




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (6): 28-38.DOI: 10.13304/j.nykjdb.2024.0409
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Jilin SUN( ), Jiaqi ZHANG, Fansen MENG, Silong SUN(
), Jiaqi ZHANG, Fansen MENG, Silong SUN( )
)
Received:2024-05-22
Accepted:2024-07-13
Online:2025-06-15
Published:2025-06-23
Contact:
Silong SUN
通讯作者:
孙思龙
作者简介:孙吉林 E-mail:3236291157@qq.com;
基金资助:CLC Number:
Jilin SUN, Jiaqi ZHANG, Fansen MENG, Silong SUN. Genome-wide Identification and Tissue Expression Pattern Analysis of HSP70 Gene Family in Thinopyrum elongatum[J]. Journal of Agricultural Science and Technology, 2025, 27(6): 28-38.
孙吉林, 张家琦, 孟凡森, 孙思龙. 二倍体长穗偃麦草HSP70基因家族鉴定及表达模式分析[J]. 中国农业科技导报, 2025, 27(6): 28-38.
基因名称 Gene name | 基因ID Gene ID | 蛋白质长度Protein length/aa | 分子质量Molecular weight/kD | 等电点pI | 不稳定系数 Instability index | 脂肪系数Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| TeHSP70-01 | GWHGABKY001532 | 887 | 97.70 | 5.45 | 44.50 | 82.33 | -0.458 |
| TeHSP70-02 | GWHGABKY001533 | 835 | 92.56 | 5.07 | 45.59 | 75.13 | -0.513 |
| TeHSP70-03 | GWHGABKY001646 | 740 | 82.25 | 5.28 | 42.34 | 79.97 | -0.380 |
| TeHSP70-04 | GWHGABKY001898 | 734 | 81.38 | 5.43 | 50.39 | 88.26 | -0.333 |
| TeHSP70-05 | GWHGABKY003413 | 686 | 75.38 | 5.23 | 38.34 | 77.07 | -0.503 |
| TeHSP70-06 | GWHGABKY011778 | 683 | 74.88 | 5.31 | 36.86 | 80.85 | -0.427 |
| TeHSP70-07 | GWHGABKY012581 | 680 | 74.83 | 5.18 | 33.92 | 85.50 | -0.381 |
| TeHSP70-08 | GWHGABKY012586 | 690 | 73.56 | 5.01 | 29.29 | 86.19 | -0.270 |
| TeHSP70-09 | GWHGABKY016707 | 665 | 73.27 | 5.06 | 25.54 | 86.50 | -0.479 |
| TeHSP70-10 | GWHGABKY017407 | 683 | 73.26 | 5.60 | 41.92 | 84.17 | -0.325 |
| TeHSP70-11 | GWHGABKY022426 | 684 | 73.14 | 5.04 | 30.07 | 87.09 | -0.310 |
| TeHSP70-12 | GWHGABKY022433 | 665 | 72.93 | 5.04 | 23.51 | 86.78 | -0.371 |
| TeHSP70-13 | GWHGABKY022437 | 678 | 72.86 | 5.29 | 36.04 | 84.04 | -0.310 |
| TeHSP70-14 | GWHGABKY022852 | 678 | 72.84 | 5.23 | 35.63 | 84.04 | -0.303 |
| TeHSP70-15 | GWHGABKY024486 | 684 | 72.77 | 5.78 | 35.28 | 87.49 | -0.268 |
| TeHSP70-16 | GWHGABKY024546 | 677 | 72.55 | 5.36 | 38.85 | 85.48 | -0.292 |
| TeHSP70-17 | GWHGABKY024596 | 657 | 71.80 | 5.17 | 36.09 | 79.15 | -0.446 |
| TeHSP70-18 | GWHGABKY025756 | 653 | 71.65 | 5.21 | 34.54 | 79.62 | -0.458 |
| TeHSP70-19 | GWHGABKY026068 | 667 | 71.63 | 5.06 | 27.89 | 88.22 | -0.296 |
| TeHSP70-20 | GWHGABKY027854 | 640 | 71.35 | 5.44 | 31.47 | 99.83 | -0.088 |
| TeHSP70-21 | GWHGABKY030282 | 660 | 71.31 | 5.07 | 27.51 | 88.53 | -0.319 |
| TeHSP70-22 | GWHGABKY032027 | 648 | 71.19 | 5.08 | 34.79 | 83.4 | -0.394 |
| TeHSP70-23 | GWHGABKY034860 | 648 | 71.02 | 5.13 | 33.73 | 81.16 | -0.415 |
| TeHSP70-24 | GWHGABKY035578 | 651 | 70.85 | 7.82 | 47.67 | 92.78 | -0.118 |
| TeHSP70-25 | GWHGABKY035973 | 620 | 69.08 | 6.25 | 41.66 | 81.1 | -0.402 |
| TeHSP70-26 | GWHGABKY042123 | 617 | 67.68 | 8.84 | 33.40 | 87.07 | -0.173 |
| TeHSP70-27 | GWHGABKY042494 | 582 | 64.25 | 5.08 | 34.38 | 83.14 | -0.417 |
| TeHSP70-28 | GWHGABKY043121 | 583 | 63.91 | 5.10 | 35.91 | 84.19 | -0.390 |
| TeHSP70-29 | GWHGABKY043154 | 544 | 60.80 | 7.58 | 39.85 | 83.46 | -0.388 |
Table 1 Physicochemical properties of TeHSP70s protein
基因名称 Gene name | 基因ID Gene ID | 蛋白质长度Protein length/aa | 分子质量Molecular weight/kD | 等电点pI | 不稳定系数 Instability index | 脂肪系数Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| TeHSP70-01 | GWHGABKY001532 | 887 | 97.70 | 5.45 | 44.50 | 82.33 | -0.458 |
| TeHSP70-02 | GWHGABKY001533 | 835 | 92.56 | 5.07 | 45.59 | 75.13 | -0.513 |
| TeHSP70-03 | GWHGABKY001646 | 740 | 82.25 | 5.28 | 42.34 | 79.97 | -0.380 |
| TeHSP70-04 | GWHGABKY001898 | 734 | 81.38 | 5.43 | 50.39 | 88.26 | -0.333 |
| TeHSP70-05 | GWHGABKY003413 | 686 | 75.38 | 5.23 | 38.34 | 77.07 | -0.503 |
| TeHSP70-06 | GWHGABKY011778 | 683 | 74.88 | 5.31 | 36.86 | 80.85 | -0.427 |
| TeHSP70-07 | GWHGABKY012581 | 680 | 74.83 | 5.18 | 33.92 | 85.50 | -0.381 |
| TeHSP70-08 | GWHGABKY012586 | 690 | 73.56 | 5.01 | 29.29 | 86.19 | -0.270 |
| TeHSP70-09 | GWHGABKY016707 | 665 | 73.27 | 5.06 | 25.54 | 86.50 | -0.479 |
| TeHSP70-10 | GWHGABKY017407 | 683 | 73.26 | 5.60 | 41.92 | 84.17 | -0.325 |
| TeHSP70-11 | GWHGABKY022426 | 684 | 73.14 | 5.04 | 30.07 | 87.09 | -0.310 |
| TeHSP70-12 | GWHGABKY022433 | 665 | 72.93 | 5.04 | 23.51 | 86.78 | -0.371 |
| TeHSP70-13 | GWHGABKY022437 | 678 | 72.86 | 5.29 | 36.04 | 84.04 | -0.310 |
| TeHSP70-14 | GWHGABKY022852 | 678 | 72.84 | 5.23 | 35.63 | 84.04 | -0.303 |
| TeHSP70-15 | GWHGABKY024486 | 684 | 72.77 | 5.78 | 35.28 | 87.49 | -0.268 |
| TeHSP70-16 | GWHGABKY024546 | 677 | 72.55 | 5.36 | 38.85 | 85.48 | -0.292 |
| TeHSP70-17 | GWHGABKY024596 | 657 | 71.80 | 5.17 | 36.09 | 79.15 | -0.446 |
| TeHSP70-18 | GWHGABKY025756 | 653 | 71.65 | 5.21 | 34.54 | 79.62 | -0.458 |
| TeHSP70-19 | GWHGABKY026068 | 667 | 71.63 | 5.06 | 27.89 | 88.22 | -0.296 |
| TeHSP70-20 | GWHGABKY027854 | 640 | 71.35 | 5.44 | 31.47 | 99.83 | -0.088 |
| TeHSP70-21 | GWHGABKY030282 | 660 | 71.31 | 5.07 | 27.51 | 88.53 | -0.319 |
| TeHSP70-22 | GWHGABKY032027 | 648 | 71.19 | 5.08 | 34.79 | 83.4 | -0.394 |
| TeHSP70-23 | GWHGABKY034860 | 648 | 71.02 | 5.13 | 33.73 | 81.16 | -0.415 |
| TeHSP70-24 | GWHGABKY035578 | 651 | 70.85 | 7.82 | 47.67 | 92.78 | -0.118 |
| TeHSP70-25 | GWHGABKY035973 | 620 | 69.08 | 6.25 | 41.66 | 81.1 | -0.402 |
| TeHSP70-26 | GWHGABKY042123 | 617 | 67.68 | 8.84 | 33.40 | 87.07 | -0.173 |
| TeHSP70-27 | GWHGABKY042494 | 582 | 64.25 | 5.08 | 34.38 | 83.14 | -0.417 |
| TeHSP70-28 | GWHGABKY043121 | 583 | 63.91 | 5.10 | 35.91 | 84.19 | -0.390 |
| TeHSP70-29 | GWHGABKY043154 | 544 | 60.80 | 7.58 | 39.85 | 83.46 | -0.388 |
蛋白名称 Protein name | 亚细胞定位 Subcelluar localization | α-螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β-折叠 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|---|
| TeHSP70-01 | 周质Periplasmic | 43.29 | 13.19 | 2.59 | 40.92 |
| TeHSP70-02 | 细胞质Cytoplasmic | 43.11 | 14.37 | 2.75 | 39.76 |
| TeHSP70-03 | 细胞质Cytoplasmic | 45.41 | 14.46 | 2.70 | 37.43 |
| TeHSP70-04 | 细胞质Cytoplasmic | 48.91 | 15.12 | 3.13 | 32.83 |
| TeHSP70-05 | 细胞质Cytoplasmic | 42.71 | 17.20 | 6.27 | 33.82 |
| TeHSP70-06 | 细胞质Cytoplasmic | 42.90 | 17.72 | 6.59 | 32.80 |
| TeHSP70-07 | 细胞质Cytoplasmic | 40.74 | 18.53 | 6.62 | 34.12 |
| TeHSP70-08 | 外膜Outermembrane | 41.30 | 19.13 | 6.23 | 33.33 |
| TeHSP70-09 | 细胞质Cytoplasmic | 45.11 | 18.05 | 6.62 | 30.23 |
| TeHSP70-10 | 细胞质Cytoplasmic | 43.48 | 19.62 | 6.73 | 30.16 |
| TeHSP70-11 | 细胞质Cytoplasmic | 38.45 | 20.03 | 7.75 | 33.77 |
| TeHSP70-12 | 细胞质Cytoplasmic | 43.46 | 18.50 | 6.92 | 31.13 |
| TeHSP70-13 | 细胞质Cytoplasmic | 38.20 | 23.16 | 8.11 | 30.53 |
| TeHSP70-14 | 细胞质Cytoplasmic | 44.10 | 19.76 | 7.82 | 28.32 |
| TeHSP70-15 | 外膜Outermembrane | 44.88 | 18.86 | 6.58 | 29.68 |
| TeHSP70-16 | 细胞质Cytoplasmic | 44.90 | 19.65 | 7.98 | 27.47 |
| TeHSP70-17 | 细胞质Cytoplasmic | 41.55 | 18.57 | 6.39 | 33.49 |
| TeHSP70-18 | 细胞质Cytoplasmic | 42.57 | 18.68 | 6.28 | 32.47 |
| TeHSP70-19 | 细胞质Cytoplasmic | 44.08 | 18.59 | 8.40 | 28.94 |
| TeHSP70-20 | 细胞质Cytoplasmic | 42.34 | 22.66 | 6.56 | 28.44 |
| TeHSP70-21 | 细胞质Cytoplasmic | 43.03 | 18.64 | 8.03 | 30.30 |
| TeHSP70-22 | 细胞质Cytoplasmic | 41.67 | 19.44 | 6.79 | 32.10 |
| TeHSP70-23 | 细胞质Cytoplasmic | 41.20 | 16.98 | 6.48 | 35.34 |
| TeHSP70-24 | 细胞质Cytoplasmic | 31.18 | 21.35 | 6.14 | 41.32 |
| TeHSP70-25 | 细胞质Cytoplasmic | 43.39 | 19.03 | 7.10 | 30.48 |
| TeHSP70-26 | 细胞质Cytoplasmic | 40.36 | 19.77 | 6.97 | 32.90 |
| TeHSP70-27 | 细胞质Cytoplasmic | 45.70 | 16.84 | 6.87 | 30.58 |
| TeHSP70-28 | 细胞质Cytoplasmic | 44.08 | 16.12 | 6.69 | 33.10 |
| TeHSP70-29 | 细胞质Cytoplasmic | 46.32 | 18.20 | 6.99 | 28.49 |
Table 2 Subcellular localization and secondary structure of TeHSP70s protein
蛋白名称 Protein name | 亚细胞定位 Subcelluar localization | α-螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β-折叠 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|---|
| TeHSP70-01 | 周质Periplasmic | 43.29 | 13.19 | 2.59 | 40.92 |
| TeHSP70-02 | 细胞质Cytoplasmic | 43.11 | 14.37 | 2.75 | 39.76 |
| TeHSP70-03 | 细胞质Cytoplasmic | 45.41 | 14.46 | 2.70 | 37.43 |
| TeHSP70-04 | 细胞质Cytoplasmic | 48.91 | 15.12 | 3.13 | 32.83 |
| TeHSP70-05 | 细胞质Cytoplasmic | 42.71 | 17.20 | 6.27 | 33.82 |
| TeHSP70-06 | 细胞质Cytoplasmic | 42.90 | 17.72 | 6.59 | 32.80 |
| TeHSP70-07 | 细胞质Cytoplasmic | 40.74 | 18.53 | 6.62 | 34.12 |
| TeHSP70-08 | 外膜Outermembrane | 41.30 | 19.13 | 6.23 | 33.33 |
| TeHSP70-09 | 细胞质Cytoplasmic | 45.11 | 18.05 | 6.62 | 30.23 |
| TeHSP70-10 | 细胞质Cytoplasmic | 43.48 | 19.62 | 6.73 | 30.16 |
| TeHSP70-11 | 细胞质Cytoplasmic | 38.45 | 20.03 | 7.75 | 33.77 |
| TeHSP70-12 | 细胞质Cytoplasmic | 43.46 | 18.50 | 6.92 | 31.13 |
| TeHSP70-13 | 细胞质Cytoplasmic | 38.20 | 23.16 | 8.11 | 30.53 |
| TeHSP70-14 | 细胞质Cytoplasmic | 44.10 | 19.76 | 7.82 | 28.32 |
| TeHSP70-15 | 外膜Outermembrane | 44.88 | 18.86 | 6.58 | 29.68 |
| TeHSP70-16 | 细胞质Cytoplasmic | 44.90 | 19.65 | 7.98 | 27.47 |
| TeHSP70-17 | 细胞质Cytoplasmic | 41.55 | 18.57 | 6.39 | 33.49 |
| TeHSP70-18 | 细胞质Cytoplasmic | 42.57 | 18.68 | 6.28 | 32.47 |
| TeHSP70-19 | 细胞质Cytoplasmic | 44.08 | 18.59 | 8.40 | 28.94 |
| TeHSP70-20 | 细胞质Cytoplasmic | 42.34 | 22.66 | 6.56 | 28.44 |
| TeHSP70-21 | 细胞质Cytoplasmic | 43.03 | 18.64 | 8.03 | 30.30 |
| TeHSP70-22 | 细胞质Cytoplasmic | 41.67 | 19.44 | 6.79 | 32.10 |
| TeHSP70-23 | 细胞质Cytoplasmic | 41.20 | 16.98 | 6.48 | 35.34 |
| TeHSP70-24 | 细胞质Cytoplasmic | 31.18 | 21.35 | 6.14 | 41.32 |
| TeHSP70-25 | 细胞质Cytoplasmic | 43.39 | 19.03 | 7.10 | 30.48 |
| TeHSP70-26 | 细胞质Cytoplasmic | 40.36 | 19.77 | 6.97 | 32.90 |
| TeHSP70-27 | 细胞质Cytoplasmic | 45.70 | 16.84 | 6.87 | 30.58 |
| TeHSP70-28 | 细胞质Cytoplasmic | 44.08 | 16.12 | 6.69 | 33.10 |
| TeHSP70-29 | 细胞质Cytoplasmic | 46.32 | 18.20 | 6.99 | 28.49 |
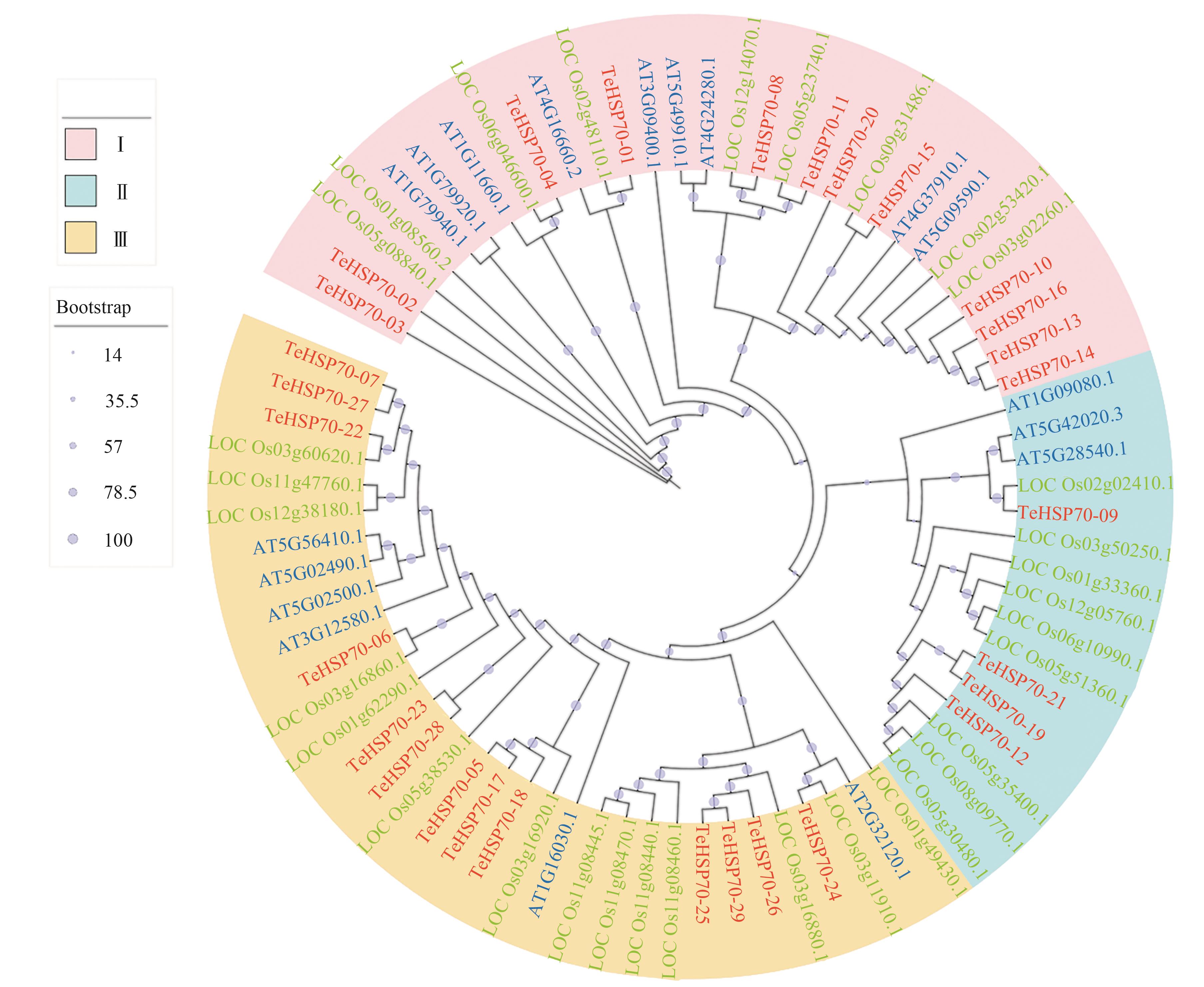
Fig. 5 Phylogenetic tree of TeHSP70 proteins with HSP70 proteins in Oryza sativa L. and Arabidopsis thalianaNote:Red, green and blue letters indicate Thinopyrum elongatum, Oryza sativa L. and Arabidopsis thaliana, respectively.
| 1 | HENDRICK J P, HARTL F U. Molecular chaperone functions of heat-shock proteins [J]. Annu. Rev. Biochem., 1993, 62(1):349-384. |
| 2 | SHAN Q, MA F T, WEI J Y, et al.. Physiological functions of heat shock proteins [J]. Curr. Protein Pept. Sci., 2020, 21(8):751-760. |
| 3 | WANG W, VINOCUR B, SHOSEYOV O, et al.. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response [J]. Trends Plant Sci., 2004, 9(5):244-252. |
| 4 | KIANG J G, TSOKOS G C. Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology [J]. Pharmacol. Ther., 1998, 80(2):183-201. |
| 5 | DAVOUDI M, CHEN J, LOU Q. Genome-wide identification and expression analysis of heat shock protein 70 (HSP70) gene family in pumpkin (Cucurbita moschata) rootstock under drought stress suggested the potential role of these chaperones in stress tolerance [J/OL]. Int. J. Mol. Sci., 2022, 23(3):1918 [2024-06-13]. . |
| 6 | DRAGOVIC Z, BROADLEY S A, SHOMURA Y, et al.. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s [J]. EMBO J., 2006, 25(11):2519-2528. |
| 7 | KARLIN S, BROCCHIERI L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution [J]. J. Mol. Evol., 1998, 47(5): 565-577. |
| 8 | HARTL F U, BRACHER A, HAYER-HARTL M. Molecular chaperones in protein folding and proteostasis [J]. Nature, 2011, 475(7356):324-332. |
| 9 | JACOB P, HIRT H, BENDAHMANE A. The heat-shock protein/chaperone network and multiple stress resistance [J]. Plant Biotechnol. J., 2017, 15(4): 405-414. |
| 10 | KAMPINGA H H, CRAIG E A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity [J]. Nat. Rev. Mol. Cell Biol., 2010, 11(8): 579-592. |
| 11 | CHEN B, FEDER M E, KANG L. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress [J]. Mol. Ecol., 2018, 27(15): 3040-3054. |
| 12 | MADAEVA I M, KURASHOVA N A, SEMENOVA N V, et al.. Heat shock protein HSP70 in oxidative stress in apnea patients [J]. Bull. Exp. Biol. Med., 2020, 169(5):695-697. |
| 13 | PAN X, ZHENG Y, LEI K, et al.. Systematic analysis of Heat Shock Protein 70 (HSP70) gene family in radish and potential roles in stress tolerance [J/OL]. BMC Plant Biol., 2024,24(1):2 [2024-06-13]. . |
| 14 | 吴毅,冯雅岚,王添宁,等.小麦及其祖先物种HSP70基因家族鉴定与表达分析 [J].草业学报,2024,33(7):53-67. |
| WU Y, FENG Y l, WANG T N, et al.. Genome-wide identification and expression analysis of the HSP70 gene family in wheat and its ancestral species [J]. Acta Pratac. Sin., 2024, 33(7):53-67. | |
| 15 | MAO H, JIANG C, TANG C, et al.. Wheat adaptation to environmental stresses under climate change: molecular basis and genetic improvement [J]. Mol. Plant., 2023, 16(10):1564-1589. |
| 16 | ZENG J, ZHOU C, HE Z, et al.. Disomic Substitution of 3D chromosome with its homoeologue 3e in tetraploid thinopyrum elongatum enhances wheat seedlings tolerance to salt stress [J/OL]. Int. J. Mol. Sci., 2023, 24(2):1609 [2024-06-13]. . |
| 17 | BAKER L, GREWAL S, YANG C Y, et al.. Exploiting the genome of Thinopyrum elongatum to expand the gene pool of hexaploid wheat [J]. Theor. Appl. Genet., 2020, 133(7):2213-2226. |
| 18 | WANG H, SUN S, GE W, et al.. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat [J/OL]. Science, 2020, 368(6493):5435 [2024-06-13]. . |
| 19 | UL H S, KHAN A, ALI M, et al.. Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses [J/OL]. Int. J. Mol. Sci., 2019, 20(21):5321 [2024-06-13]. . |
| 20 | BERARDINI T Z, REISER L, LI D, et al.. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome [J]. Genesis, 2015, 53(8):474-485. |
| 21 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al.. Pfam: the protein families database in 2021 [J]. Nucl. Acids Res., 2021, 49(D1): D412-D419. |
| 22 | POTTER S C, LUCIANI A, EDDY S R, et al.. HMMER web server: 2018 update [J]. Nucleic Acids Res., 2018, 46(W1):200-204. |
| 23 | LETUNIC I, KHEDKAR S, BORK P. SMART: recent updates, new developments and status in 2020 [J]. Nucleic Acids Res., 2021, 49(D1):458-460. |
| 24 | PAYSAN-LAFOSSE T, BLUM M, CHUGURANSKY S, et al.. InterPro in 2022 [J]. Nucleic Acids Res., 2023, 51(D1): 418-427. |
| 25 | DUVAUD S, GABELLA C, LISACEK F, et al.. Expasy, the Swiss bioinformatics resource portal, as designed by its users [J]. Nucleic Acids Res., 2021, 49(W1):216-227. |
| 26 | YU C S, LIN C J, HWANG J K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions [J]. Protein Sci., 2004, 13(5):1402-1406. |
| 27 | DELEAGE G. ALIGNSEC: viewing protein secondary structure predictions within large multiple sequence alignments [J]. Bioinformatics, 2017, 33(24):3991-3992. |
| 28 | BAILEY T L, JOHNSON J, GRANT C E, et al.. The MEME suite [J]. Nucleic Acids Res., 2015, 43(W1):39-49. |
| 29 | LESCOT M, DEHAIS P, THIJS G, et al.. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002, 30(1):325-327. |
| 30 | CHEN C, WU Y, LI J, et al.. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol. Plant., 2023, 16(11):1733-1742. |
| 31 | KAWAHARA Y, DE LA BASTIDE M, HAMILTON J P, et al.. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data [J/OL]. Rice (N Y)., 2013, 6(1):4 [2024-06-13]. . |
| 32 | ALAUX M, ROGERS J, LETELLIER T, et al.. Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data [J/OL]. Genome Biol., 2018,19(1):111 [2024-06-13]. . |
| 33 | ROZEWICKI J, LI S, AMADA K M, et al.. MAFFT-DASH: integrated protein sequence and structural alignment [J]. Nucleic Acids Res., 2019, 47(W1):5-10. |
| 34 | MINH B Q, SCHMIDT H A, CHERNOMOR O, et al.. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era [J]. Mol. Biol. Evol., 2020, 37(5):1530-1534. |
| 35 | LETUNIC I, BORK P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation [J]. Nucleic Acids Res., 2021, 49(W1):293-296. |
| 36 | SZKLARCZYK D, KIRSCH R, KOUTROULI M, et al.. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest [J]. Nucleic Acids Res., 2023, 51(D1):638-646. |
| 37 | SHANNON P, MARKIEL A, OZIER O, et al.. Cytoscape: a software environment for integrated models of biomolecular interaction networks [J]. Genome Res., 2003, 13(11):2498-2504. |
| 38 | KIM D, PAGGI J M, PARK C, et al.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype [J]. Nat. Biotechnol., 2019, 37(8):907-915. |
| 39 | ANDERS S, PYL P T, HUBER W. HTSeq—a python framework to work with high-throughput sequencing data [J]. Bioinformatics, 2015, 31(2):166-169. |
| 40 | USMAN M G, RAFII M Y, MARTINI M Y, et al.. Molecular analysis of HSP70 mechanisms in plants and their function in response to stress [J]. Biotechnol. Genet. Eng. Rev., 2017, 33(1):26-39. |
| 41 | LEISTER D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene [J]. Trends Genet., 2004,20(3):116-122. |
| 42 | SHANER L, MORANO K A. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity [J]. Cell Stress Chaperones, 2007,12(1):1-8. |
| 43 | ROSENZWEIG R, NILLEGODA N B, MAYER M P, et al.. The Hsp70 chaperone network [J]. Nat. Rev. Mol. Cell Biol., 2019, 20(11):665-680. |
| 44 | AHMED M, TOTH Z, DECSI K. The impact of salinity on crop yields and the confrontational behavior of transcriptional regulators, nanoparticles, and antioxidant defensive mechanisms under stressful conditions: a review [J/OL]. Int. J. Mol. Sci., 2024, 25(5):2654 [2024-06-13]. . |
| 45 | PENG Z, WANG Y, GENG G, et al.. Comparative analysis of physiological, enzymatic, and transcriptomic responses revealed mechanisms of salt tolerance and recovery in Tritipyrum [J/OL]. Front. Plant Sci., 2021, 12:800081 [2024-06-13]. . |
| [1] | Bei MA, Jie GONG, Yinke DU, Yuwei GAN, Rong CHENG, Bo ZHU, Lixia YI, Jinxiu MA, Shiqing GAO. Identification and Expression Analysis of TaINP1 Gene Related to Pollen Pore Development in Wheat [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 22-35. |
| [2] | Zhiduo DONG, Qiuping FU, Jian HUANG, Tong QI, Yanbo FU, Kuerban Kaisaier. Analysis of Salt Tolerance Capacity of Xinjiang Cotton Guring Germination [J]. Journal of Agricultural Science and Technology, 2025, 27(4): 57-67. |
| [3] | Tingting MA, Yanrong ZHAO, Yuqing WEI, Yuejuan WANG, Xuefei WANG, Erdong ZHANG. Dynamic Characteristics of Stem Growth and Sugar Accumulation of Sweet Sorghum at Late Growth Stage Under Soil Salt Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(2): 42-50. |
| [4] | Shixu QU, Yu SUN, Yizhen SUO, Haipeng YUAN, Yuhong ZHANG. Effects of Exogenous Calcium on Physiological Characteristics and Secondary Metabolites of Cannabis sativa L. Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2025, 27(2): 80-88. |
| [5] | Xuewen XU, Xingpeng WANG, Hongbo WANG, Zhenxi CAO. Physiological Regulation of Growth of Cotton Seedlings Under Salt Stress by Rhamnolipids [J]. Journal of Agricultural Science and Technology, 2025, 27(1): 72-79. |
| [6] | Xiaorong GUO, Ying LIU, Jiazhen FAN, Tao HUANG, Rong ZHOU. Bioinformatics Analysis of Porcine CREBRF Gene and Its Expression Pattern [J]. Journal of Agricultural Science and Technology, 2024, 26(9): 44-53. |
| [7] | Fulin ZHANG, Rui XI, Yuxiang LIU, Zhaolong CHEN, Qinghui YU, Ning LI. Genome-wide Identification and Expression Analysis of Tomato BURP Structural Domain Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(8): 51-62. |
| [8] | Xinyue BAO, Hongmin CHEN, Weiwei WANG, Yimiao TANG, Zhaofeng FANG, Jinxiu MA, Dezhou WANG, Jinghong ZUO, Zhanjun YAO. Cloning and Expression Analysis of Wheat TaCOBL-5 Genes [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 11-21. |
| [9] | Mingdi CHEN, Guihua HU, Haiwen ZHANG, Wangtian WANG. Bioinformatics and Expression Pattern Analysis of Rice RR Gene Family [J]. Journal of Agricultural Science and Technology, 2024, 26(5): 20-29. |
| [10] | Shuang LI, Aiying WANG, Zhen JIAO, Qing CHI, Hao SUN, Tao JIAO. Physiological and Chemical Characteristics and Transcriptome Analysis of Different Type of Wheat Seedlings Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2024, 26(2): 20-32. |
| [11] | Nigela Abuduwaili, Xiuzhen WEI, Jingjie MEN, Lingna CHEN, Yongkun CHEN, Gengqing HUANG. Bioinformatics and Expression Pattern Analysis of JAZ and MYC2-like Gene Family in Lavandula angustifolia [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 79-90. |
| [12] | Xuemin JIANG, Xiangqian CHEN, Hongyan LI, Qiyan JIANG. Metabolomic Analysis of Wheat Response to Salt Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(9): 43-56. |
| [13] | Yulu WU, Jiaxin HU, Yuxi CHEN, Bingsong ZHENG, DaoLiang YAN. Effects of External Application of α-Ketoglutarate on Growth, Carbon, Nitrogen and Phosphorus Accumulation and Their Stoichiometric Relationships in Kosteletzkya virginica Under Salt Stress [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 170-177. |
| [14] | Zhonghua MA, Juan CHEN, Na WU, Benju MAN, Xiaogang WANG, Yongqing ZHE, Jili LIU. Effects of Salt Stress and Phosphorus Supply on Photosynthetic Characteristics and Total Biomass of Switchgrass at Seedling Stage [J]. Journal of Agricultural Science and Technology, 2023, 25(6): 190-200. |
| [15] | Fan ZHANG, Hong WANG, Xuebing ZHANG, Jianjun CHEN. Physiological and Biochemical Responses of Peach Self-rooted Rootstock to NaCl and Analysis of Salt Tolerance Threshold [J]. Journal of Agricultural Science and Technology, 2023, 25(11): 70-79. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号