




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (11): 79-90.DOI: 10.13304/j.nykjdb.2024.0423
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Nigela Abuduwaili, Xiuzhen WEI, Jingjie MEN, Lingna CHEN, Yongkun CHEN( ), Gengqing HUANG(
), Gengqing HUANG( )
)
Received:2024-05-23
Accepted:2024-06-21
Online:2024-11-15
Published:2024-11-19
Contact:
Yongkun CHEN,Gengqing HUANG
尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤( ), 黄耿青(
), 黄耿青( )
)
通讯作者:
陈永坤,黄耿青
作者简介:第一联系人:尼格拉•阿布都外力 E-mail: 1755877085@qq.com
基金资助:CLC Number:
Nigela Abuduwaili, Xiuzhen WEI, Jingjie MEN, Lingna CHEN, Yongkun CHEN, Gengqing HUANG. Bioinformatics and Expression Pattern Analysis of JAZ and MYC2-like Gene Family in Lavandula angustifolia[J]. Journal of Agricultural Science and Technology, 2024, 26(11): 79-90.
尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤, 黄耿青. 薰衣草 JAZ和MYC2-like基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(11): 79-90.
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数量 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAZ1 | La01G01624 | Chr1 | 14 191 132 | 14 192 638 | 334 | 35.531 14 | 9.73 |
| LaJAZ2 | La01G02122 | Chr1 | 22 315 757 | 22 318 426 | 279 | 29.241 02 | 7.64 |
| LaJAZ3 | La01G02748 | Chr1 | 29 933 237 | 29 935 361 | 233 | 25.500 76 | 9.58 |
| LaJAZ4 | La01G02775 | Chr1 | 30 304 246 | 30 305 952 | 270 | 29.719 64 | 8.74 |
| LaJAZ5 | La02G01205 | Chr2 | 27 444 500 | 27 446 515 | 252 | 26.659 79 | 9.34 |
| LaJAZ6 | La04G00443 | Chr4 | 23 907 703 | 2 908 702 | 207 | 21.913 95 | 8.46 |
| LaJAZ7 | La10G01957 | Chr10 | 37 391 837 | 37 394 501 | 255 | 27.540 92 | 9.12 |
| LaJAZ8 | La11G01369 | Chr11 | 30 835 601 | 30 838 263 | 297 | 31.243 32 | 8.31 |
| LaJAZ9 | La12G01015 | Chr12 | 20 257 088 | 20 259 717 | 275 | 29.305 02 | 9.43 |
| LaJAZ10 | La14G00862 | Chr14 | 9 169 268 | 9 170 127 | 211 | 22.548 44 | 9.44 |
| LaJAZ11 | La15G00121 | Chr15 | 866 936 | 870 632 | 345 | 37.302 14 | 8.46 |
| LaJAZ12 | La15G00153 | Chr15 | 1 075 151 | 1 079 126 | 212 | 22.390 18 | 9.80 |
| LaJAZ13 | La15G00189 | Chr15 | 1 330 626 | 1 333 013 | 283 | 30.562 52 | 9.45 |
| LaJAZ14 | La15G01076 | Chr15 | 25 198 964 | 25 199 826 | 213 | 22.829 80 | 9.93 |
| LaJAZ15 | La16G00168 | Chr16 | 1 435 513 | 1 440 135 | 156 | 17.603 23 | 10.38 |
| LaJAZ16 | La17G00791 | Chr17 | 17 083 687 | 17 085 577 | 226 | 24.465 89 | 9.93 |
| LaJAZ17 | La18G00414 | Chr18 | 4 272 331 | 4 275 830 | 330 | 34.990 72 | 9.87 |
| LaJAZ18 | La19G01387 | Chr19 | 22 164 844 | 22 167 383 | 136 | 15.788 10 | 9.52 |
| LaJAZ19 | La20G01484 | Chr20 | 16 672 832 | 16 673 901 | 214 | 22.483 34 | 9.08 |
| LaJAZ20 | La20G02795 | Chr20 | 29 446 294 | 29 450 179 | 190 | 20.995 00 | 10.19 |
| LaJAZ21 | La21G00568 | Chr21 | 6 010 126 | 6 014 598 | 413 | 45.401 64 | 8.20 |
| LaJAZ22 | La21G00790 | Chr21 | 16 075 757 | 16 078 401 | 278 | 29.511 56 | 9.81 |
| LaJAZ23 | La24G00748 | Chr24 | 6 890 348 | 6 890 710 | 121 | 13.522 38 | 9.31 |
| LaJAZ24 | La00G00334 | scaffold171 | 144 787 | 145 649 | 213 | 22.829 80 | 9.93 |
| LaJAZ25 | La00G00563 | scaffold214 | 58 478 | 62 950 | 413 | 45.391 54 | 8.27 |
| LaJAZ26 | La00G03185 | scaffold362 | 51 131 | 53 153 | 244 | 25.608 65 | 10.10 |
Table 1 Basic information of JAZ genes in Lavandula angustifolia
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数量 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAZ1 | La01G01624 | Chr1 | 14 191 132 | 14 192 638 | 334 | 35.531 14 | 9.73 |
| LaJAZ2 | La01G02122 | Chr1 | 22 315 757 | 22 318 426 | 279 | 29.241 02 | 7.64 |
| LaJAZ3 | La01G02748 | Chr1 | 29 933 237 | 29 935 361 | 233 | 25.500 76 | 9.58 |
| LaJAZ4 | La01G02775 | Chr1 | 30 304 246 | 30 305 952 | 270 | 29.719 64 | 8.74 |
| LaJAZ5 | La02G01205 | Chr2 | 27 444 500 | 27 446 515 | 252 | 26.659 79 | 9.34 |
| LaJAZ6 | La04G00443 | Chr4 | 23 907 703 | 2 908 702 | 207 | 21.913 95 | 8.46 |
| LaJAZ7 | La10G01957 | Chr10 | 37 391 837 | 37 394 501 | 255 | 27.540 92 | 9.12 |
| LaJAZ8 | La11G01369 | Chr11 | 30 835 601 | 30 838 263 | 297 | 31.243 32 | 8.31 |
| LaJAZ9 | La12G01015 | Chr12 | 20 257 088 | 20 259 717 | 275 | 29.305 02 | 9.43 |
| LaJAZ10 | La14G00862 | Chr14 | 9 169 268 | 9 170 127 | 211 | 22.548 44 | 9.44 |
| LaJAZ11 | La15G00121 | Chr15 | 866 936 | 870 632 | 345 | 37.302 14 | 8.46 |
| LaJAZ12 | La15G00153 | Chr15 | 1 075 151 | 1 079 126 | 212 | 22.390 18 | 9.80 |
| LaJAZ13 | La15G00189 | Chr15 | 1 330 626 | 1 333 013 | 283 | 30.562 52 | 9.45 |
| LaJAZ14 | La15G01076 | Chr15 | 25 198 964 | 25 199 826 | 213 | 22.829 80 | 9.93 |
| LaJAZ15 | La16G00168 | Chr16 | 1 435 513 | 1 440 135 | 156 | 17.603 23 | 10.38 |
| LaJAZ16 | La17G00791 | Chr17 | 17 083 687 | 17 085 577 | 226 | 24.465 89 | 9.93 |
| LaJAZ17 | La18G00414 | Chr18 | 4 272 331 | 4 275 830 | 330 | 34.990 72 | 9.87 |
| LaJAZ18 | La19G01387 | Chr19 | 22 164 844 | 22 167 383 | 136 | 15.788 10 | 9.52 |
| LaJAZ19 | La20G01484 | Chr20 | 16 672 832 | 16 673 901 | 214 | 22.483 34 | 9.08 |
| LaJAZ20 | La20G02795 | Chr20 | 29 446 294 | 29 450 179 | 190 | 20.995 00 | 10.19 |
| LaJAZ21 | La21G00568 | Chr21 | 6 010 126 | 6 014 598 | 413 | 45.401 64 | 8.20 |
| LaJAZ22 | La21G00790 | Chr21 | 16 075 757 | 16 078 401 | 278 | 29.511 56 | 9.81 |
| LaJAZ23 | La24G00748 | Chr24 | 6 890 348 | 6 890 710 | 121 | 13.522 38 | 9.31 |
| LaJAZ24 | La00G00334 | scaffold171 | 144 787 | 145 649 | 213 | 22.829 80 | 9.93 |
| LaJAZ25 | La00G00563 | scaffold214 | 58 478 | 62 950 | 413 | 45.391 54 | 8.27 |
| LaJAZ26 | La00G03185 | scaffold362 | 51 131 | 53 153 | 244 | 25.608 65 | 10.10 |
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaGL1 | La03G00379 | Chr3 | 2 443 649 | 2 447 074 | 537 | 59.435 64 | 4.74 |
| LaGL2 | La13G01461 | Chr13 | 28 879 003 | 28 882 769 | 564 | 62.550 64 | 5.89 |
| LaGL3 | La22G02008 | Chr22 | 19 300 412 | 19 303 804 | 615 | 68.438 72 | 6.34 |
| LaGL4 | La23G02048 | Chr23 | 15 406 101 | 15 409 527 | 615 | 68.475 87 | 7.08 |
| LaGL5 | La00G04274 | scaffold463 | 39 967 | 46 925 | 1 260 | 139.787 86 | 7.35 |
| LaJAM1 | La04G02776 | Chr4 | 19 290 825 | 19 295 838 | 488 | 54.222 49 | 6.37 |
| LaJAM2 | La04G01048 | Chr4 | 6 403 237 | 6 405 850 | 607 | 67.699 30 | 7.29 |
| LaJAM3 | La09G00555 | Chr9 | 10 822 064 | 10 824 378 | 586 | 65.382 98 | 8.34 |
| LaJAM4 | La09G02346 | Chr9 | 26 062 346 | 26 064 746 | 500 | 56.370 18 | 7.70 |
Table 2 Basic information of MYC2-like genes in Lavandula angustifolia
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaGL1 | La03G00379 | Chr3 | 2 443 649 | 2 447 074 | 537 | 59.435 64 | 4.74 |
| LaGL2 | La13G01461 | Chr13 | 28 879 003 | 28 882 769 | 564 | 62.550 64 | 5.89 |
| LaGL3 | La22G02008 | Chr22 | 19 300 412 | 19 303 804 | 615 | 68.438 72 | 6.34 |
| LaGL4 | La23G02048 | Chr23 | 15 406 101 | 15 409 527 | 615 | 68.475 87 | 7.08 |
| LaGL5 | La00G04274 | scaffold463 | 39 967 | 46 925 | 1 260 | 139.787 86 | 7.35 |
| LaJAM1 | La04G02776 | Chr4 | 19 290 825 | 19 295 838 | 488 | 54.222 49 | 6.37 |
| LaJAM2 | La04G01048 | Chr4 | 6 403 237 | 6 405 850 | 607 | 67.699 30 | 7.29 |
| LaJAM3 | La09G00555 | Chr9 | 10 822 064 | 10 824 378 | 586 | 65.382 98 | 8.34 |
| LaJAM4 | La09G02346 | Chr9 | 26 062 346 | 26 064 746 | 500 | 56.370 18 | 7.70 |
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAM5 | La11G00149 | Chr11 | 952 209 | 952 997 | 214 | 23 742.54 | 6.64 |
| LaJAM6 | La14G01835 | Chr14 | 35 018 055 | 35 020 479 | 487 | 53 926.27 | 5.73 |
| LaJAM7 | La15G00077 | Chr15 | 455 788 | 457 275 | 496 | 54 676.16 | 6.17 |
| LaJAM8 | La20G01069 | Chr20 | 13 368 557 | 13 371 145 | 488 | 54 015.28 | 6.39 |
| LaJAM9 | La23G01124 | Chr23 | 8 221 465 | 8 223 969 | 604 | 67 064.71 | 7.03 |
| LaJAM10 | La26G00764 | Chr26 | 4 697 504 | 4 699 730 | 509 | 57 161.84 | 7.48 |
| LaJAM11 | La27G00171 | Chr27 | 2 153 629 | 2 156 648 | 571 | 63 635.30 | 8.47 |
| LaJAM12 | La00G05647 | scaffold88 | 71 229 | 74 222 | 604 | 67 036.69 | 7.03 |
| LaJAM13 | La00G04428 | scaffold479 | 84 467 | 87 484 | 571 | 63 579.26 | 8.52 |
| LaMYC1 | La01G01976 | Chr1 | 19 882 771 | 19 885 227 | 659 | 71 878.19 | 5.25 |
| LaMYC2 | La11G01192 | Chr11 | 26 776 504 | 26 778 951 | 655 | 71 246.38 | 5.11 |
| LaMYC3 | La13G00765 | Chr13 | 12 863 123 | 12 864 978 | 476 | 52 637.39 | 6.80 |
| LaMYC4 | La13G00767 | Chr13 | 13 043 198 | 13 044 704 | 476 | 52 623.36 | 6.80 |
| LaMYC5 | La14G01082 | Chr14 | 19 151 335 | 19 158 157 | 678 | 75 041.97 | 6.39 |
| LaMYC6 | La14G01770 | Chr14 | 34 640 706 | 34 642 877 | 617 | 66 918.74 | 5.00 |
| LaMYC7 | La15G00833 | Chr15 | 16 057 347 | 16 059 330 | 598 | 66 439.99 | 5.93 |
| LaMYC8 | La19G01463 | Chr19 | 25 336 517 | 25 338 519 | 474 | 52 241.72 | 6.06 |
Table 2 Basic information of MYC2-like genes in Lavandula angustifolia
基因名称 Gene name | 基因ID Gene ID | 染色体 Chromosome | 起始位置 Initial position | 终止位置 Termination position | 氨基酸数 Amino acids/aa | 分子量 MW/kD | 等电点 pI |
|---|---|---|---|---|---|---|---|
| LaJAM5 | La11G00149 | Chr11 | 952 209 | 952 997 | 214 | 23 742.54 | 6.64 |
| LaJAM6 | La14G01835 | Chr14 | 35 018 055 | 35 020 479 | 487 | 53 926.27 | 5.73 |
| LaJAM7 | La15G00077 | Chr15 | 455 788 | 457 275 | 496 | 54 676.16 | 6.17 |
| LaJAM8 | La20G01069 | Chr20 | 13 368 557 | 13 371 145 | 488 | 54 015.28 | 6.39 |
| LaJAM9 | La23G01124 | Chr23 | 8 221 465 | 8 223 969 | 604 | 67 064.71 | 7.03 |
| LaJAM10 | La26G00764 | Chr26 | 4 697 504 | 4 699 730 | 509 | 57 161.84 | 7.48 |
| LaJAM11 | La27G00171 | Chr27 | 2 153 629 | 2 156 648 | 571 | 63 635.30 | 8.47 |
| LaJAM12 | La00G05647 | scaffold88 | 71 229 | 74 222 | 604 | 67 036.69 | 7.03 |
| LaJAM13 | La00G04428 | scaffold479 | 84 467 | 87 484 | 571 | 63 579.26 | 8.52 |
| LaMYC1 | La01G01976 | Chr1 | 19 882 771 | 19 885 227 | 659 | 71 878.19 | 5.25 |
| LaMYC2 | La11G01192 | Chr11 | 26 776 504 | 26 778 951 | 655 | 71 246.38 | 5.11 |
| LaMYC3 | La13G00765 | Chr13 | 12 863 123 | 12 864 978 | 476 | 52 637.39 | 6.80 |
| LaMYC4 | La13G00767 | Chr13 | 13 043 198 | 13 044 704 | 476 | 52 623.36 | 6.80 |
| LaMYC5 | La14G01082 | Chr14 | 19 151 335 | 19 158 157 | 678 | 75 041.97 | 6.39 |
| LaMYC6 | La14G01770 | Chr14 | 34 640 706 | 34 642 877 | 617 | 66 918.74 | 5.00 |
| LaMYC7 | La15G00833 | Chr15 | 16 057 347 | 16 059 330 | 598 | 66 439.99 | 5.93 |
| LaMYC8 | La19G01463 | Chr19 | 25 336 517 | 25 338 519 | 474 | 52 241.72 | 6.06 |
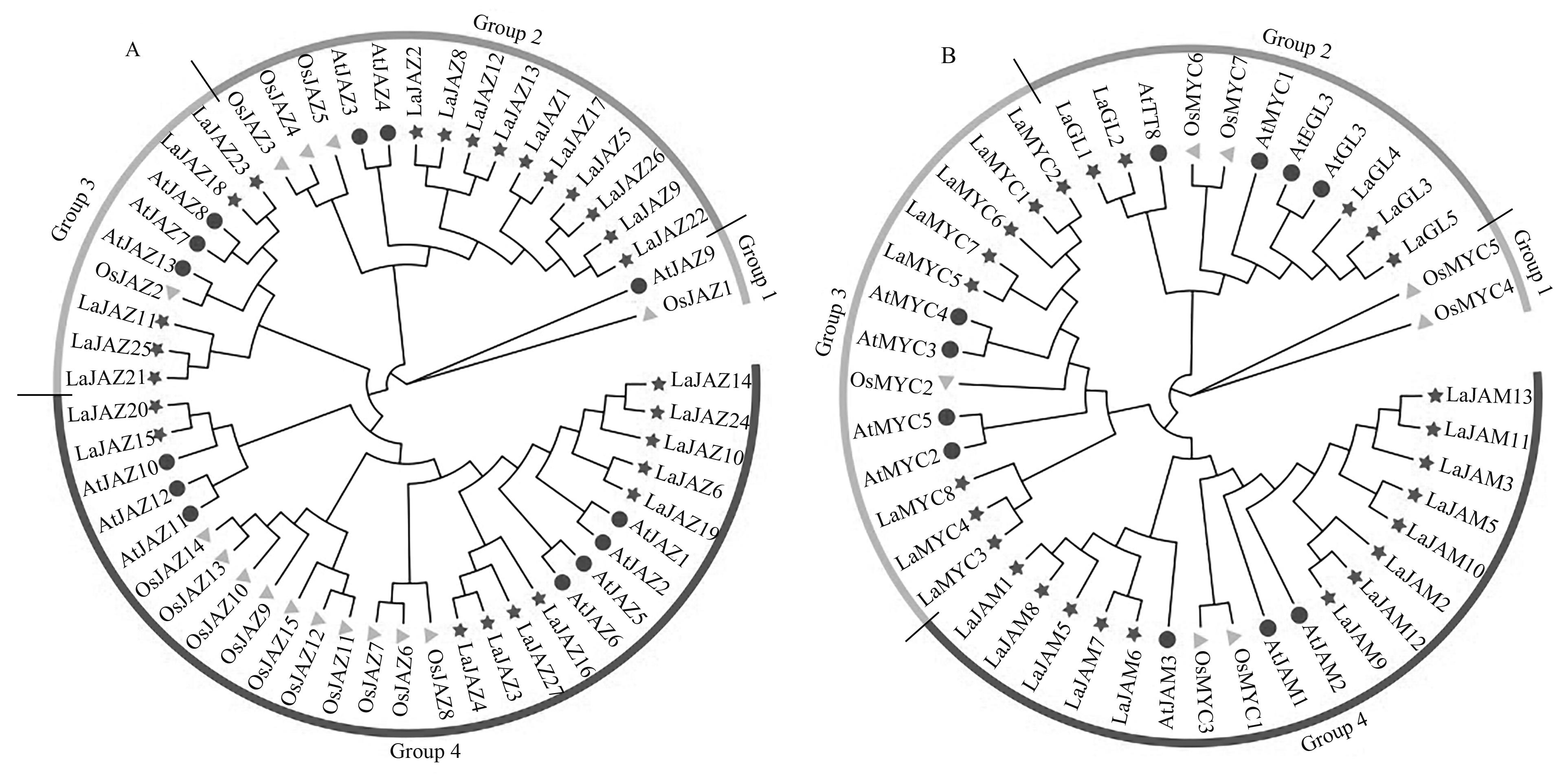
Fig. 2 Phylogenetic tree analysis of JAZ and MYC2-like proteinA: Phylogenetic tree of JAZ proteins; B: Phylogenetic tree of MYC2-like proteins; —Lavandula angustifolia; —Arabidopsis thaliana; —Oryza sativa
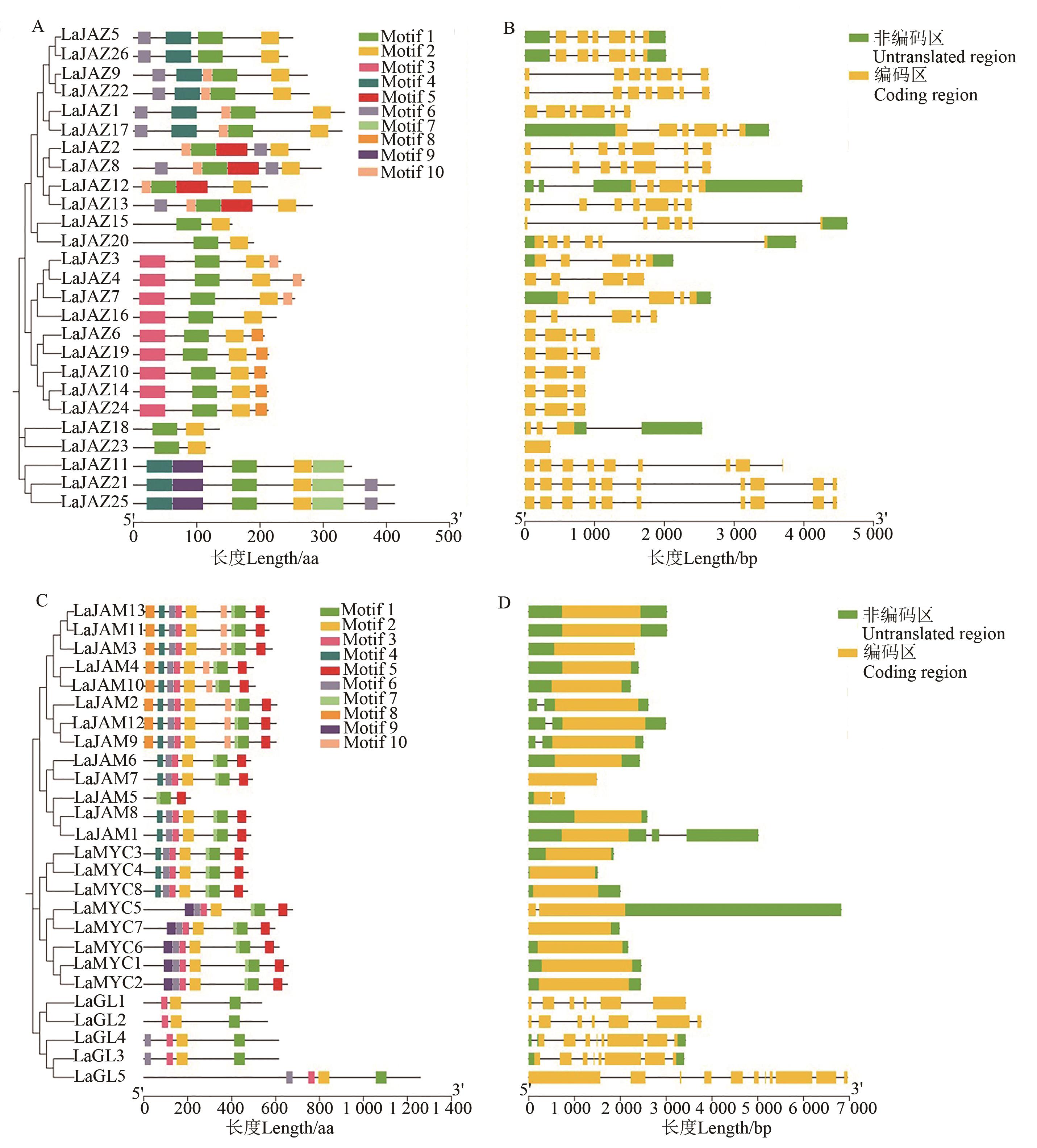
Fig. 3 Conserved motif and gene structure analysis of proteins encoded by LaJAZ and LaMYC2-like genesA: Conserved motif composition of LaJAZ proteins; B: Structure of LaJAZ genes; C: Conserved motif composition of LaMYC2-like proteins; D: Structure of LaMYC2-like genes

Fig. 4 Analysis of cis-elements in the 2 000 bp upstream promoter of LaJAZ and LaMYC2-like genesA: Distribution of cis-acting elements in the LaJAZ promoters; B: Distribution of cis-acting elements in the LaMYC2-like promoters

Fig. 5 Expression analysis of LaJAZ and LaMYC2-like genes in different tissuesNote:Petal—Deep purple petals at the flowering stage; Sepal1—Deep purple sepals at the flower bud stage; Sepal2—Deep purple sepals at the blooming stage; Sepal3—Deep purple sepals at fade stage; Stem—Stems at 2-month-old stage;Leaf—Leaves at 2-month-old stage.

Fig. 6 Expression analysis of LaJAZ and LaMYC2-like genes in different tissues of lavenderA: Different lavender tissues used for gene expression analysis; B: qRT-PCR analysis of expression of the LaJAZ and LaMYC2-like genes in different lavender tissues. 1—Deep purple petals; 2—Deep purple sepals in bud stage; 3—Deep purple sepals at blooming stage; 4—Deep purple sepals at fade stage; 5—Light purple petals; 6—Light purple sepals at bud stage; 7—Light purple sepals at blooming stage; 8—Light purple sepals at fade stage; 9—2-month-old roots; 10—2-month-old stems; 11—2-month-old leaves; different small letters indicate significant differences at P<0.05 level.
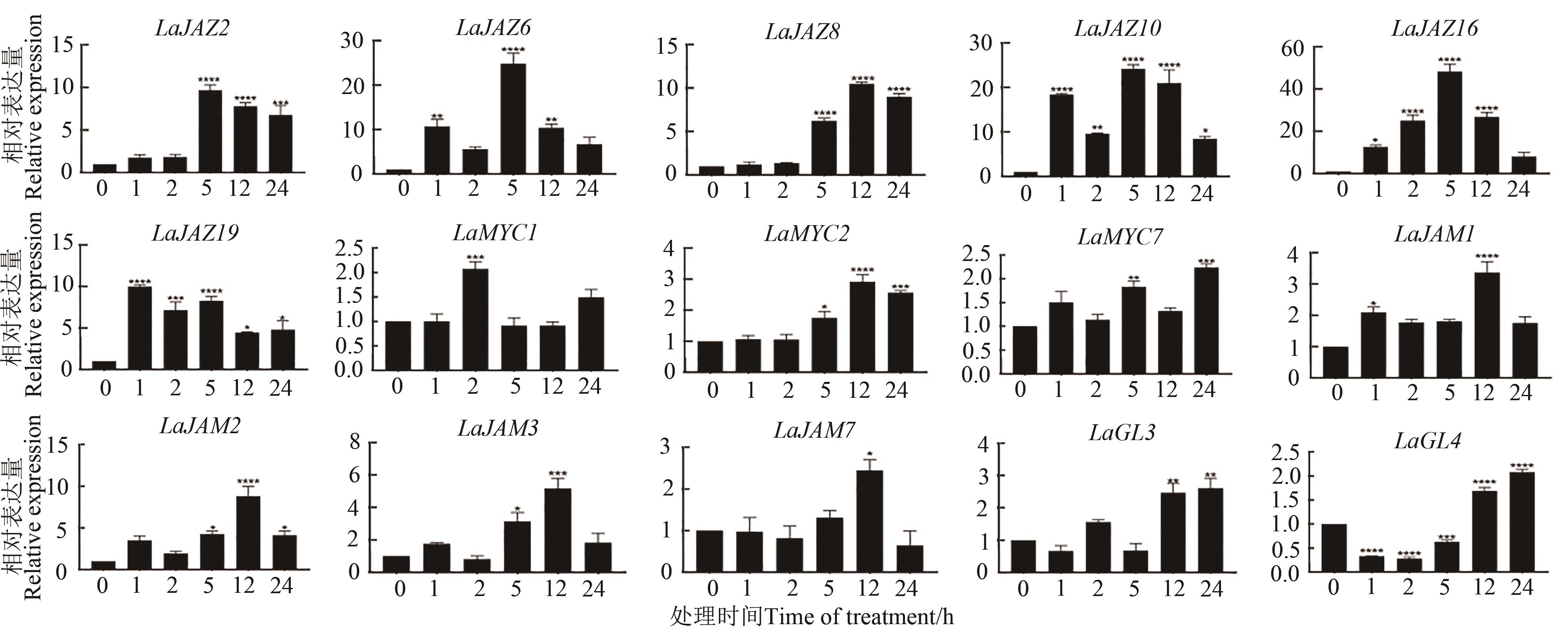
Fig. 7 Relative expression of LaJAZ and LaMYC2-like genes was observed after MeJA treatment of 2-month-old lavender leavesNote: *, **, *** and **** indicate significant differences at P<0.05, P<0.01, P<0.001 and P<0.000 1 levels, repectively.
| 1 | ZHANG W Y, LI J R, DONG Y M,et al..Genome-wide identification and expression of BAHD acyltransferase gene family shed novel insights into the regulation of linalyl acetate and lavandulyl acetate in lavender [J/OL]. J.Plant Physiol.,2024,292:154143 [2024-04-20]. . |
| 2 | SOHN S I, PANDIAN S, RAKKAMMAL K, et al.. Jasmonates in plant growth and development and elicitation of secondary metabolites:an updated overview [J/OL].Plant Sci., 2022,13:942789 [2024-04-20]. . |
| 3 | CHEN X Y, WANG D D, FANG X,et al..Plant specialized metabolism regulated by jasmonate signaling [J].Plant Cell Physiol., 2019,60(12):2638-2647. |
| 4 | CHINI A, FONSECA S, FERNÁNDEZ G, et al.. The JAZ family of repressors is the missing link in jasmonate signaling [J].Nature, 2007, 448:666-671. |
| 5 | KATSIR L, CHUNG H S, KOO A J, et al.. Jasmonate signaling:a conserved mechanism of hormone sensing [J].Curr. Opin. Plant Biol., 2008, 11(4):428-435. |
| 6 | BAI Y H, MENG Y J, HUANG D L,et al..Origin and evolutionary analysis of the plant-specific TIFY transcription factor family [J]. Genomics, 2011, 98(2):128-136. |
| 7 | YAN Y X, STOLZ S, CHÉTELAT A, et al.. A downstream mediator in the growth repression limb of the jasmonate pathway [J].Plant Cell, 2007, 19(8):2470-2483. |
| 8 | YE H Y, DU H, TANG N,et al..Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice [J]. Plant Mol. Biol., 2009, 71(3):291-305. |
| 9 | ZHANG Z B, LI X L, YU R,et al..Isolation,structural analysis,and expression characteristics of the maize TIFY gene family [J].Mol. Genet. Genomics,2015,290(5):1849-1858. |
| 10 | WANG Y, QIAO L, BAI J, et al.. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.) [J/OL].BMC Genomics,2017,18(1):152 [2024-04-20]. . |
| 11 | SASAKI-SEKIMOTO Y, JIKUMARU Y, OBAYASHI T,et al..Basic Helix-loop-Helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1),JAM2,and JAM3 are negative regulators of jasmonate responses in Arabidopsis [J].Plant Physiol., 2013, 163(1):291-304. |
| 12 | GARRIDO-BIGOTES A, FIGUEROA N E, FIGUEROA P M, et al..Jasmonate signalling pathway in strawberry:genome-wide identification,molecular characterization and expression of JAZs and MYCs during fruit development and ripening [J/OL].PLoS One,2018,13(5):e0197118 [2024-04-20].. |
| 13 | WANG S, WANG Y, YANG R, et al.. Genome-wide identification and analysis uncovers the potential role of JAZ and MYC families in potato under abiotic stress [J/OL]. Int. J. Mol. Sci., 2023,24(7):6706 [2024-04-20]. . |
| 14 | LI J R, WANG Y M, DONG Y M, et al.. The chromosome-based lavender genome provides new insights into Lamiaceae evolution and terpenoid biosynthesis [J/OL]. Hortic. Res., 2021, 8:53 [2024-04-20]. . |
| 15 | CHEN C J, CHEN H, ZHANG Y,et al..TBtools:an integrative toolkit developed for interactive analyses of big biological data [J].Mol. Plant, 2020, 13(8):1194-1202. |
| 16 | LESCOT M, DÉHAIS P, THIJS G, et al.. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002, 30(1):325-327. |
| 17 | DONG Y M, LI J R, ZHANG W Y, et al.. Exogenous application of methyl jasmonate affects the emissions of volatile compounds in lavender (Lavandula angustifolia) [J].Plant Physiol.Biochem.,2022,185:25-34. |
| 18 | HUANG G Q, GONG S Y, XU W L,et al..A fasciclin-like arabinogalactan protein,GhFLA1,is involved in fiber initiation and elongation of cotton [J]. Plant Physiol.,2013,161(3):1278-1290. |
| 19 | HONG G J, XUE X Y, MAO Y B,et al.. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression [J]. Plant Cell, 2012, 24(6):2635-2648. |
| 20 | RADHIKA V, KOST C, MITHÖFER A,et al..Regulation of extrafloral nectar secretion by jasmonates in Lima bean is light dependent [J]. Proc. Natl. Acad. Sci. USA, 2010, 107(40):17228-17233. |
| 21 | GANGAPPA S N, PRASAD V B R, CHATTOPADHYAY S.Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis [J].Plant Physiol., 2010, 154(3):1210-1219. |
| 22 | XU J S, VAN HERWIJNEN Z O, DRÄGER D B, et al.. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells [J].Plant Cell,2019,30(12):2988-3005. |
| 23 | SHEN Q, LU X, YAN T,et al..The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua [J]. New Phytol., 2016, 210(4):1269-1281. |
| 24 | DONG Y M, ZHANG W Y, LI J R,et al.. The transcription factor LaMYC4 from lavender regulates volatile terpenoid biosynthesis [J/OL].BMC Plant Biol., 2022, 22(1):289 [2024-04-20]. . |
| 25 | MA Y N, XU D B, LI L, et al.. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua [J/OL]. Sci. Adv., 2018, 4(11):eaas9357 [2024-04-20]. . |
| 26 | SHI M, ZHOU W, ZHANG J L,et al..Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins [J/OL]. Sci. Rep., 2016, 6:20919 [2024-04-20]. . |
| 27 | GUO D L, KANG K C, WANG P,et al..Transcriptome profiling of spike provides expression features of genes related to terpene biosynthesis in lavender [J/OL]. Sci.Rep., 2020, 10:6933 [2024-04-20]. . |
| 28 | KAZAN K, MANNERS J M.MYC2: the master in action [J]. Mol. Plant, 2013, 6(3):686-703. |
| [1] | Xiaorong GUO, Ying LIU, Jiazhen FAN, Tao HUANG, Rong ZHOU. Bioinformatics Analysis of Porcine CREBRF Gene and Its Expression Pattern [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 44-53. |
| [2] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [3] | Mingdi CHEN, Guihua HU, Haiwen ZHANG, Wangtian WANG. Bioinformatics and Expression Pattern Analysis of Rice RR Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 20-29. |
| [4] | Zhanqing WU, Wei CHEN, Zhan ZHAO, Hailiang XU, Haoyuan LI, Xingxing PENG, Dongxu CHEN, Mingyue ZHANG. Genome-wide Identification and Bioinformatics Analysis of GRAS Gene Family in Maize [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 15-25. |
| [5] | Bo LIU, Wangtian WANG, Li MA, Junyan WU, Yuanyuan PU, Lijun LIU, Yan FANG, Wancang SUN, Yan ZHANG, Ruimin LIU, Xiucun ZENG. Identification and Characterization of IPT Gene Family in Brassica rapa L. [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 56-66. |
| [6] | Yurong DENG, Lian HAN, Jinlong WANG, Xinghan WEI, Xudong WANG, Ying ZHAO, Xiaohong WEI, Chaozhou LI. Identification of SOD Family Genes in Chenopodium quinoa and Their Response to Mixed Saline-alkali Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 28-39. |
| [7] | Boqiong WU, Dongyao CUI, Renhe JIAO, Jian SONG, Yaoyao ZHAN, Yaqing CHANG. Cloning of Hexokinase Gene from Strongylocentrotus intermedius and Its Expression Response to High Temperature-acidification Stress [J]. Journal of Agricultural Science and Technology, 2022, 24(7): 205-217. |
| [8] | Qinqin WANG, Xiugui CHEN, Xuke LU, Shuai WANG, Yuexin ZHANG, Yapeng FAN, Quanjia CHEN, Wuwei YE. Bioinformatics Analysis and Functional Verification of GhPKE1 inUpland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(1): 38-45. |
| [9] | GUAN Sijing, WANG Nan, XU Rongrong, GE Tiantian, GAO Jing, YAN Yonggang, ZHANG Gang, CHEN Ying, ZHANG Mingying. [J]. Journal of Agricultural Science and Technology, 2021, 23(12): 66-75. |
| [10] | GE Chuan1, YANG Rong2, LI Liujun2, ZHANG Jiancheng2, ZHENG Xingwei2*. Genome-Wide Identification and Characterization of the YABBY Family Genes of Wheat (Trticum aestivum L.) [J]. Journal of Agricultural Science and Technology, 2019, 21(8): 11-18. |
| [11] | LI Xia1,2, FAN Xin2, LIANG Chengzhen2, WANG Yuan2, ZHANG Rui2, MENG Zhigang2*, LIU Xiaodong1*, SUN Guoqing2*. Functional Analysis of BS2 Gene in Tobacco [J]. Journal of Agricultural Science and Technology, 2019, 21(11): 43-50. |
| [12] | YANG Xiaomin, RUI Cun, ZHANG Yuexin, WANG Delong, WANG Junjuan, LU Xuke, CHEN Xiugui, GUO Lixue, WANG Shuai, CHEN Chao, YE Wuwei*. Cloning and Stress Resistance Analysis of Cotton DNA Methyltransferase GhDMT9 Gene [J]. Journal of Agricultural Science and Technology, 2019, 21(10): 12-19. |
| [13] | ZHAO Yan1, SHENG Yunlong1, SONG Yafei1, ZHANG Jiakuo1, WENG Qiaoyun1, YUAN Jincheng1, ZHAO Zhihai2*, LIU Yinghui1*. Genome-wide Identification and Bio-informatics Analysis of Superoxide Dismutase Gene Family in Setaria italica [J]. Journal of Agricultural Science and Technology, 2018, 20(8): 1-6. |
| [14] | MA Yanzhi. The Effects of MeJA and CS Mixture on Seed Germination of Bupleurum Chinense DC. [J]. Journal of Agricultural Science and Technology, 2017, 19(4): 38-44. |
| [15] | LIU Yun1,2, LU Hai-qiang2, SHI Peng-jun2, XIE Xiang-ming1*. Thermostability Improvement of the Mannanase Man5Xz8 from Achaetomium sp.Xz8 [J]. , 2015, 17(3): 49-55. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号