2023, 49(1): 119-128. | |
| 3 | 郭梁,张振华,庄杰云.水稻抽穗期QTL及其与产量性状遗传控制的关系[J].中国水稻科学,2012,26(2):235-245. |
| GUO L, ZHANG Z H, ZHUANG J Y. Quantitative trait loci for heading date and their relationship with the genetic control of yield traits in rice (Oryza sativa L.) [J]. Chin. J. Rice Sci., 2012,26(2): 235-245. | |
| 4 | 胡时开,苏岩,叶卫军,等.水稻抽穗期遗传与分子调控机理研究进展[J].中国水稻科学, 2012,26(3):373-382. |
| HU S K, SU Y, YE W J, et al.. Advances in genetic analysis and molecular regulation mechanism of heading date in rice (Oryza sativa L.) [J]. Chin. J. Rice Sci., 2012, 26(3): 373-382. | |
| 5 | 林鸿宣,钱惠荣,熊振民.几个水稻品种抽穗期主效基因与微效基因的定位研究[J].遗传学报,1996(3):205-213. |
| LIN H X, QIAN H R, XIONG Z M. Mapping of major genes and minor genes for heading date in several rice varieties (Oryza sativa L.) [J]. J. Genet. Genomics, 1996 (3): 205-213. | |
| 6 | YANO M, HARUSHIMA Y, NAGAMURA Y, et al.. Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map [J]. Theor. Appl. Genet., 1997, 95(7): 1025-1032. |
| 7 | XUE W, XING Y, WENG X, et al.. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice [J]. Nat. Genet., 2008, 40(6): 761-767. |
| 8 | YAN W H, WANG P, CHEN H X, et al.. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice [J]. Mol. Plant, 2011, 4(2): 319-330. |
| 9 | MICHELMORE R W, PARAN I, KESSELI R V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations [J]. Proc. Natl. Acad. Sci. USA, 1991, 88(21): 9828-9832. |
| 10 | 朱玉贤,李毅,郑晓峰,等.现代分子生物学 [M].北京:高等教育出版社,2019:1-304. |
| 11 | ZHENG Y, XU F, LI Q, et al.. QTL mapping combined with bulked segregant analysis identify SNP markers linked to leaf shape traits in Pisum sativum using SLAF sequencing [J/OL]. Front. Genet., 2018, 9: 615 [2024-05-24]. https:doi.org/10.3389/fgene.2018.00615. |
| 12 | 孙亚倩,陈士亮,褚佳豪,等.基于BSA-seq结合连锁分析发掘大豆荚粒性状QTLs及候选基因[J].中国农业科技导报,2023,25(7):29-42. |
| SUN Y Q, CHEN S L, CHU J H, et al.. Mining of QTLs and candidate genes for pod and seed traits via combining BSA-seq and linkage mapping in soybean [J]. J. Agric. Sci. Technol., 2023, 25(7): 29-42. | |
| 13 | 宋夏夏,王利民,张建平,等.胡麻株高QTL定位与候选基因功能分析[J].中国农业科技导报,2020,22(6):26-32. |
| SONG X X, WANG L M, ZHANG J P, et al.. QTL mapping and function analysis of candidate genes related to plant height in flax [J]. J. Agric. Sci. Technol., 2020, 22(6): 26-32. | |
| 14 | 王雪彬,张健,韦燕燕,等.基于BSA-seq的水稻籽粒耐陈化QTL定位分析[J].分子植物育种,2023,21(16):5337-5347. |
| WANG X B, ZHANG J, WEI Y Y, et al.. QTLs mapping analysis of rice grain aging tolerance based on BSA-seq [J]. Mol. Plant Breed., 2023, 21(16): 5337-5347. | |
| 15 | 孙家臣,蒋辅燕,尹兴福,等.基于BSA的玉米花期相关性状基因定位[J].玉米科学,2023, 31(3):48-57. |
| SUN J C, JIANG F Y, YIN X F, et al.. Gene mapping of maize flowering time related traits based on BSA [J]. J. Maize Sci., 2023,31(3):48-57. | |
| 16 | 吴元明,林佳怡,柳雨汐,等.基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J].生物技术通报, 2023,39(8):173-184. |
| WU Y M, LIN J Y, LIU Y X, et al.. Identification of rice plant height associated QTL using BSA-seq and RNA-seq [J]. Biotechnol. Bull., 2023. 39(8): 173-184. | |
| 17 | 杨锟.水稻穗发芽和抽穗期的QTL定位[D].南京:南京农业大学, 2019. |
| YANG K. Mapping of QTL for pre-harvest sprouting and heading date in rice (Oryza sativa L.) [D]. Nanjing:Nanjing Agricultural University, 2019. | |
| 18 | WANG C C, YU H, HUANG J, et al.. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0 [J]. Plant Biotechnol. J., 2020, 18(1): 14-16. |
| 19 | 吴心成,桂金鑫,刘瑫,等.水稻穗长全基因组关联分析[J/OL].分子植物育种,1-13[2024-05-22].. |
| WU X C, GUI J X, LIU T, et al.. Genome-wide association study for panicle length in rice [J/OL]. Mol. Plant Breeding,2024: 1-13 [2024-05-22]. . | |
| 20 | TAKAGI H, ABE A, YOSHIDA K, et al.. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations [J]. Plant J., 2013, 74(1): 174-183. |
| 21 | 蒋丹,洪广成,陈倩,等.水稻抽穗期分子调控研究进展[J].分子植物育种, 2019,17(21): 7071-7077. |
| JIANG D, HONG G C, CHEN Q, et al.. Research progress in molecular regulation of heading date in rice (Oryza sativa) [J]. Mol. Plant Breeding, 2019, 17(21): 7071-7077. | |
| 22 | WAADT R, SELLER C A, HSU P-K, et al.. Plant hormone regulation of abiotic stress responses [J]. Nat. Rev. Mol. Cell Biol., 2022, 23(10): 680-694. |
| 23 | 闫晓峰,胡渊,黄晓龙,等.水稻抽穗期基因OsFKF1的克隆和互作蛋白筛选[J].南京农业大学学报,2023,46(3):429-437. |
| YAN X F, HU Y, HUANG X L, et al.. Cloning of rice heading date gene OsFKF1 and screening of interacting proteins [J]. J. Nanjing Agric. Univ., 2023, 46(3):429-437. | |
| 24 | 曲丽君,张宏军,项超,等.杂交稻"大青棵"现象遗传基础剖析[J].中国水稻科学,2013,27(6):559-568. |
| QU L J, ZHANG H J, XIANG C, Study on the genetic basis for abnormal heading in hybrid rice [J]. Chin. J. Rice Sci.,2013,27(6):559-568. | |
| 25 | JIAO Z L, ZHANG Q M, XU W J, et al..The putative obtusifoliol 14α-demethylase OsCYP51H3 affects multiple aspects of rice growth and development [J]. Physiol. Plant, 174(5), e13764. |
| 26 | YAMORI W, SAKATA N, SUZUKI Y, et al.. Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice [J]. Plant J., 2011, 68(6): 966-976. |
| 27 | ZHANG G Z, JIN S H, LI P, et al.. Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis [J]. Plant Cell Rep., 2017,36(12): 1995-2006. |
| 28 | 于安东,刘琳,龙瑞才,等.植物UDP-糖基转移酶(UGT)的功能及应用前景[J].植物生理学报,2022,58(4):631-642. |
| YU A D, LIU L, LONG R C,et al..Function and application prospect of plant UDP-glycosyltransferase (UGT) [J]. Plant Physiol. J.,2022,58(4):631-642. | |
| 29 | HONG Y, ZHANG Y, SINUMPORN S, et al.. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice [J]. Plant J., 2018, 95(5): 877-891. |
| [1] | 孙亮, 徐益, 蔡沁, 郭靖豪, 赵灿, 郭保卫, 邢志鹏, 霍中洋, 张洪程, 胡雅杰. 中微量元素对水稻产量和品质的影响研究进展[J]. 中国农业科技导报, 2024, 26(8): 9-19. |
| [2] | 刘大为, 秦锋, 廖骞, 王修善, 谢方平, 李铁辉. 南方籼稻热风干燥特性及其工艺参数优化[J]. 中国农业科技导报, 2024, 26(8): 93-102. |
| [3] | 岳伟, 王晖, 陈曦, 占新春, 阮新民. 安徽省稻米品质综合评价方法研究[J]. 中国农业科技导报, 2024, 26(6): 141-147. |
| [4] | 陈明迪, 胡桂花, 张海文, 王旺田. 水稻RR基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(5): 20-29. |
| [5] | 曾建光, 刘桃李, 孙林娟, 袁定阳, 黄钰博, 金晨钟, 谭炎宁. 水稻矮秆迟抽穗突变体d534的性状及其对赤霉素的敏感性分析[J]. 中国农业科技导报, 2024, 26(3): 7-14. |
| [6] | 钱政, 杨孙哲, 张国卿, 郭紫微, 张林朋, 万家兴, 杨红云. 基于卷积神经网络的水稻氮素营养诊断[J]. 中国农业科技导报, 2023, 25(9): 113-121. |
| [7] | 李慧君, 张伟健, 吴伟健, 李高洋, 陈艺杰, 黄枫城, 黄永相, 蔺中, 甄珍. 种植海水稻对滨海盐土化学性质和微生物群落影响[J]. 中国农业科技导报, 2023, 25(9): 147-156. |
| [8] | 邵社刚, 李婷, 柳勇, 林兰稳, 张东, 倪栋, 李俊杰, 朱立安. 外源菌剂对稻秆腐解及微生物群落结构的影响[J]. 中国农业科技导报, 2023, 25(9): 166-177. |
| [9] | 单莉莉. 孕穗期低温对水稻叶片生理、产量的影响及外源褪黑素缓解效应[J]. 中国农业科技导报, 2023, 25(9): 23-33. |
| [10] | 孙亚倩, 陈士亮, 褚佳豪, 李喜焕, 张彩英. 基于BSA-seq结合连锁分析发掘大豆荚粒性状QTLs及候选基因[J]. 中国农业科技导报, 2023, 25(7): 29-42. |
| [11] | 张冬梦, 姚栋萍, 吴俊, 罗秋红, 庄文, 刘雄伦, 邓启云, 柏斌. 灌浆期田间自然低温对稻米蒸煮食味品质的影响[J]. 中国农业科技导报, 2023, 25(6): 144-153. |
| [12] | 肖芬芳, 张从合, 王慧, 叶亚峰, 张道林, 汪和廷, 李波, 吴跃进, 刘斌美. 杂交水稻花粉收集装置气力输送系统仿真优化[J]. 中国农业科技导报, 2023, 25(4): 110-122. |
| [13] | 尹林江, 李威, 赵卫权, 赵祖伦, 吕思思, 孙小琼. 水稻多时相植被指数特征及覆盖度提取研究[J]. 中国农业科技导报, 2023, 25(2): 83-98. |
| [14] | 谷英楠, 刘鑫, 王敬元, 段新宇, 黄莹, 林庆娟, 刘艳霞, 毕洪文. 黑龙江省水稻产业发展现状及展望[J]. 中国农业科技导报, 2023, 25(12): 17-25. |
| [15] | 杨文俊, 朱雨婷, 张杰, 徐凯祥, 韦聪敏, 陈全家. 棉花苗期耐盐相关性状QTL元分析[J]. 中国农业科技导报, 2023, 25(12): 26-34. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号