




















Journal of Agricultural Science and Technology ›› 2024, Vol. 26 ›› Issue (2): 46-55.DOI: 10.13304/j.nykjdb.2022.0818
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Lihua LI( ), Zhengwen SUN, Huifeng KE, Qishen GU, Liqiang WU, Yan ZHANG, Guiyin ZHANG, Xingfen WANG(
), Zhengwen SUN, Huifeng KE, Qishen GU, Liqiang WU, Yan ZHANG, Guiyin ZHANG, Xingfen WANG( )
)
Received:2022-09-25
Accepted:2022-10-18
Online:2024-02-15
Published:2024-02-04
Contact:
Xingfen WANG
李丽花( ), 孙正文, 柯会锋, 谷淇深, 吴立强, 张艳, 张桂寅, 王省芬(
), 孙正文, 柯会锋, 谷淇深, 吴立强, 张艳, 张桂寅, 王省芬( )
)
通讯作者:
王省芬
作者简介:李丽花 E-mail:13473229312@163.com;
基金资助:CLC Number:
Lihua LI, Zhengwen SUN, Huifeng KE, Qishen GU, Liqiang WU, Yan ZHANG, Guiyin ZHANG, Xingfen WANG. Development and Effect Evaluation of KASP Markers for Fiber Strength in Gossypium hirsutum L.[J]. Journal of Agricultural Science and Technology, 2024, 26(2): 46-55.
李丽花, 孙正文, 柯会锋, 谷淇深, 吴立强, 张艳, 张桂寅, 王省芬. 陆地棉纤维强度KASP-SNP标记的开发及效应评价[J]. 中国农业科技导报, 2024, 26(2): 46-55.
| 标记 Marker | 等位碱基 Allele | 引物名称 Primer name | 序列 Sequence (5’-3’) |
|---|---|---|---|
| FS-15 | G/T | FS-15-FAM | TTGTTCTTCCAGACACCTTTGAAC |
| FS-15-HEX | CTTTGTTCTTCCAGACACCTTTGAAA | ||
| FS-15-Reverse | AGCTAAAAAATTGGAGGCTGAGAGGAAT | ||
| FS-16 | A/T | FS-15-FAM | AAATTGAGCAAATTCAGGATGGAACCT |
| FS-16-HEX | AAATTGAGCAAATTCAGGATGGAACCA | ||
| FS-15-Reverse | ACATCAAAACAATCTGCTTGTACTGGCTT | ||
| FS-29 | A/G | FS-29-FAM | ACGGAGCTGAAGGATGCAAGTAA |
| FS-29-HEX | CGGAGCTGAAGGATGCAAGTAG | ||
| FS-29-Reverse | GCAAACTCTCGATTTCAAATCCAAAGGTT |
Table 1 Primer sequences used for KASP assays
| 标记 Marker | 等位碱基 Allele | 引物名称 Primer name | 序列 Sequence (5’-3’) |
|---|---|---|---|
| FS-15 | G/T | FS-15-FAM | TTGTTCTTCCAGACACCTTTGAAC |
| FS-15-HEX | CTTTGTTCTTCCAGACACCTTTGAAA | ||
| FS-15-Reverse | AGCTAAAAAATTGGAGGCTGAGAGGAAT | ||
| FS-16 | A/T | FS-15-FAM | AAATTGAGCAAATTCAGGATGGAACCT |
| FS-16-HEX | AAATTGAGCAAATTCAGGATGGAACCA | ||
| FS-15-Reverse | ACATCAAAACAATCTGCTTGTACTGGCTT | ||
| FS-29 | A/G | FS-29-FAM | ACGGAGCTGAAGGATGCAAGTAA |
| FS-29-HEX | CGGAGCTGAAGGATGCAAGTAG | ||
| FS-29-Reverse | GCAAACTCTCGATTTCAAATCCAAAGGTT |
| 年份Year | 环境 Environment | 最大值Max/(cN·tex-1) | 最小值Min/ (cN·tex-1) | 平均值Mean/ (cN·tex-1) | 标准差 SD | 偏度 Skew | 峰度 Kurt | 变异系数 CV/% |
|---|---|---|---|---|---|---|---|---|
| 2014 | E1:保定Baoding | 35.80 | 23.30 | 29.38 | 2.51 | 0.11 | -0.045 | 8.54 |
| E2:河间Hejian | 34.80 | 24.20 | 29.17 | 2.24 | 0.31 | -0.26 | 7.68 | |
| E3:辛集Xinji | 36.30 | 25.20 | 30.80 | 2.21 | 0.068 | -0.004 | 7.18 | |
| E4:青县Qing county | 36.80 | 24.80 | 30.08 | 2.16 | 0.16 | 0.066 | 7.18 | |
| E5:崖城Yacheng | 36.30 | 23.30 | 28.47 | 2.12 | 0.34 | 0.39 | 7.45 | |
| 2015 | E6:辛集Xinji | 37.47 | 23.23 | 31.01 | 2.01 | 0.059 | 0.89 | 6.48 |
| E7:青县Qing county | 35.51 | 19.91 | 28.39 | 2.34 | 0.044 | 0.29 | 8.24 | |
| E8:崖城Yacheng | 31.61 | 22.70 | 25.61 | 1.76 | 0.68 | 0.30 | 6.87 |
Table 2 Phenotypic variation for fiber strength of 376 accessions in 8 environments
| 年份Year | 环境 Environment | 最大值Max/(cN·tex-1) | 最小值Min/ (cN·tex-1) | 平均值Mean/ (cN·tex-1) | 标准差 SD | 偏度 Skew | 峰度 Kurt | 变异系数 CV/% |
|---|---|---|---|---|---|---|---|---|
| 2014 | E1:保定Baoding | 35.80 | 23.30 | 29.38 | 2.51 | 0.11 | -0.045 | 8.54 |
| E2:河间Hejian | 34.80 | 24.20 | 29.17 | 2.24 | 0.31 | -0.26 | 7.68 | |
| E3:辛集Xinji | 36.30 | 25.20 | 30.80 | 2.21 | 0.068 | -0.004 | 7.18 | |
| E4:青县Qing county | 36.80 | 24.80 | 30.08 | 2.16 | 0.16 | 0.066 | 7.18 | |
| E5:崖城Yacheng | 36.30 | 23.30 | 28.47 | 2.12 | 0.34 | 0.39 | 7.45 | |
| 2015 | E6:辛集Xinji | 37.47 | 23.23 | 31.01 | 2.01 | 0.059 | 0.89 | 6.48 |
| E7:青县Qing county | 35.51 | 19.91 | 28.39 | 2.34 | 0.044 | 0.29 | 8.24 | |
| E8:崖城Yacheng | 31.61 | 22.70 | 25.61 | 1.76 | 0.68 | 0.30 | 6.87 |
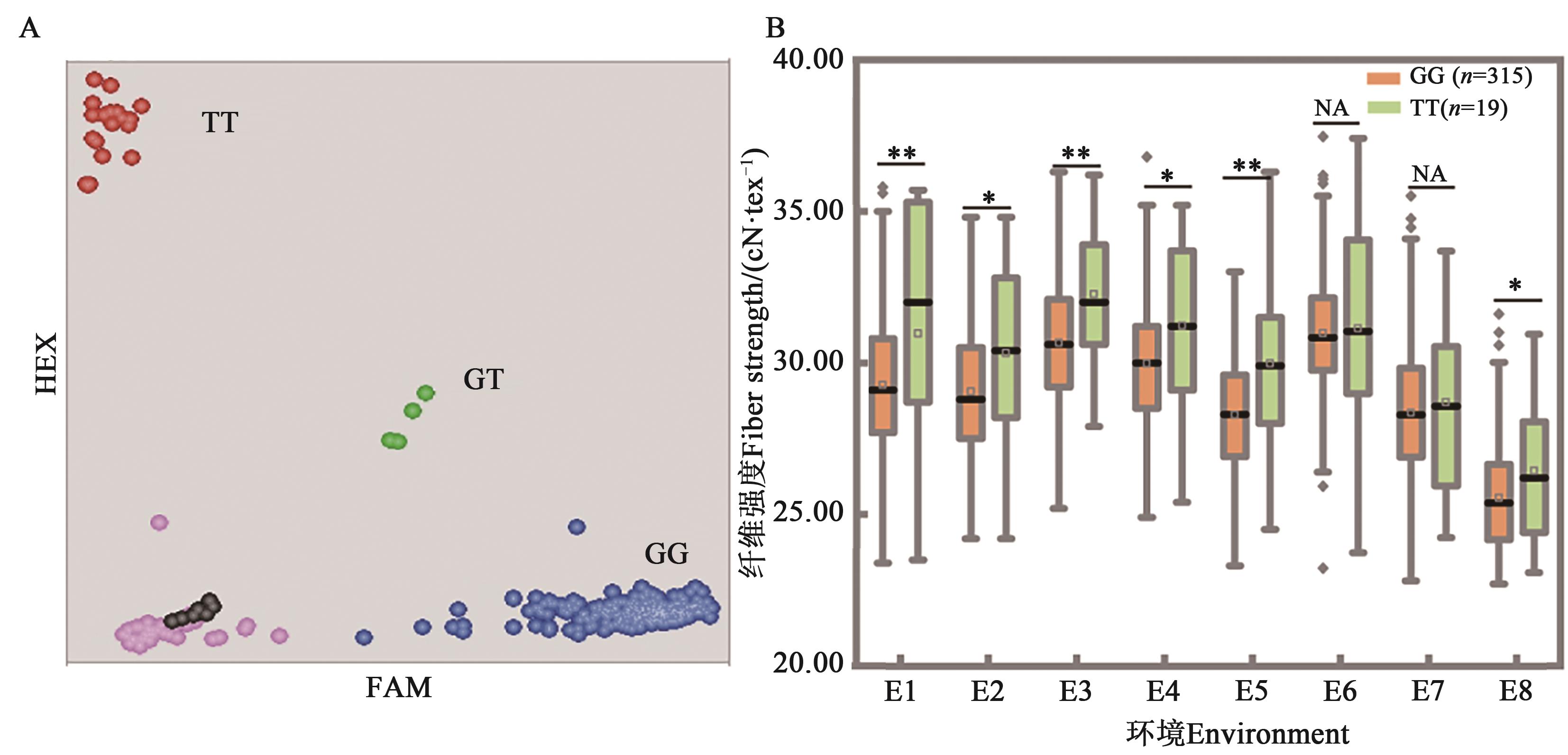
Fig. 2 Genotyping results of FS-15 in the population and allelic effects on fiber strengthA: Genotyping results of accessions, each dot represents 1 material,the red and blue represent homozygous, green represents heterozygous, pink represents failed data and black represents the non-template control, respectively; B: Allele effects, * and ** indicate significant difference at P<0.05 and P<0.01 levels, respectively, and NA indicates no significant difference; code of environment is same as Table 2
| 年份Year | 环境 Environment | FS-15/ (cN·tex-1) | 效应值 Allele effect G<T | 概率值 P-value | FS-16/ (cN·tex-1) | 效应值 Allele effect A<T | 概率值 P-value | FS-29/ (cN·tex-1) | 效应值 Allele effect G<A | 概率值 P-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | TT | AA | TT | AA | GG | ||||||||
| 2014 | E1:保定Baoding | 29.27 | 30.97 | 1.70 | 0.003 9 | 29.26 | 30.80 | 1.54 | 0.008 2 | 29.49 | 28.91 | 0.58 | 0.082 8 |
| E2:河间Hejian | 29.06 | 30.32 | 1.26 | 0.017 8 | 29.12 | 30.21 | 1.08 | 0.036 0 | 29.27 | 28.83 | 0.44 | 0.139 7 | |
| E3:辛集Xinji | 30.67 | 32.27 | 1.61 | 0.002 0 | 30.74 | 32.11 | 1.36 | 0.006 7 | 30.81 | 30.63 | 0.18 | 0.543 5 | |
| E4:青县Qing county | 29.99 | 31.23 | 1.24 | 0.014 4 | 30.00 | 31.15 | 1.15 | 0.021 0 | 30.24 | 29.37 | 0.87 | 0.002 1 | |
| E5:崖城Yacheng | 28.29 | 29.98 | 1.69 | 0.000 5 | 28.38 | 29.84 | 1.46 | 0.002 9 | 28.58 | 27.93 | 0.65 | 0.020 9 | |
| 2015 | E6:辛集Xinji | 30.98 | 31.14 | 0.15 | 0.753 9 | 30.97 | 31.09 | 0.12 | 0.798 9 | 31.03 | 30.92 | 0.11 | 0.693 7 |
| E7:青县Qing county | 28.36 | 28.70 | 0.35 | 0.538 9 | 28.34 | 28.68 | 0.34 | 0.527 4 | 28.58 | 27.83 | 0.75 | 0.015 8 | |
| E8:崖城Yacheng | 25.54 | 26.44 | 0.90 | 0.031 4 | 25.56 | 26.39 | 0.83 | 0.041 7 | 25.76 | 24.98 | 0.78 | 0.000 7 | |
Table 3 Allelic effects of KASP-SNP markers on fiber strength
| 年份Year | 环境 Environment | FS-15/ (cN·tex-1) | 效应值 Allele effect G<T | 概率值 P-value | FS-16/ (cN·tex-1) | 效应值 Allele effect A<T | 概率值 P-value | FS-29/ (cN·tex-1) | 效应值 Allele effect G<A | 概率值 P-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | TT | AA | TT | AA | GG | ||||||||
| 2014 | E1:保定Baoding | 29.27 | 30.97 | 1.70 | 0.003 9 | 29.26 | 30.80 | 1.54 | 0.008 2 | 29.49 | 28.91 | 0.58 | 0.082 8 |
| E2:河间Hejian | 29.06 | 30.32 | 1.26 | 0.017 8 | 29.12 | 30.21 | 1.08 | 0.036 0 | 29.27 | 28.83 | 0.44 | 0.139 7 | |
| E3:辛集Xinji | 30.67 | 32.27 | 1.61 | 0.002 0 | 30.74 | 32.11 | 1.36 | 0.006 7 | 30.81 | 30.63 | 0.18 | 0.543 5 | |
| E4:青县Qing county | 29.99 | 31.23 | 1.24 | 0.014 4 | 30.00 | 31.15 | 1.15 | 0.021 0 | 30.24 | 29.37 | 0.87 | 0.002 1 | |
| E5:崖城Yacheng | 28.29 | 29.98 | 1.69 | 0.000 5 | 28.38 | 29.84 | 1.46 | 0.002 9 | 28.58 | 27.93 | 0.65 | 0.020 9 | |
| 2015 | E6:辛集Xinji | 30.98 | 31.14 | 0.15 | 0.753 9 | 30.97 | 31.09 | 0.12 | 0.798 9 | 31.03 | 30.92 | 0.11 | 0.693 7 |
| E7:青县Qing county | 28.36 | 28.70 | 0.35 | 0.538 9 | 28.34 | 28.68 | 0.34 | 0.527 4 | 28.58 | 27.83 | 0.75 | 0.015 8 | |
| E8:崖城Yacheng | 25.54 | 26.44 | 0.90 | 0.031 4 | 25.56 | 26.39 | 0.83 | 0.041 7 | 25.76 | 24.98 | 0.78 | 0.000 7 | |

Fig. 3 Genotyping results of FS-16 in the population and allelic effects on fiber strengthA: Genotyping results of accessions, each dot represents 1 material,the red and blue represent homozygous, green represents heterozygous, pink represents failed data and black represents the non-template control, respectively; B: Allele effects, * and ** indicate significant difference at P<0.05 and P<0.01 levels, respectively, and NA indicates no significant difference; code of environment is same as Table 2
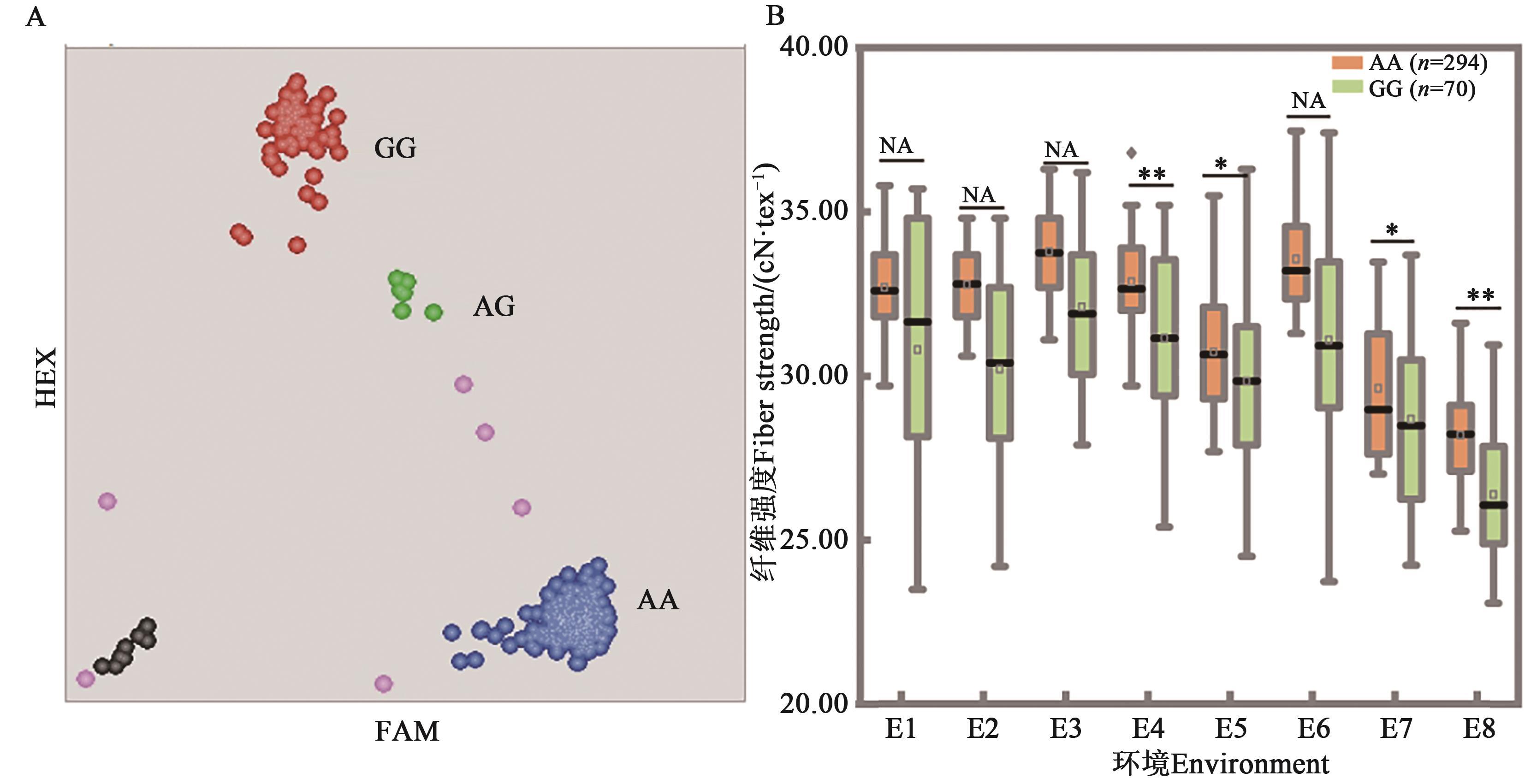
Fig. 4 Genotyping results of FS-29 in the population and allelic effects on fiber strengthA: Genotyping results of accessions, each dot represents 1 material,the red and blue represent homozygous, green represents heterozygous, pink represents failed data and black represents the non-template control, respectively; B: Allele effects, * and ** indicate significant difference at P<0.05 and P<0.01 levels, respectively, and NA indicates no significant difference; code of environment is same as Table 2
基因 Gene | 标记 Marker | 等位基因(样本数) Allele (number) | 平均值Average/ (cN·tex-1) | 等位效应 Allele effect | 选择率(样本数) Selection rate/%(number) | 概率值 P value |
|---|---|---|---|---|---|---|
| Gh_A07G1766(G/T) | FS-15 | G(315) T(19) | 29.12 30.21 | 1.09 | 50.5(159) 63.2(12) | 0.006 9 |
| Gh_A07G1765 (A/T) | FS-16 | A(336) T(20) | 29.15 30.10 | 0.95 | 51.8(174) 60.0(12) | 0.016 0 |
| Gh_D11G1931 (A/G) | FS-29 | A(294) G(70) | 29.35 28.77 | 0.57 | 56.1(165) 40.0(28) | 0.010 5 |
Table 4 Allelic effects of 3 markers in the population of 376 accessions
基因 Gene | 标记 Marker | 等位基因(样本数) Allele (number) | 平均值Average/ (cN·tex-1) | 等位效应 Allele effect | 选择率(样本数) Selection rate/%(number) | 概率值 P value |
|---|---|---|---|---|---|---|
| Gh_A07G1766(G/T) | FS-15 | G(315) T(19) | 29.12 30.21 | 1.09 | 50.5(159) 63.2(12) | 0.006 9 |
| Gh_A07G1765 (A/T) | FS-16 | A(336) T(20) | 29.15 30.10 | 0.95 | 51.8(174) 60.0(12) | 0.016 0 |
| Gh_D11G1931 (A/G) | FS-29 | A(294) G(70) | 29.35 28.77 | 0.57 | 56.1(165) 40.0(28) | 0.010 5 |
| 标记Marker | Hap1 | Hap2 | Hap3 | Hap4 |
|---|---|---|---|---|
| FS-15(G/T) | G | G | T | T |
| FS-16(A/T) | A | A | T | T |
| FS-29(A/G) | A | G | A | G |
Table 5 Haplotype combination
| 标记Marker | Hap1 | Hap2 | Hap3 | Hap4 |
|---|---|---|---|---|
| FS-15(G/T) | G | G | T | T |
| FS-16(A/T) | A | A | T | T |
| FS-29(A/G) | A | G | A | G |
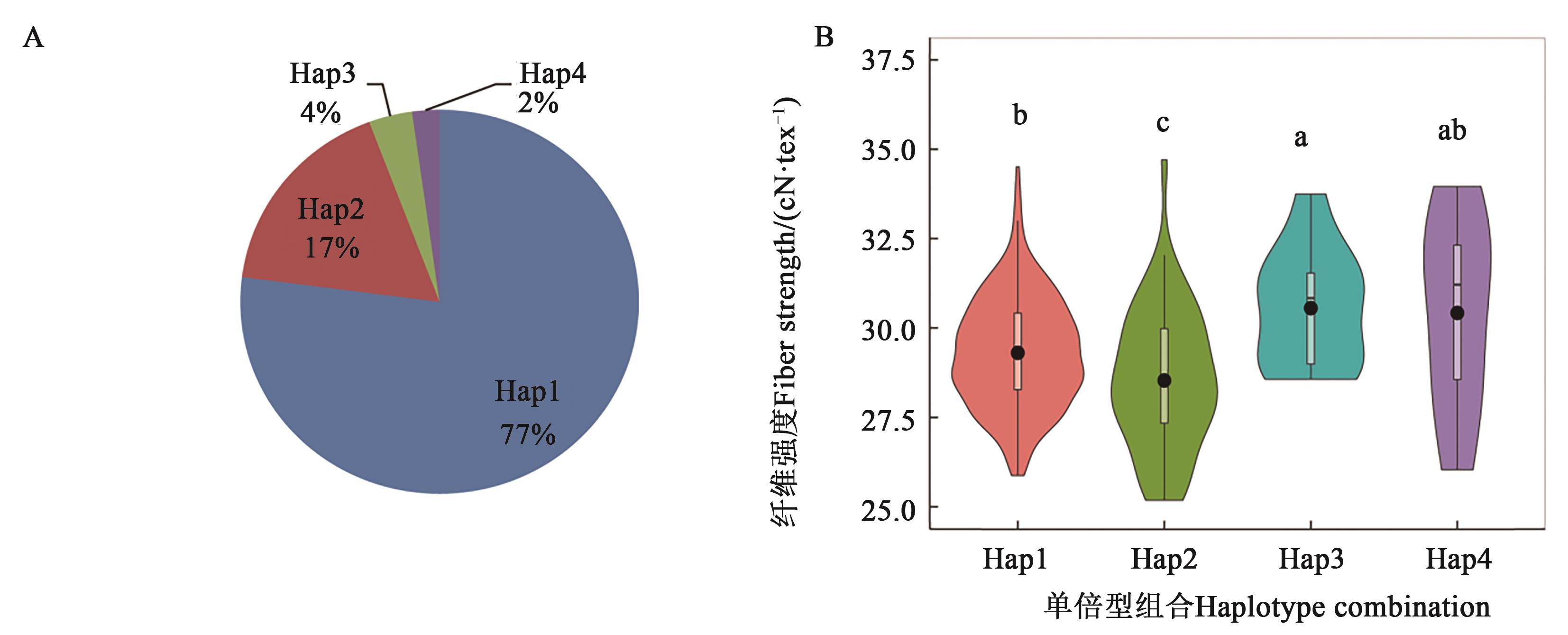
Fig. 5 Effects of different haplotype combination on fiber strength based on 3 KASP markersA: Frequency of four mainly haplotype combination in 376 accessions; B: Effect of different haplotype combianation on fiber strength, different lowercase letters represent Duncan’s multiple comparison at P<0.05 level
单倍型组合(样本数) Haplotype combination (number) | 纤维强度 Fiber strength/(cN·tex-1) | 选择率(样本数) Selection rate /% (number) | 概率值 P value |
|---|---|---|---|
| Hap1(242) | 29.30±1.54 b | 55.9(133) | 1.62×10-4 |
| Hap2(54) | 28.53±1.65 c | 38.9(21) | |
| Hap3(11) | 30.56±1.73 a | 72.7(8) | |
| Hap4(7) | 30.42±2.79 ab | 57.1(4) |
Table 6 Effect of different haplotypes combination on accessions with high fiber strength
单倍型组合(样本数) Haplotype combination (number) | 纤维强度 Fiber strength/(cN·tex-1) | 选择率(样本数) Selection rate /% (number) | 概率值 P value |
|---|---|---|---|
| Hap1(242) | 29.30±1.54 b | 55.9(133) | 1.62×10-4 |
| Hap2(54) | 28.53±1.65 c | 38.9(21) | |
| Hap3(11) | 30.56±1.73 a | 72.7(8) | |
| Hap4(7) | 30.42±2.79 ab | 57.1(4) |
单倍型组合(样本数) Haplotype combination (number) | 纤维长度 FL/mm | 概率值 P value | 铃重 BW/g | 概率值 P-value | 籽指 SI/g | 概率值 P-value | 衣指 LI/g | 概率值 P value |
|---|---|---|---|---|---|---|---|---|
| Hap1(242) | 29.18±0.99 b | 5.45×10-7 | 6.1±0.48 ab | 0.028 | 11.1±0.83 ab | 0.016 | 7.26±0.27 ab | 0.021 |
| Hap2(54) | 28.37±1.14 c | 5.93±0.43 b | 10.91±0.90 b | 7.08±0.67 b | ||||
| Hap3(11) | 29.71±0.90 a | 6.29±0.42 a | 11.59±1.04 a | 7.74±1.01 a | ||||
| Hap4(7) | 29.3±1.30 ab | 5.92±0.34 b | 11.63±0.92 a | 7.09±0.41 b |
Table 7 Effects ofdifferent haplotype combinationon other traits
单倍型组合(样本数) Haplotype combination (number) | 纤维长度 FL/mm | 概率值 P value | 铃重 BW/g | 概率值 P-value | 籽指 SI/g | 概率值 P-value | 衣指 LI/g | 概率值 P value |
|---|---|---|---|---|---|---|---|---|
| Hap1(242) | 29.18±0.99 b | 5.45×10-7 | 6.1±0.48 ab | 0.028 | 11.1±0.83 ab | 0.016 | 7.26±0.27 ab | 0.021 |
| Hap2(54) | 28.37±1.14 c | 5.93±0.43 b | 10.91±0.90 b | 7.08±0.67 b | ||||
| Hap3(11) | 29.71±0.90 a | 6.29±0.42 a | 11.59±1.04 a | 7.74±1.01 a | ||||
| Hap4(7) | 29.3±1.30 ab | 5.92±0.34 b | 11.63±0.92 a | 7.09±0.41 b |
| 1 | KUMAR V, SINGH B, SINGH S K, et al.. Role of GhHDA5 in H3K9 deacetylation and fiber initiation in Gossypium hirsutum [J]. Plant J., 2018, 95(6): 1069-1083. |
| 2 | SMITH W C. Cotton: Origin, History, Technology, and Production [M]. New York: Wiley, 1999: 269-288. |
| 3 | ZHANG T Z, YUAN Y L, YU J, et al.. Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection [J]. Theor. Appl. Genet., 2003, 106:262-268. |
| 4 | BASRA A S. Cotton fibers—developmental biology, quality improvement, and textile processing [M]. New York: Food Products Press, 1999: 183-229. |
| 5 | MEREDITH W R. Minimum number of genes controlling cotton fiber strength in a backcross population [J]. Crop Sci., 2005, 45:1114-1119. |
| 6 | PATERSON A H, SARANGA Y, MENZ M, et al.. QTL analysis of genotype × environment interactions affecting cotton fiber quality [J]. Theor. Appl. Genet., 2003, 106(3): 384-396. |
| 7 | WILKINS T A, ARPAT A B. The cotton fiber transcriptome [J]. Physiol. Plan., 2005, 124(3): 295-300. |
| 8 | PATERSON A H, Damon S, Hewitt J D, et al.. Mendelian factors underlying quantitative traits in tomato: comparison across species generations and environments [J]. Genetics, 1991,127: 181-197. |
| 9 | SHEN X, GUO W, LU Q, et al.. Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton [J]. BMC Genomics, 2007, 155(3): 371-380. |
| 10 | TAN Z Y, FANG X M, TANG S Y, et al.. Genetic map and QTL controlling fiber quality traits in upland cotton (Gossypium hirsutum L.) [J]. Euphytica, 2015, 203:615-628. |
| 11 | FANG X, LIU X, WANG X, et al.. Fine-mapping qFS07.1 controlling fiber strength in upland cotton (Gossypium hirsutum L.) [J]. Theor. Appl. Genet., 2017, 130(4): 795-806. |
| 12 | SU C F, WANG W, QIU X M, et al.. Fine-mapping a fiber strength QTL QFS-D11-1 on cotton chromosome 21 using introgressed lines [J]. Plant Breeding, 2013, 132:725-730. |
| 13 | ZHANG T, YUAN Y, YU J, et al.. Molecular tagging of a major QTL for fiber strength in upland cotton and its marker-assisted selection [J]. Theor. Appl. Genet., 2003, 106(2): 262-268. |
| 14 | 袁有禄,张天真,郭旺珍,等.棉花高品质纤维性状QTLs的分子标记筛选及其定位[J].遗传学报,2001, 28(12):1151-1161. |
| YUAN Y L, ZHANG T Z, GUO W Z, et al.. Screening and mapping of molecular markers for QTLs of high quality cotton fiber traits [J]. Acta Genetica Sinica, 2001, 28(12):1151-1161. | |
| 15 | ZHANG Z, SHANG H, SHI Y, et al.. Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to quantitative trait loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum) [J/OL]. BMC Plant Biol., 2016, 16:79 [2022-11-28]. . |
| 16 | GANAL M W, ALTMANN T, RÖDER M S. SNP identification in crop plants [J]. Curr. Opin. Plant Biol., 2009, 12(2): 211-217. |
| 17 | ZHANG Z, GE Q, LIU A, et al.. Construction of a high-density genetic map and its application to QTL identification for fiber strength in upland cotton [J]. Crop Sci., 2017, 57(2): 774-788. |
| 18 | MA Z, HE S, WANG X, et al.. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield [J]. Nat. Genet., 2018, 50(6): 803-813. |
| 19 | FANG L, WANG Q, HU Y, et al.. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits [J]. Nat. Genet., 2017, 49(7): 1089-1098. |
| 20 | LI C, FU Y, SUN R, et al.. Single-locus and multi-locus genome-wide association studies in the genetic dissection of fiber quality traits in upland cotton (Gossypium hirsutum L.) [J/OL]. Front. Plant Sci., 2018, 9:1083 [2022-10-05]. . |
| 21 | LI P, SU T, WANG H, et al.. Development of a core set of KASP markers for assaying genetic diversity in Brassica rapa subsp. chinensis Makino [J]. Plant Breeding, 2019, 138(3): 309-324. |
| 22 | YANG G, CHEN S, CHEN L, et al.. Development of a core SNP arrays based on the KASP method for molecular breeding of rice [J]. Rice, 2019, 12(1): 1-18. |
| 23 | CHANDRA S, SINGH D, PATHAK J, et al.. SNP discovery from next-generation transcriptome sequencing data and their validation using KASP assay in wheat (Triticum aestivum L.) [J]. Mol. Breeding, 2017, 37(7): 1-15. |
| 24 | OBENG-BIO E, BADU-APRAKU B, ELORHOR IFIE B, et al.. Phenotypic characterization and validation of provitamin A functional genes in early maturing provitamin a-quality protein maize (Zea mays) inbred lines [J]. Plant Breeding, 2020, 139(3): 575-588. |
| 25 | ZHANG J, STEWART J. Economical and rapid method for extracting cotton genomic DNA [J]. Cotton Sci., 2000, 4(3): 193-201. |
| 26 | ZHANG T, HU Y, JIANG W, et al.. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement [J]. Nat. Biotechnol., 2015, 33(5): 531-537. |
| 27 | ZILL P, BUTTNER A, EISENMENGER W, et al.. Single nucleotide polymorphism and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene in suicide victims [J]. Biol. Psychiat., 2004, 56(8):581-586. |
| 28 | KUMP K L, BRADBURY P J, WISSER R J, et al.. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population [J]. Nat. Genet., 2011, 43(2): 163-168. |
| 29 | WANG X, WANG H, LIU S, et al.. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings [J]. Nat. Genet., 2016, 48(10): 1233-1241. |
| 30 | HUANG X, SANG T, ZHAO Q, et al.. Genome-wide association studies of 14 agronomic traits in rice landraces [J]. Nat. Genet., 2010, 42(11): 961-967. |
| 31 | BAGGE M, XIA X, LÜBBERSTEDT T. Functional markers in wheat [J]. Curr. Opin. Plant Biol., 2007, 10(2): 211-216. |
| 32 | PERUMALSAMY S, BHARANI M, SUDHA M, et al.. Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa L.). [J]. Plant Breeding, 2010, 129(4): 400-406. |
| 33 | LIU Y, WANG F, ZHANG A, et al.. Development and validation of functional markers (tetra-primer ARMS and KASP) for the bacterial blight resistance gene xa5 in rice [J]. Plant Pathol., 2021, 50(3): 323-327. |
| 34 | FANG T, LEI L, LI G, et al.. Development and deployment of KASP markers for multiple alleles of Lr34 in wheat [J]. Theor. Appl. Genet., 2020, 133(7): 2183-2195. |
| 35 | RASHEED A, WEN W, GAO F, et al.. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat [J]. Theor. Appl. Genet., 2016, 129(10): 1843-1860. |
| 36 | AKEY J, JIN L, XIONG M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? [J]. Eu. J. Hum. Gene., 2001, 9(4): 291-300. |
| 37 | DALY M J, RIOUX J D, SCHAFFNER S F, et al.. High-resolution haplotype structure in the human genome [J]. Nat. Genet., 2001, 29(2): 229-232. |
| 38 | MILLER P A, WILLIAMS Jr J C, ROBINSON H F, et al.. Estimates of genotypic and environmental variances and covariances in upland cotton and their implications in selection [J]. Agron. J., 1958, 50(3): 126-131. |
| 39 | LI F, FAN G, LU C, et al.. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution [J]. Nat. Biotechnol., 2015, 33(5): 524-530. |
| 40 | SHEN X, GUO W, ZHU X, et al.. Molecular mapping of QTLs for fiber qualities in three diverse lines in upland cotton using SSR markers [J]. Mol. Breeding, 2005, 15(2): 169-181. |
| 41 | 潘家驹.棉花育种学[M]. 北京:中国农业出版社,1998:204. |
| PAN J. Cotton Breeding [M]. Beijing: China Agriculture Press, 1998:204. |
| [1] | Deyou ZHENG, Dongyun ZUO, Qiaolian WANG, Limin LYU, Hailiang CHENG, Aixing GU, Guoli SONG. Screening of Combination of Flumetralin and Fungicide to Control Cotton Fusarium wilt [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 119-124. |
| [2] | Shengmei LI, Bo PANG, Shiwei GENG, Wu SONG, Hongmei LI, Maosen MA, Ru ZHANG, Xinyan WANG, Wenwei GAO. Photosynthetic and Physiological Characteristics of Gossypium hirsutum L. × Gossypium barbadense L. Backross Populations in Full Boll Stage [J]. Journal of Agricultural Science and Technology, 2024, 26(1): 40-51. |
| [3] | Wei WANG, Qiang ZHAO, Abuduaini Munire·, Alimu·Amuli, Xinxin LI, Yangqing TIAN. Effects of Different Exogenous Substances on Chemical Capping and Yield and Quality of Cotton [J]. Journal of Agricultural Science and Technology, 2023, 25(9): 57-68. |
| [4] | Pengfei LIU, Xiaoshuang LU, Dilimurat Reheman, Tangnur Slay, Yanying QU, Quanjia CHEN, Xiaojuan DENG. Genetic Variation Analysis of Main Quality Traits and Agronomic Traits in Upland Cotton Seed [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 22-32. |
| [5] | Liting CHEN, Yuanyuan YAN. Investigation of Regulatory Mechanism of Floral Integrators in Upland Cotton [J]. Journal of Agricultural Science and Technology, 2023, 25(6): 11-21. |
| [6] | Zhengran SUN, Cuiping ZHANG, Jinli ZHANG, Hao WU, Xiuyan LIU, Zhenkai WANG, Yuzhen YANG, Daohua HE. Effects of Chemical Detopping on Cotton Plant Growth in Guanzhong Cotton Region [J]. Journal of Agricultural Science and Technology, 2023, 25(4): 167-177. |
| [7] | Wenjun YANG, Yuting ZHU, Jie ZHANG, Kaixiang XU, Congmin WEI, Quanjia CHEN. Meta-analysis of QTL for Salt Tolerance-related Traits at Seeding Stage in Cotton [J]. Journal of Agricultural Science and Technology, 2023, 25(12): 26-34. |
| [8] | Da CHEN, Jisheng JU, Qi MA, Shouzhen XU, Juanjuan LIU, Wenmin YUAN, Jilian LI, Caixiang WANG, Junji SU. Effects of FeNPs on Cotton Roots Growth and Its Response to Drought Stress at Seedling Stage [J]. Journal of Agricultural Science and Technology, 2023, 25(11): 49-57. |
| [9] | Man ZHANG, Jin ZHANG, Xinyu ZHANG, Guoning WANG, Xingfen WANG, Yan ZHANG. Cloning and Functional Analysis of GhNAC1 in Upland Cotton Involved in Verticillium Wilt Resistance [J]. Journal of Agricultural Science and Technology, 2023, 25(10): 35-44. |
| [10] | Guoqing LU, Caixia MA, Guoqing SUN, Huiming GUO, Hongmei CHENG. Molecular Characterization and Inheritance Stability Analysis of Herbicide-resistant Cotton GV-2 [J]. Journal of Agricultural Science and Technology, 2023, 25(1): 42-49. |
| [11] | Yan LIU, Hongshuai BAO, Hongyan SHANG, Guoning WANG, Yan ZHANG, Xingfen WANG, Zhiying MA, Jinhua WU. Selection of Cotton Fusarium Wilt and Culture Conditions [J]. Journal of Agricultural Science and Technology, 2022, 24(8): 124-132. |
| [12] | Ling LI, Helin DONG, Pengcheng LI, Liwen TIAN, Chunmei LI, Yunzhen MA, Na ZHANG, Fang WANG, Wenxiu XU. Effects of Machine Harvesting Planting Methods on Photosynthetic Characteristics and Dry Matter Accumulation of Different Plant Types of Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(8): 172-181. |
| [13] | Zhengwen SUN, Qishen GU, Yan ZHANG, Xingfen WANG, Zhiying MA. Research Progress on Cotton Gene Discovery and Molecular Breeding [J]. Journal of Agricultural Science and Technology, 2022, 24(7): 32-38. |
| [14] | Chengchuan YAN, Qingtao ZENG, Qin CHEN, Jincheng FU, Tingwei WANG, Quanjia CHEN, Yanying QU. Screening and Evaluation of Drought Resistance Indicators at Flowering and Boll Stage of Upland Cotton [J]. Journal of Agricultural Science and Technology, 2022, 24(7): 46-57. |
| [15] | Lijuan ZHANG, Yukun QIN, Huihuang CHENG, Yongqi LI, Haihua LUO. Research on Characteristics of Nitrogen and Phosphorus Loss from Surface Runoff of Cotton Field in Northern Jiangxi Province of Poyang Lake Region [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 166-175. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号