




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (7): 30-43.DOI: 10.13304/j.nykjdb.2024.0621
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Yi HU1( ), Jie GONG2(
), Jie GONG2( ), Wei ZHAO2, Rong CHENG2, Zhongyu LIU1(
), Wei ZHAO2, Rong CHENG2, Zhongyu LIU1( ), Shiqing GAO2(
), Shiqing GAO2( ), Yazhen YANG1(
), Yazhen YANG1( )
)
Received:2024-08-02
Accepted:2024-09-18
Online:2025-07-15
Published:2025-07-11
Contact:
Zhongyu LIU,Shiqing GAO,Yazhen YANG
胡懿1( ), 公杰2(
), 公杰2( ), 赵玮2, 程蓉2, 柳忠玉1(
), 赵玮2, 程蓉2, 柳忠玉1( ), 高世庆2(
), 高世庆2( ), 杨亚珍1(
), 杨亚珍1( )
)
通讯作者:
柳忠玉,高世庆,杨亚珍
作者简介:胡懿 E-mail:2813092866@qq.com基金资助:CLC Number:
Yi HU, Jie GONG, Wei ZHAO, Rong CHENG, Zhongyu LIU, Shiqing GAO, Yazhen YANG. Identification of PHY Gene Family in Wheat (Triticum aestivum L.)and Their Expression Analysis Under Heat Stress[J]. Journal of Agricultural Science and Technology, 2025, 27(7): 30-43.
胡懿, 公杰, 赵玮, 程蓉, 柳忠玉, 高世庆, 杨亚珍. 小麦PHY基因家族鉴定及热胁迫下表达分析[J]. 中国农业科技导报, 2025, 27(7): 30-43.
| 基因 Gene | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaPHYA1 | CTCTCCTGTTGGAAGTTCTGTC | CCCTGGTGCTTGATCCTAAG |
| TaPHYA2 | GACAAAGTATGGCGTCTGGG | GATCTTAGCCACTGCCATCC |
| TaPHYA3 | AGCTGCTATGCAATACTGTGG | CTTGCGCGGACATCACAAA |
| TaPHYB1 | TCTTCAGATTTGCCTGTCCTG | CTCCGCTCTGACTCTCTGAT |
| TaPHYB2 | TTCAGATTTGCCTGTCCTGG | CTGATGTACTGCACCTCACC |
| TaPHYB3 | GAAGGCATTGGACTAAGCGT | AGCTCCGTTCCCTACTTTCT |
| TaPHYC1 | CGAGCATGGTGAGGTCATTG | CACAGGACTTGCAGCACAAT |
| TaPHYC2 | ATGTGATATGCTCCTCCGGG | GGTAACACAATGCTGCACCA |
| TaPHYC3 | GGTAGATCTCGTGGAGGGTG | TGCCCTGTCAAATCTTGTGC |
| 18S rRNA | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
Table 1 qRT-PCR primers
| 基因 Gene | 正向引物 Forward primer (5’-3’) | 反向引物 Reverse primer (5’-3’) |
|---|---|---|
| TaPHYA1 | CTCTCCTGTTGGAAGTTCTGTC | CCCTGGTGCTTGATCCTAAG |
| TaPHYA2 | GACAAAGTATGGCGTCTGGG | GATCTTAGCCACTGCCATCC |
| TaPHYA3 | AGCTGCTATGCAATACTGTGG | CTTGCGCGGACATCACAAA |
| TaPHYB1 | TCTTCAGATTTGCCTGTCCTG | CTCCGCTCTGACTCTCTGAT |
| TaPHYB2 | TTCAGATTTGCCTGTCCTGG | CTGATGTACTGCACCTCACC |
| TaPHYB3 | GAAGGCATTGGACTAAGCGT | AGCTCCGTTCCCTACTTTCT |
| TaPHYC1 | CGAGCATGGTGAGGTCATTG | CACAGGACTTGCAGCACAAT |
| TaPHYC2 | ATGTGATATGCTCCTCCGGG | GGTAACACAATGCTGCACCA |
| TaPHYC3 | GGTAGATCTCGTGGAGGGTG | TGCCCTGTCAAATCTTGTGC |
| 18S rRNA | CGCGCGCTACGGCTTTGACCTA | CGGCAGATTCCCACGCGTTACG |
指标 Index | 基因名称 Gene name | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TaPHYA1 | TaPHYA2 | TaPHYA3 | TaPHYB1 | TaPHYB2 | TaPHYB3 | TaPHYC1 | TaPHYC2 | TaPHYC3 | |
基因ID号 Gene ID | TraesCS4A02G262900 | TraesCS4B02G052000 | TraesCS4D02G052200 | TraesCS4A02G122500 | TraesCS4B02G182400 | TraesCS4D02G183400 | TraesCS5A02G391300 | TraesCS5B02G396200 | TraesCS5D02G401000 |
| 染色体位置 Chromosome location | 4A:575 025 345~575 033 010(+) | 4B:40 780 124~40 786 295(-) | 4D:28 456 337~28 461 004(-) | 4A:151 945 624~151 953 947(+) | 4B:399 939 743~399 948 265(+) | 4D:320 592 499~320 600 764(-) | 5A:586 594 493~586 599 825(+) | 5B: 573 216 965~ 573 221 131(+) | 5D: 466 220 578~466 225 769(+) |
| 亚细胞定位 Subcellular localization | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus |
氨基酸数目 Number of amino acids/aa | 1 130 | 885 | 1 134 | 1 166 | 1 166 | 1 164 | 1 164 | 1 152 | 1 130 |
分子量 Molecular weight/Da | 124 881.26 | 97 690.63 | 125 617.12 | 127 679.74 | 127 695.74 | 127 593.73 | 128 636.61 | 126 939.97 | 126 015.48 |
| 理论等电点 Theoretical pI | 5.52 | 5.37 | 5.51 | 5.71 | 5.71 | 5.75 | 5.79 | 5.69 | 5.79 |
负电荷残基数 (Asp+Glu ) Number of negatively charged residues (Asp+Glu ) | 146 | 115 | 148 | 137 | 137 | 137 | 139 | 143 | 138 |
正电荷残基数 (Arg+Lys) Number of positively charged residues (Arg+Lys) | 113 | 86 | 114 | 113 | 113 | 114 | 114 | 115 | 113 |
总原子数 Total number of atoms | 17 552 | 13 707 | 17 633 | 17 898 | 17 899 | 17 894 | 18 095 | 17 802 | 17 710 |
分子式 Formula | |||||||||
不稳定指数 Instability index | 52.41 | 52.47 | 51.94 | 48.98 | 48.98 | 48.89 | 49.63 | 49.39 | 48.81 |
脂溶指数 Aliphatic index | 94.67 | 92.33 | 93.31 | 88.94 | 88.85 | 89.09 | 94.74 | 90.76 | 93.32 |
| 亲水性 GRAVY | -0.094 | -0.101 | -0.113 | -0.127 | -0.130 | -0.127 | -0.122 | -0.209 | -0.147 |
Table 2 Prediction of physicochemical properties and subcellular localization of wheat PHY gene family proteins
指标 Index | 基因名称 Gene name | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TaPHYA1 | TaPHYA2 | TaPHYA3 | TaPHYB1 | TaPHYB2 | TaPHYB3 | TaPHYC1 | TaPHYC2 | TaPHYC3 | |
基因ID号 Gene ID | TraesCS4A02G262900 | TraesCS4B02G052000 | TraesCS4D02G052200 | TraesCS4A02G122500 | TraesCS4B02G182400 | TraesCS4D02G183400 | TraesCS5A02G391300 | TraesCS5B02G396200 | TraesCS5D02G401000 |
| 染色体位置 Chromosome location | 4A:575 025 345~575 033 010(+) | 4B:40 780 124~40 786 295(-) | 4D:28 456 337~28 461 004(-) | 4A:151 945 624~151 953 947(+) | 4B:399 939 743~399 948 265(+) | 4D:320 592 499~320 600 764(-) | 5A:586 594 493~586 599 825(+) | 5B: 573 216 965~ 573 221 131(+) | 5D: 466 220 578~466 225 769(+) |
| 亚细胞定位 Subcellular localization | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus | 细胞核Nucleus |
氨基酸数目 Number of amino acids/aa | 1 130 | 885 | 1 134 | 1 166 | 1 166 | 1 164 | 1 164 | 1 152 | 1 130 |
分子量 Molecular weight/Da | 124 881.26 | 97 690.63 | 125 617.12 | 127 679.74 | 127 695.74 | 127 593.73 | 128 636.61 | 126 939.97 | 126 015.48 |
| 理论等电点 Theoretical pI | 5.52 | 5.37 | 5.51 | 5.71 | 5.71 | 5.75 | 5.79 | 5.69 | 5.79 |
负电荷残基数 (Asp+Glu ) Number of negatively charged residues (Asp+Glu ) | 146 | 115 | 148 | 137 | 137 | 137 | 139 | 143 | 138 |
正电荷残基数 (Arg+Lys) Number of positively charged residues (Arg+Lys) | 113 | 86 | 114 | 113 | 113 | 114 | 114 | 115 | 113 |
总原子数 Total number of atoms | 17 552 | 13 707 | 17 633 | 17 898 | 17 899 | 17 894 | 18 095 | 17 802 | 17 710 |
分子式 Formula | |||||||||
不稳定指数 Instability index | 52.41 | 52.47 | 51.94 | 48.98 | 48.98 | 48.89 | 49.63 | 49.39 | 48.81 |
脂溶指数 Aliphatic index | 94.67 | 92.33 | 93.31 | 88.94 | 88.85 | 89.09 | 94.74 | 90.76 | 93.32 |
| 亲水性 GRAVY | -0.094 | -0.101 | -0.113 | -0.127 | -0.130 | -0.127 | -0.122 | -0.209 | -0.147 |
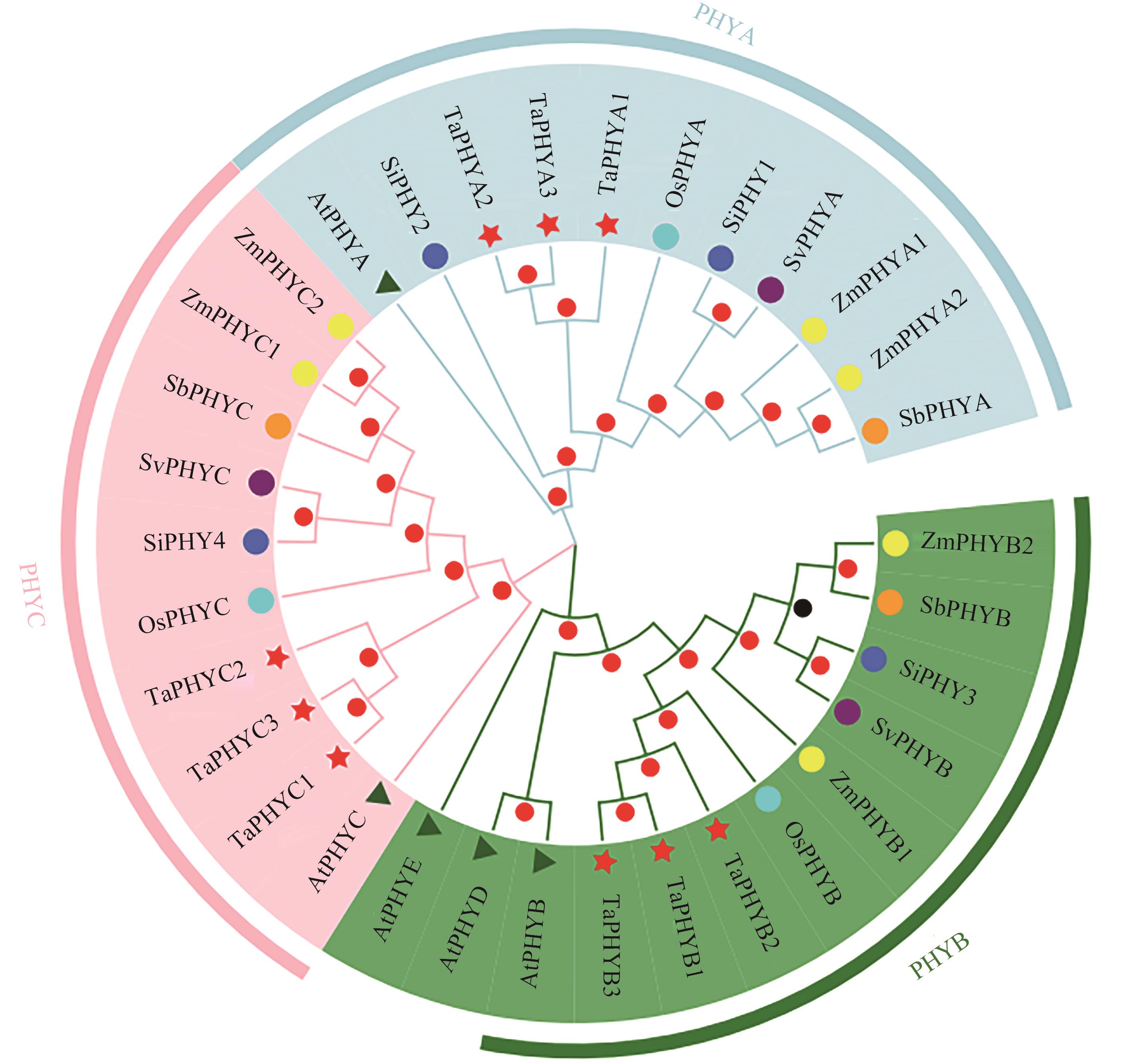
Fig. 2 Phylogeny analysis of TaPHY proteinsNote:At—Arabidopsis thaliana; Os—Oryza sativa L.; Ta—Triticum aestivum L.; Zm—Zea mays L.; Si —Setaria italica L.; Sb—Sorghum bicolor L.; Sv—Setaria viridis L.
蛋白名称 Protein name | α螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β转角 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| TaPHYA1 | 48.05 | 14.16 | 5.49 | 32.30 |
| TaPHYA2 | 45.54 | 14.24 | 5.65 | 34.58 |
| TaPHYA3 | 47.80 | 14.11 | 5.20 | 32.89 |
| TaPHYB1 | 46.91 | 13.89 | 5.40 | 33.79 |
| TaPHYB2 | 46.57 | 14.24 | 4.89 | 34.31 |
| TaPHYB3 | 47.16 | 14.60 | 5.07 | 33.16 |
| TaPHYC1 | 46.13 | 16.07 | 5.24 | 32.56 |
| TaPHYC2 | 45.49 | 13.54 | 5.56 | 35.42 |
| TaPHYC3 | 47.50 | 14.22 | 5.88 | 32.40 |
Table 3 Secondary structure of TaPHY proteins ant its percentage
蛋白名称 Protein name | α螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β转角 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|
| TaPHYA1 | 48.05 | 14.16 | 5.49 | 32.30 |
| TaPHYA2 | 45.54 | 14.24 | 5.65 | 34.58 |
| TaPHYA3 | 47.80 | 14.11 | 5.20 | 32.89 |
| TaPHYB1 | 46.91 | 13.89 | 5.40 | 33.79 |
| TaPHYB2 | 46.57 | 14.24 | 4.89 | 34.31 |
| TaPHYB3 | 47.16 | 14.60 | 5.07 | 33.16 |
| TaPHYC1 | 46.13 | 16.07 | 5.24 | 32.56 |
| TaPHYC2 | 45.49 | 13.54 | 5.56 | 35.42 |
| TaPHYC3 | 47.50 | 14.22 | 5.88 | 32.40 |

Fig. 6 TaPHY expression database heat map analysisA~C: Based on transcriptome data of WheatOmics 1.0, Z10—1 leaf stage, Z13—3 leaves stage, Z23—Early tillering stage, Z30—Standing stage, Z32—Early jointing stage, Z39—Late jointing stage, Z65—Middle flowering stage, Z71—2 d after flowering, Z75—10 d after flowering, Z85—30 d after flowering; D~F: Based on wheat expression browser database, S1—Roots in seedling stage, S2—Stems and leaves in seedling stage, V1—Stems and leaves in vegetative stage, V2—Roots in vegetative stage, V3—Panicles in vegetative stage, R1—Roots and leaves in reproductive growth stage, R2—Roots in reproductive growth stage, R3—Panicles in reproductive growth stage, R4—Grain in reproductive growth stage
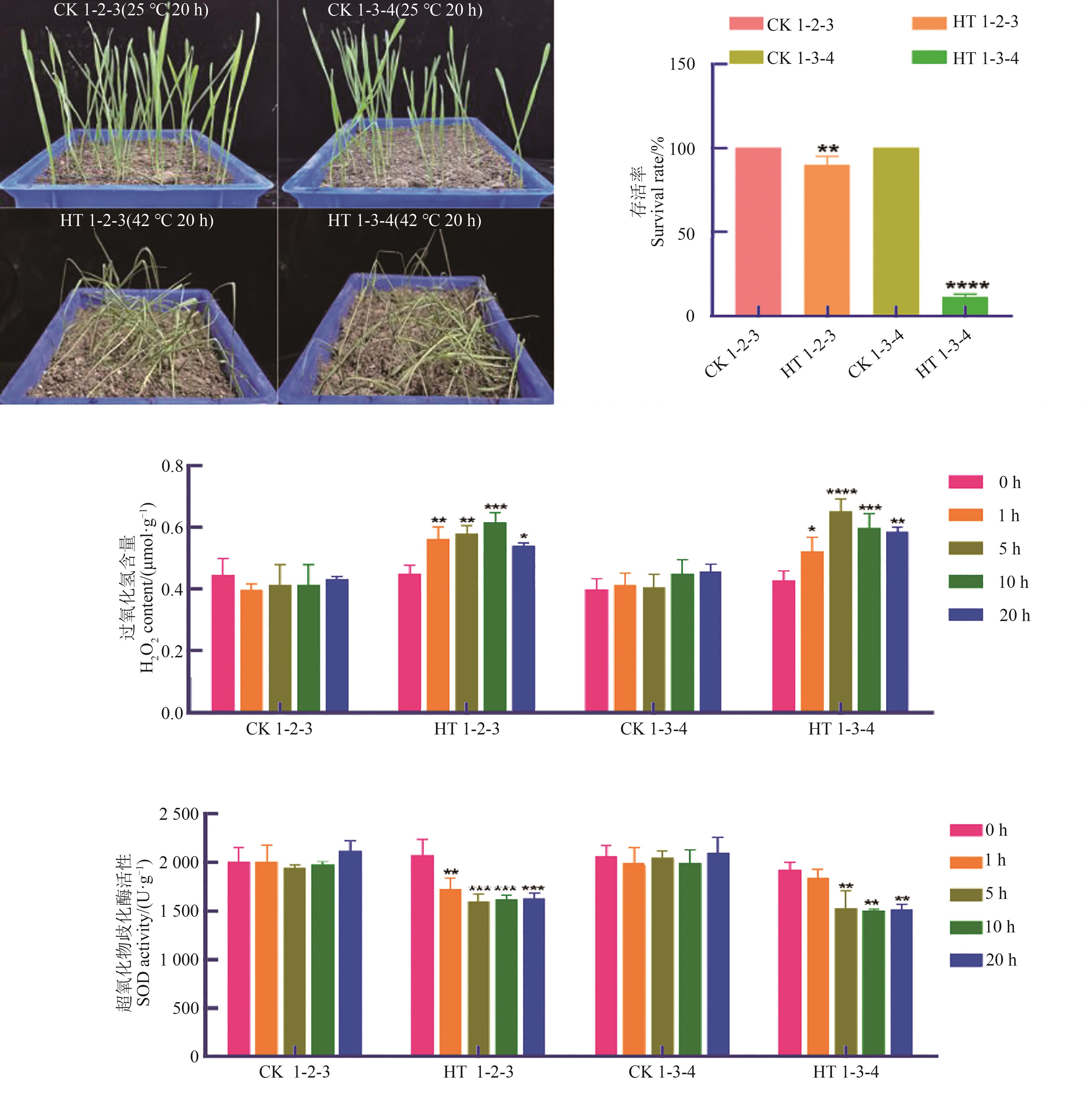
Fig. 7 Survival rate and physiological indexes of 1-2-3 and 1-3-4 under heat stressNote:*,**, *** and **** indicate significant at P<0.05,P<0.01, P<0.001 and P<0.000 1 levels, respectively.

Fig. 8 Relative expression of TaPHY genes under the heat stress conditionNote:Different lowercase letters indicate significant differences between different times at P<0.05 level.
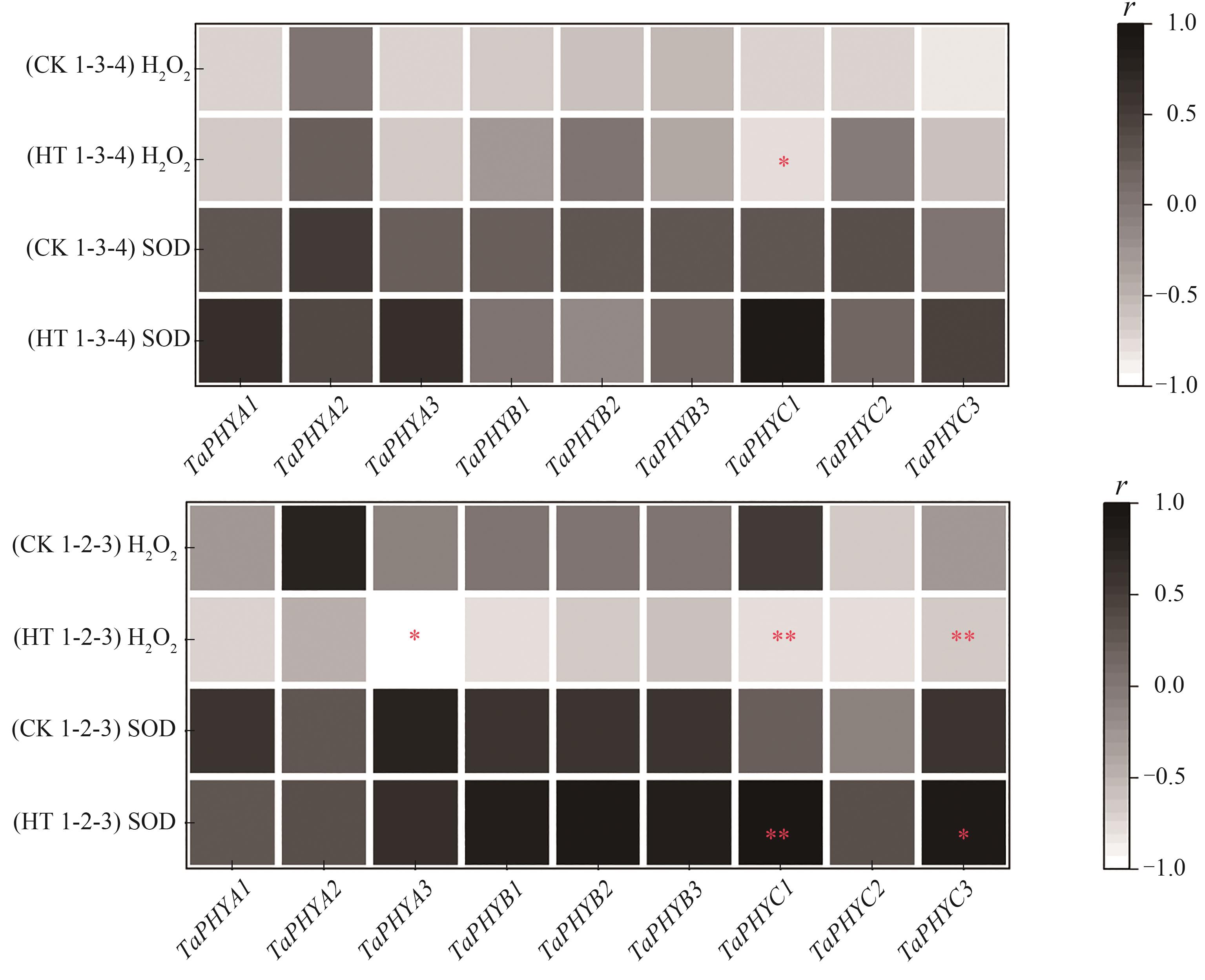
Fig. 9 Correlation analysis of H2O2 content, SOD activity and relative expression of TaPHYsNote:* and ** indicate significant correlations at P<0.05 and P<0.01 levels, respectively.
| [1] | 王富刚, 张静, 张雄, 等. 光敏色素与植物的光形态建成 [J]. 基因组学与应用生物学, 2017, 36(8):3167-3171. |
| WANG F G, ZHANG J, ZHANG X, et al.. Phytochromes and plant photomorphogenesis [J]. Genomics Appl. Biol., 2017, 36(8):3167-3171. | |
| [2] | 王静,王艇.高等植物光敏色素的分子结构、生理功能和进化特征[J].植物学通报,2007,24(5):649-658. |
| WANG J, WANG T. Molecular structure,physiological function and evolution of phytochrome in higher plants [J]. Chin. Bull.Bot., 2007,24(5):649-658. | |
| [3] | SONG B, ZHAO H, DONG K,et al..Phytochrome A inhibits shade avoidance responses under strong shade through repressing the brassinosteroid pathway in Arabidopsis [J]. Plant J., 2020,104(6):1520-1534. |
| [4] | KIPPES N, VANGESSEL C, HAMILTON J, et al.. Effect of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods [J/OL]. BMC Plant Biol., 2020,20(1):197 [2024-07-10].. |
| [5] | XU P, LIAN H, XU F,et al..Phytochrome B and AGB1 coordinately regulate photomorphogenesis by antagonistically modulating PIF3 stability in Arabidopsis [J]. Mol. Plant, 2019, 12(2):229-247. |
| [6] | YANG L, JIANG Z, JING Y,et al..PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis [J].J.Integr.Plant Biol.,2020,62(9):1372-1384. |
| [7] | LIU S, YANG L, LI J, et al.. FHY3 interacts with phytochrome B and regulates seed dormancy and germination [J]. Plant Physiol., 2021,187(1):289-302. |
| [8] | 张芳,张晓枫,王进征,等.植物光敏色素PHYA、PHYB研究进展[J].生物学通报,2011,46(1):11-14. |
| ZHANG F, ZHANG X F, WANG J Z, et al.. Research advances on the phytochromes A and B [J]. Bull. Biol., 2011,46(1):11-14. | |
| [9] | SONG J, LIU Q, HU B, et al.. Photoreceptor PhyB involved in Arabidopsis temperature perception and heat-tolerance formation [J/OL]. Int. J. Mol. Sci., 2017,18(6):E1194 [2024-07-10]. . |
| [10] | JIANG B C, SHI Y T, PENG Y, et al.. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by sstabilizing the phyB thermosensor in Arabidopsis [J]. Mol. Plant, 2020, 13(6):894-906. |
| [11] | LEGRIS M, KLOSE C, BURGIE E S, et al.. Phytochrome B integrates light and temperature signals in Arabidopsis [J].Science, 2016,354(6314):897-900. |
| [12] | JUNG J H, DOMIJAN M, KLOSE C, et al.. Phytochromes function as thermosensors in Arabidopsis [J]. Science, 2016,354(6314):886-889. |
| [13] | HE Y, LI Y, CUI L, et al.. Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in rice [J/OL].Front. Plant Sci., 2016,7:1963 [2024-07-10]. . |
| [14] | GAO Y, JIANG W, DAI Y,et al..A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice [J]. Plant Mol. Biol., 2015,87(4/5):413-428. |
| [15] | 李敏, 苏慧, 李阳阳, 等. 黄淮海麦区小麦耐热性分析及其鉴定指标的筛选 [J]. 中国农业科学, 2021, 54 (16): 3381-3393. |
| LI M, SU H, LI Y Y, et al.. Analysis of heat tolerance of wheat with different genotypes and screening of identification indexes in Huang-Huai-Hai region [J]. Sci. Agric. Sin., 2021, 54 (16): 3381-3393. | |
| [16] | REI S, MICAEL P, VIEIRA C, et al..Fold change in regucalcin expression after chill-coma recovery (ChCR) obtained by qRT-PCR using the 2-ΔΔCT method [J]. Methods, 2001, 25 (4):402-408. |
| [17] | 王璐璐. 玉米光敏色素基因的克隆与苗期光暗条件下表达分析 [D]. 沈阳:沈阳农业大学, 2017. |
| WANG L L. The cloning of phytochrome gene in maize and the expression analysis in the light and dark conditionsat the seeding stage [D]. Shenyang: Shenyang Agricultural University, 2017. | |
| [18] | AUKERMAN M J, HIRSCHFELD M, WESTER L,et al..A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing [J]. Plant Cell,1997,9(8):1317-1326. |
| [19] | 李建平,杨建平,宋梅芳,等.拟南芥光敏色素D(AtphyD)在不同光质下的表达特性分析[J].新疆农业科学,2014,51(2):305-310. |
| LI J P, YANG J P, SONG M F,et al..Differential patterns of expression of the Arabidopsis phytochrome D under different light conditions [J]. Xinjiang Agric. Sci., 2014,51(2):305-310. | |
| [20] | 陈由, 王亚琴. 水稻光敏色素 A基因克隆及其在光形态建成中的作用 [J]. 华南师范大学学报(自然科学版), 2014, 46(1):71-76. |
| CHEN Y, WANG Y Q. Cloning and Function Analysis of rice Phytochrome A gene in photomorphogenesis [J]. J. South China Norm. Univ. (Nat. Sci.), 2014, 46(1):71-76. | |
| [21] | 丁武思,陈士瞻,刘磊,等.2个玉米光敏色素C基因的克隆及功能验证[J].河南农业科学,2021,50(1):16-26. |
| DING W S, CHEN S Z, LIU L,et al..Cloning and function verification of two phytochrome C genes in Zea mays [J]. J.Henan Agric. Sci., 2021,50(1):16-26. | |
| [22] | SÁNCHEZ-LAMAS M, LORENZO C D, CERDÁN P D.Bottom-up assembly of the phytochrome network [J/OL]. PLoS Genet., 2016,12(11):e1006413 [2024-07-10]. . |
| [23] | CHEN A, LI C X, HU W, et al.. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod [J]. Proc. Natl. Acad. Sci. USA, 2014,111(28):10037-10044. |
| [24] | PEARCE S, KIPPES N, CHEN A, et al.. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways [J/OL]. BMC Plant Biol., 2016,16(1):141 [2024-07-10]. . |
| [25] | 李丹阳, 张晓花, 刘灵芝, 等. 光敏色素互作因子OsPIL15基因对水稻磷、氮吸收利用的影响[J].河南农业科学,2021,50(5):24-31. |
| LI D Y, ZHANG X H, LIU L Z, et al.. Effects of phytochrome interacting factor OsPIL15 gene on absorption and utilization of phosphorus and nitrogen in rice [J]. J. Henan Agric. Sci., 2021, 50(5):24-31. | |
| [26] | SUN J J, LU J, BAI M J, et al.. Phytochrome-interacting factors interact with transcription factor CONSTANS to suppress flowering in rose [J].Plant Physiol.,2021,186(2):1186-1201. |
| [27] | QIU Y J, LI M N, KIM R J A,et al..Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA [J/OL].Nat.Commun., 2019,10(1):140 [2024-07-10]. . |
| [28] | BURKO Y, WILLIGE B C, SELUZICKI A, et al.. PIF7 is a master regulator of thermomorphogenesis in shade [J/OL].Nat. Commun.,2022,13(1):4942 [2024-07-10]. . |
| [1] | Qiang ZHU, Zongxian CHE, Heng CUI, Jiudong ZHANG, Xingguo BAO. Effect of Green Manure Replacing Nitrogen Fertilizer on Greenhouse Gases in Wheat Fields [J]. Journal of Agricultural Science and Technology, 2025, 27(7): 182-189. |
| [2] | Sile HU, Yulong BAO, Tubuxinbayaer, Jifeng TAO, Enliang GUO. Chlorophyll Content Inversion of Spring Wheat Based on Unmanned Aerial Vehicle Hyperspectral and Integrated Learning [J]. Journal of Agricultural Science and Technology, 2025, 27(6): 93-103. |
| [3] | Shuo SHI, Yu FENG, Liang LI, Rui MENG, Yanze ZHANG, Xiurong YANG. Transcriptome Analysis of Resistance to Sharp Eyespot of Wheat Mediated by Piriformospora indica and Key Genes Screening [J]. Journal of Agricultural Science and Technology, 2025, 27(5): 133-145. |
| [4] | Bei MA, Jie GONG, Yinke DU, Yuwei GAN, Rong CHENG, Bo ZHU, Lixia YI, Jinxiu MA, Shiqing GAO. Identification and Expression Analysis of TaINP1 Gene Related to Pollen Pore Development in Wheat [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 22-35. |
| [5] | Pengyang SHAO, Yuzhu SHA, Xiu LIU, Guoshun CHEN, Caiye ZHU, Jiqing WANG, Fanxiong WANG, Xiaowei CHEN, Wenxin YANG. Effects of Astragalus Feed Additive on Growth Performance, Serum Ig, Rumen Fermentation and Microbiota of Lambs [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 83-94. |
| [6] | Zhenyu XUE, Kangkang ZHANG, Yuanyuan ZHANG, Qiangqiang YAN, Lirong YAO, Hong ZHANG, Yaxiong MENG, Erjing SI, Baochun LI, Xiaole MA, Huajun WANG, Juncheng WANG. Screening and Functional Gene Detection of High-quality and Drought-resistant Wheat Germplasms [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 35-49. |
| [7] | Xianyin SUN, Qiuhuan MU, Yong MI, Guangde LYU, Xiaolei QI, Yingying SUN, Xundong YIN, Ruixia WANG, Ke WU, Zhaoguo QIAN, Yan ZHAO, Minggang GAO. Classification and Evaluation of New Wheat Lines Based on GT Biplot [J]. Journal of Agricultural Science and Technology, 2024, 26(7): 14-24. |
| [8] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [9] | Yangyang DU, Yuanyuan BAO, Xiangyu LIU, Xinyong ZHANG. Effects of Tartary Buckwheat Rotation on Enzyme Activities and Microorganisms in Rhizosphere Soil of Cultivated Potato in Yunnan Province [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 192-200. |
| [10] | Meng LIU, Chenyi LIN, Rui WU, Shuang CAO, Zhihao LIANG, Ruonan ZHANG. Heat Stress Response and Comprehensive Evaluation of Heat Resistance of Lentinula edodes Mycelia [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 90-100. |
| [11] | Gang ZHAO, Shuying WANG, Shangzhong LI, Jianjun ZHANG, Yi DANG, Lei WANG, Xingmao LI, Wanli CHENG, Gang ZHOU, Shengli NI, Tinglu FAN. Effects of Precipitation on Yield and Water Consumption of Winter Wheat in Loess Plateau in Recent 40 Years [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 164-173. |
| [12] | Erhao ZHANG, Panpan LIU, Ping HE, Yue JIAN, Yuting XU, Chengxin CHEN, Yazhou LU, Xiaozhong LAN, Sangmu SUOLANG. Physiochemical Properties and Microbial Community Structure in Rhizosphere Soil of Dracocephalum tanguticum [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 201-213. |
| [13] | Hong ZHANG, Weiguo LI, Xiaodong ZHANG, Bihui LU, Chengcheng ZHANG, Wei LI, Tinghuai MA. Extraction of Winter Wheat Planting Area Based on Fusion Features of HJ-1 and GF-1 Image [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 109-119. |
| [14] | Jingyun ZHANG, Feng GUAN, Bo SHI, Xinjian WAN. Effects of Wheat Root Exudates on Bitter Gourd Seeding Growth and Soil Environment [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 181-190. |
| [15] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号