




















中国农业科技导报 ›› 2024, Vol. 26 ›› Issue (8): 51-62.DOI: 10.13304/j.nykjdb.2023.0311
张福林1,2( ), 奚瑞1,2, 刘宇翔1,2, 陈兆龙1,2, 余庆辉1, 李宁1(
), 奚瑞1,2, 刘宇翔1,2, 陈兆龙1,2, 余庆辉1, 李宁1( )
)
收稿日期:2023-04-19
接受日期:2023-08-25
出版日期:2024-08-15
发布日期:2024-08-12
通讯作者:
李宁
作者简介:张福林E-mail:18095998056@163.com;
基金资助:
Fulin ZHANG1,2( ), Rui XI1,2, Yuxiang LIU1,2, Zhaolong CHEN1,2, Qinghui YU1, Ning LI1(
), Rui XI1,2, Yuxiang LIU1,2, Zhaolong CHEN1,2, Qinghui YU1, Ning LI1( )
)
Received:2023-04-19
Accepted:2023-08-25
Online:2024-08-15
Published:2024-08-12
Contact:
Ning LI
摘要:
BURP结构域蛋白为植物所特有,在植物生长发育和胁迫响应中起着重要作用。为鉴定番茄BURP结构域基因(SlBURP)及分析其表达,从番茄基因组中鉴定出12个SlBURP基因,系统进化分析将其分为7个亚族,不同亚族的保守基序和基因结构有明显差异。SlBURPs启动子序列中广泛存在胁迫、生长、激素、光响应元件,表明该家族基因在对非生物胁迫的响应中有重要作用。共线性分析表明,番茄SlBURP基因和拟南芥BURP基因家族成员来自同一祖先。RT-qPCR表明,SlBURP6、SlBURP7、SlBURP8、SlBURP12在叶中大量表达;SlBURP2、SlBURP6、SlBURP7、SlBURP8在盐处理后转录水平上升;在植物调节剂处理下SlBURP5、SlBURP8、SlBURP9、SlBURP12基因在果实发育期的转录水平均有上升。综上所述,SlBURP基因在番茄生长发育和逆境胁迫中起着重要作用。
中图分类号:
张福林, 奚瑞, 刘宇翔, 陈兆龙, 余庆辉, 李宁. 番茄BURP结构域基因家族全基因组鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(8): 51-62.
Fulin ZHANG, Rui XI, Yuxiang LIU, Zhaolong CHEN, Qinghui YU, Ning LI. Genome-wide Identification and Expression Analysis of Tomato BURP Structural Domain Gene Family[J]. Journal of Agricultural Science and Technology, 2024, 26(8): 51-62.
处理 Treatment | 试剂 Reagent |
|---|---|
| CK | 蒸馏水Distilled water |
| EBR0.05 | 0.05 mg·L-1 EBR |
| EBR0.10 | 0.10 mg·L-1 EBR |
| EBR0.20 | 0.20 mg·L-1 EBR |
| MT50 | 50 μmol·L-1 MT |
| MT100 | 100 μmol·L-1 MT |
| MT150 | 150 μmol·L-1 MT |
| NAA10 | 10 mg·L-1 NAA |
| NAA20 | 20 mg·L-1 NAA |
| NAA30 | 30 mg·L-1 NAA |
表 1 植物生长调节剂试验处理
Table 1 Treatments with different concentration of plant growth regulators
处理 Treatment | 试剂 Reagent |
|---|---|
| CK | 蒸馏水Distilled water |
| EBR0.05 | 0.05 mg·L-1 EBR |
| EBR0.10 | 0.10 mg·L-1 EBR |
| EBR0.20 | 0.20 mg·L-1 EBR |
| MT50 | 50 μmol·L-1 MT |
| MT100 | 100 μmol·L-1 MT |
| MT150 | 150 μmol·L-1 MT |
| NAA10 | 10 mg·L-1 NAA |
| NAA20 | 20 mg·L-1 NAA |
| NAA30 | 30 mg·L-1 NAA |
基因名称 Gene name | 正向引物序列 Forward primer sequence (5’-3’) | 反向引物序列 Reverse primer sequence (5’-3’) |
|---|---|---|
| SlBURP2 | GCGGAGGCAATGCTTGATT | GCCACCATCTTGGGAGTTTCTA |
| SlBURP3 | CGGTGTTGATGGAACTAAGGTG | TGACAAATAGAGGGAGCACCAG |
| SlBURP5 | ACAGAGGAAGAGATTCGTGAGC | GAAGGGAATGGTTGATTGGC |
| SlBURP6 | GCAATGCGTGGCGTCTATT | TGATTTCGTCACTCTTCCGC |
| SlBURP7 | GGCAATACGCCTGTCCAAA | GACCAGCATTTCCATCGTTTC |
| SlBURP8 | GGTCGTAATGTGGTGGTTCG | CGCTTCGTAGACTCGGACTTT |
| SlBURP9 | GGTGAGACGAAACGATGTGTT | CCACCGTTGATTCCTTTGACT |
| SlBURP10 | AGACGGTGTTGTTCCTGAGC | GTATCGTTGGGATGCTTGAGTT |
| SlBURP11 | TCAAGCCAAGGTCTATCCAGC | AATGAGCGATGAGGGAACG |
| SlBURP12 | GTTGGAGGCAATAAAGGCG | AACAAACGGTGAGGGTCCA |
| SlActin | CAGGGTGTTCTTCAGGAGCAA | GGTGTTATGGTCGGAATGGG |
表2 用于RT-qPCR的引物序列
Table 2 Primers sequences used for RT-qPCR
基因名称 Gene name | 正向引物序列 Forward primer sequence (5’-3’) | 反向引物序列 Reverse primer sequence (5’-3’) |
|---|---|---|
| SlBURP2 | GCGGAGGCAATGCTTGATT | GCCACCATCTTGGGAGTTTCTA |
| SlBURP3 | CGGTGTTGATGGAACTAAGGTG | TGACAAATAGAGGGAGCACCAG |
| SlBURP5 | ACAGAGGAAGAGATTCGTGAGC | GAAGGGAATGGTTGATTGGC |
| SlBURP6 | GCAATGCGTGGCGTCTATT | TGATTTCGTCACTCTTCCGC |
| SlBURP7 | GGCAATACGCCTGTCCAAA | GACCAGCATTTCCATCGTTTC |
| SlBURP8 | GGTCGTAATGTGGTGGTTCG | CGCTTCGTAGACTCGGACTTT |
| SlBURP9 | GGTGAGACGAAACGATGTGTT | CCACCGTTGATTCCTTTGACT |
| SlBURP10 | AGACGGTGTTGTTCCTGAGC | GTATCGTTGGGATGCTTGAGTT |
| SlBURP11 | TCAAGCCAAGGTCTATCCAGC | AATGAGCGATGAGGGAACG |
| SlBURP12 | GTTGGAGGCAATAAAGGCG | AACAAACGGTGAGGGTCCA |
| SlActin | CAGGGTGTTCTTCAGGAGCAA | GGTGTTATGGTCGGAATGGG |
基因ID Gene ID | 基因名称 Gene name | 染色体 Chromosome | 氨基酸数量 Number of amino acids | 分子量 Molecular weight/Da | 等电点 pI | 脂肪指数 Aliphatic index | 亲水性 Hydrophilicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| Solyc01g004131.1 | SlBURP1 | 1 | 813 | 92 342.76 | 6.17 | 79.68 | -0.299 | 叶绿体Chloroplast |
| Solyc01g004134.1 | SlBURP2 | 1 | 1 200 | 131 593.70 | 8.28 | 57.20 | -0.589 | 叶绿体Chloroplast |
| Solyc02g000714.2 | SlBURP3 | 2 | 365 | 39 549.89 | 7.26 | 73.04 | -0.438 | 胞外Extracellular |
| Solyc02g000715.1 | SlBURP4 | 2 | 405 | 45 924.69 | 6.09 | 75.11 | -0.547 | 细胞质Cytoplasmic |
| Solyc02g000716.1 | SlBURP5 | 2 | 264 | 30 034.04 | 7.10 | 87.54 | -0.031 | 胞外Extracellular |
| Solyc03g002688.1 | SlBURP6 | 3 | 322 | 36 324.05 | 8.54 | 84.72 | -0.248 | 细胞核Nucleus |
| Solyc05g000048.2 | SlBURP7 | 5 | 337 | 38 173.93 | 7.65 | 84.69 | -0.217 | 细胞核Nucleus |
| Solyc05g000050.1 | SlBURP8 | 5 | 325 | 37 217.51 | 6.05 | 76.46 | -0.303 | 胞外Extracellular |
| Solyc05g000051.1 | SlBURP9 | 5 | 611 | 66 805.31 | 6.16 | 51.72 | -0.689 | 细胞核Nucleus |
| Solyc05g001003.1 | SlBURP10 | 5 | 629 | 69 069.40 | 7.93 | 58.46 | -0.632 | 胞外Extracellular |
| Solyc06g002169.1 | SlBURP11 | 6 | 693 | 76 193.18 | 9.22 | 61.07 | -0.470 | 叶绿体Chloroplast |
| Solyc08g001592.1 | SlBURP12 | 8 | 408 | 45 869.40 | 9.02 | 76.45 | -0.262 | 细胞质Cytoplasmic |
表3 番茄BURP基因家族成员鉴定及理化性质
Table 3 Identification and characteristics of the SlBURP gene family in tomato
基因ID Gene ID | 基因名称 Gene name | 染色体 Chromosome | 氨基酸数量 Number of amino acids | 分子量 Molecular weight/Da | 等电点 pI | 脂肪指数 Aliphatic index | 亲水性 Hydrophilicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| Solyc01g004131.1 | SlBURP1 | 1 | 813 | 92 342.76 | 6.17 | 79.68 | -0.299 | 叶绿体Chloroplast |
| Solyc01g004134.1 | SlBURP2 | 1 | 1 200 | 131 593.70 | 8.28 | 57.20 | -0.589 | 叶绿体Chloroplast |
| Solyc02g000714.2 | SlBURP3 | 2 | 365 | 39 549.89 | 7.26 | 73.04 | -0.438 | 胞外Extracellular |
| Solyc02g000715.1 | SlBURP4 | 2 | 405 | 45 924.69 | 6.09 | 75.11 | -0.547 | 细胞质Cytoplasmic |
| Solyc02g000716.1 | SlBURP5 | 2 | 264 | 30 034.04 | 7.10 | 87.54 | -0.031 | 胞外Extracellular |
| Solyc03g002688.1 | SlBURP6 | 3 | 322 | 36 324.05 | 8.54 | 84.72 | -0.248 | 细胞核Nucleus |
| Solyc05g000048.2 | SlBURP7 | 5 | 337 | 38 173.93 | 7.65 | 84.69 | -0.217 | 细胞核Nucleus |
| Solyc05g000050.1 | SlBURP8 | 5 | 325 | 37 217.51 | 6.05 | 76.46 | -0.303 | 胞外Extracellular |
| Solyc05g000051.1 | SlBURP9 | 5 | 611 | 66 805.31 | 6.16 | 51.72 | -0.689 | 细胞核Nucleus |
| Solyc05g001003.1 | SlBURP10 | 5 | 629 | 69 069.40 | 7.93 | 58.46 | -0.632 | 胞外Extracellular |
| Solyc06g002169.1 | SlBURP11 | 6 | 693 | 76 193.18 | 9.22 | 61.07 | -0.470 | 叶绿体Chloroplast |
| Solyc08g001592.1 | SlBURP12 | 8 | 408 | 45 869.40 | 9.02 | 76.45 | -0.262 | 细胞质Cytoplasmic |
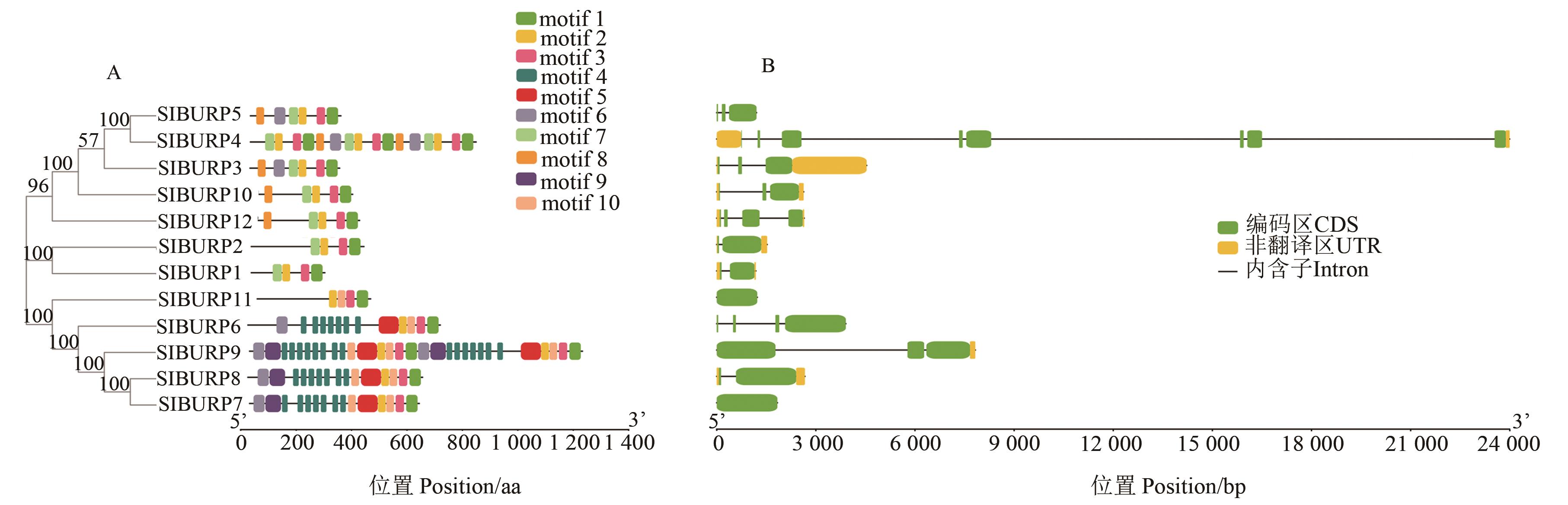
图2 番茄SlBURP家族保守基序分布A:编码蛋白保守基序分析;B:基因结构分析
Fig. 2 Distribution of conserved motifs in SlBURP familyA: Conserved motifs analysis of protens coded;C:Gene structure analysis motifs

图5 BURP基因种间共线性分析注:At—拟南芥;Sl—番茄;St—马铃薯;Os—水稻;Zm—玉米。
Fig. 5 Interspecies colinearity analysis of BURP genesNote: At—Arabidopsis thaliana;Sl—Solanum lycopersicum;St—Solanum tuberosum;Os—Oryza sativa;Zm—Zea mays.
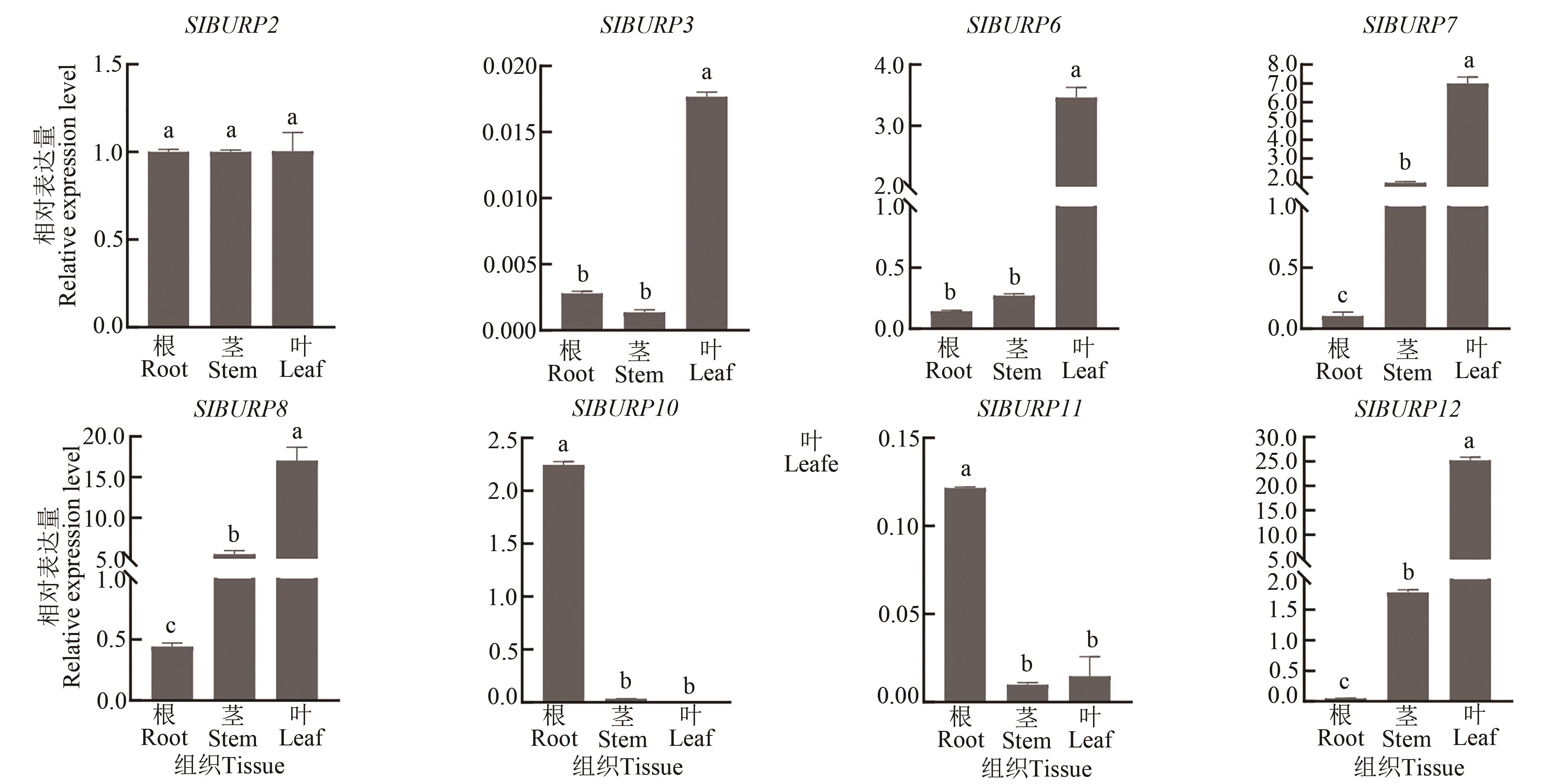
图6 SlBURP基因在根、茎、叶中的表达注:不同小写字母表示不同器官间在P<0.05水平差异显著。
Fig. 6 Expression of SlBURPs gene in root, stem and leafNote: Different lowercase letters indicate significant differences between different tissues at P<0.05 level.

图7 盐胁迫SlBURP基因在叶中的表达注:不同小写字母表示在P<0.05水平差异显著。
Fig. 7 Expression analysis of SlBURP gene in leaves under salt stressNote: Different lowercase letters indicate significant differences at P<0.05 level.

图8 不同植物生长调节剂下SlBURP基因在果实发育时的表达注不同小写字母表示同一时期不同处理间在P<0.05水平差异显著。
Fig. 8 Expression of SlBURP gene during fruit development under plant growth regulators stressNote: Different lowercase letters indicate significant differences between different treatments of same stage at P<0.05 level.
| 1 | PHILLIPS K, LUDIDI N. Drought and exogenous abscisic acid alter hydrogen peroxide accumulation and differentially regulate the expression of two maize RD22-like genes [J/OL]. Sci. Rep., 2017, 7(1):8821 [2023-07-31]. . |
| 2 | VAN SON L, TIEDEMANN J, RUTTEN T, et al.. The BURP domain protein AtUSPL1 of Arabidopsis thaliana is destined to the protein storage vacuoles and overexpression of the cognate gene distorts seed development [J]. Plant Mol. Biol., 2009, 71(4):319-329. |
| 3 | JIN J, JEMAA E, XU Z, et al.. Arabidopsis ETHYLENE INSENSITIVE 3 directly regulates the expression of PG1β-like family genes in response to aluminum stress [J/OL]. J. Exp. Bot., 2022, 14:161 [2023-07-31]. . |
| 4 | DING X P, HOU X, XIE K B, et al.. Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses [J]. Planta, 2009, 230(1):149-163. |
| 5 | KOMATSU S, NANJO Y, NISHIMURA M. Proteomic analysis of the flooding tolerance mechanism in mutant soybean [J]. J. Proteomics, 2013, 79:231-250. |
| 6 | TANG Y L, LI X J, ZHONG Y T, et al.. Functional analysis of soybean SALI3-2 in yeast [J]. Shenzhen Univ. Sci. Eng., 2007, 24:324-330. |
| 7 | GAN D, JIANG H, ZHANG J, et al.. Genome-wide analysis of BURP domain-containing genes in maize and sorghum [J]. Mol. Biol. Rep., 2011, 38(7):4553-4563. |
| 8 | YU S W, ZHANG L D, ZUO K J, et al.. Isolation and characterization of a BURP domain-containing gene BnBDC1 from Brassica napus involved in abiotic and biotic stress [J]. Physiol. Plantarum, 2004, 122(2):210-218. |
| 9 | THAMMEGOWDA H V, LE V S, CHRISTIANE S, et al.. AtRD22 and AtUSPL 1, members of the plant-specific BURP domain family involved in Arabidopsis thaliana drought tolerance [J/OL]. PLoS One, 2014, 9(10):e110065 [2023-07-31]. . |
| 10 | TEERAWANICHPAN P, XIA Q, CALDWELL S J, et al.. Protein storage vacuoles of Brassica napus zygotic embryos accumulate a BURP domain protein and perturbation of its production distorts the PSV [J]. Plant Mol. Biol., 2009, 71(4):331-343. |
| 11 | WANG A M, XIA Q, XIE W S, et al.. The classical ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development [J]. Proc. Natl. Acad. Sci. USA, 2003, 100:14487-14492. |
| 12 | XU X Y, YANG H K, SINGH S P, et al.. Genetic manipulation of non-classic oilseed plants for enhancement of their potential as a biofactory for triacylglycerol production [J/OL]. Engineering, 2018, 4(4):11 [2023-07-31]. . |
| 13 | XU B, GOU J Y, LI F G, et al.. A cotton BURP domain protein interacts with α-expansin and their co-expression promotes plant growth and fruit production [J]. Mol. Plant, 2013, 6(3):945-958. |
| 14 | MATUS J T, AQUEA F, ESPINOZA C, et al.. Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses [J/OL]. PLoS One, 2014, 9(10):e110372 [2023-07-31]. . |
| 15 | 宋单单,史梦雪,高金汇,等.复配型转光棚膜光学性能及其对日光温室番茄生长及果实品质的影响[J].植物生理学报,2022,58(11):2218-2226. |
| SONG D D, SHI M X, GAO J H, et al.. Optical properties of complex-type of light conversion greenhouse films and their effects on the growth and fruit qualities of tomato in solar greenhouse [J]. J. Plant Physiol., 2022, 58(11):2218-2226. | |
| 16 | 刘向蕾,尹亚红,高玉迪,等.番茄SlNPF68基因参与缺氮条件下根系觅食反应[J].植物生理学报,2022,58(7):1212-1220. |
| LIU X L, YIN Y H, GAO Y D, et al.. SlNPF68 gene involved in root foraging response under nitrogen deficiency in tomato [J]. J. Plant Physiol., 2022, 58(7):1212-1220. | |
| 17 | 万玉通,裴茂松,韦同路,等.甲基化修饰影响果实生长发育的研究进展[J].植物生理学报,2022,57(3):531-541. |
| WAN Y T, PEI M S, WEI T L, et al.. Progress of methylation modification on fruit development [J]. J. Plant Physiol., 2022, 57(3):531-541. | |
| 18 | SUN H R, WEI H L, WANG H T, et al.. Genome-wide identification and expression analysis of the BURP domain-containing genes in Gossypium hirsutum [J/OL]. BMC Genomics, 2019, 20:558 [2023-07-31]. . |
| 19 | XU H, LI Y, YAN Y, et al.. Genome-scale identification of soybean BURP domain-containing genes and their expression under stress treatments [J]. BMC Plant Biol., 2010, 10(1):1-16. |
| 20 | JEONG H Y, NGUYEN H P, EOM S H, et al.. Integrative analysis of pectin methylesterase (PME) and PME inhibitors in tomato (Solanum lycopersicum): identification, tissue-specific expression, and biochemical characterization [J]. Plant Physiol. Biochem., 2018, 132:557-565. |
| 21 | XU Q, DUNBRACK R L. Assignment of protein sequences to existing domain and family classification systems: Pfam and the PDB [J]. Bioinformatics, 2012, 28(21):2763-2772. |
| 22 | SONNHAMMER E, EDDY S R, DURBIN R. Pfam: a comprehensive database of protein domain families based on seed alignments [J]. Proteins, 2015, 28(3):405-420. |
| 23 | SCHULTZ J, COPLEY R R, TOBIAS D, et al.. SMART: a web-based tool for the study of genetically mobile domains [J]. Nucl. Acids Res., 2000, 28:231-234. |
| 24 | ARTIMO P, JONNALAGEDDA M, ARNOLD K, et al.. ExPASy: SIB bioinformatics resource portal [J]. Nucl. Acids Res.. 2012, 40:597-603. |
| 25 | HORTON P, PARK K J, OBAYASHI T, et al.. WoLF PSORT: protein localization predictor [J]. Nucl. Acids Res., 2007, 35:585-587. |
| 26 | KUMAR S, STECHER G, TAMURA K. MEGA7 molecular evolutionary genetics analysis version 7.0 for bigger datasets [J]. Mol. Biol. Evol., 2015, 33:1870-1874. |
| 27 | CHEN C, CHEN H, ZHANG Y, et al.. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J/OL]. Mol. Plant, 2020, 13(8):009 [2023-07-31] . |
| 28 | ROMBAUTS, DEHAIS, MONTAGU V, et al.. PlantCARE, a plant cis-acting regulatory element database [J]. Nucl. Acids Res., 1999, 27:295-296. |
| 29 | LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔ CT [J]. Methods, 2001, 25:492-508. |
| 30 | 张中保,吴忠义,魏建华.玉米BURP家族基因的鉴定和分析[J].玉米科学,2014,22(3):36-42. |
| ZHANG Z B, WU Z Y, WEI J H. Genome-wide analysis and identification of ZmBURP genes family in maize [J]. J. Maize Sci., 2014, 22(3):36-42. | |
| 31 | SHAO Y H, WEI G, WANG L, et al.. Genome-wide analysis of BURP domain-containing genes in Populus trichocarpa [J]. J. Integr. Plant Biol., 2011, 53(9):743-755. |
| 32 | GRANGER C, CORYELL V, KHANNA A, et al.. Identification, structure, and differential expression of members of a BURP domain containing protein family in soybean [J/OL]. Genome, 2002, 45(4):693 [2023-07-31]. . |
| 33 | LI Y, CHEN X, CHEN Z, et al.. Identification and expression analysis of BURP domain-containing genes in Medicago truncatula [J/OL]. Front. Plant Sci., 2016, 7:485 [2023-07-31]. . |
| 34 | YU S W, ZHANG L D, ZUO K J, et al.. Isolation and characterization of a BURP domain-containing gene BnBDC1 from Brassica napus involved in abiotic and biotic stress [J]. Physiol. Plantarum, 2004, 100:210-218. |
| 35 | WANG H M, ZHOU L, FU Y P, et al.. Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress [J]. Plant Cell Environ., 2012, 35(11):1932-4197. |
| 36 | 米子岚,钟活权,江年琼,等.BURP蛋白家族与植物对非生物胁迫的响应[J].中国细胞生物学学报,2015, 37(9):1302-1308. |
| MI Z L, ZHONG H Q, JIANG N Q, et al.. BURP Proteins Family and the Response of Plant to Abiotic Stress [J]. J. Cell Biol., 2015, 37(9):1302-1308. | |
| 37 | WANG L H, WU N N, ZHU Y, et al.. The divergence and positive selection of the plant-specific BURP-containing protein family [J]. Ecol. Evol., 2015, 5(22):5394-5412. |
| [1] | 张俊蕾, 盖晓彤, 赵正婷, 刘弟, 王金凤, 姜宁, 刘雅婷. 烟草番茄斑萎病毒RT-LAMP检测体系的建立及优化[J]. 中国农业科技导报, 2024, 26(8): 140-150. |
| [2] | 王子凡, 李燕, 张庆银, 王丹丹, 师建华, 耿晓彬, 田东良, 钟增明, 赵晓明, 齐连芬. 微生物菌剂对设施番茄主要病害及土壤微生物群落的影响[J]. 中国农业科技导报, 2024, 26(6): 102-112. |
| [3] | 李名博, 刘玉乐, 穆志民, 郭俊旺, 卫勇, 任东悦, 贾济深, 卫泽中, 栗宇红. 基于YOLOX-L-TN模型的番茄果实识别[J]. 中国农业科技导报, 2024, 26(4): 97-105. |
| [4] | 李双, 王爱英, 焦浈, 池青, 孙昊, 焦涛. 盐胁迫下不同抗性小麦幼苗生理生化特性及转录组分析[J]. 中国农业科技导报, 2024, 26(2): 20-32. |
| [5] | 邓玉荣, 韩联, 王金龙, 韦兴翰, 王旭东, 赵颖, 魏小红, 李朝周. 藜麦SOD家族基因的鉴定及其对混合盐碱胁迫的响应[J]. 中国农业科技导报, 2024, 26(1): 28-39. |
| [6] | 姜雪敏, 陈向前, 李红燕, 姜奇彦. 小麦盐胁迫响应的代谢组学分析[J]. 中国农业科技导报, 2023, 25(9): 43-56. |
| [7] | 卢登洋, 王鑫, 唐章虎, 吴翠云, 蒲云峰, 闫敏, 鲍荆凯, 姜喜. 梨果实发育过程中酚类物质组成及抗氧化活性比较[J]. 中国农业科技导报, 2023, 25(9): 97-104. |
| [8] | 吴雨露, 扈嘉鑫, 陈宇熙, 郑炳松, 闫道良. 外施α-酮戊二酸对盐胁迫下海滨锦葵生长、碳氮磷养分积累及其计量关系的影响[J]. 中国农业科技导报, 2023, 25(7): 170-177. |
| [9] | 麻仲花, 陈娟, 吴娜, 满本菊, 王晓港, 者永清, 刘吉利. 盐胁迫与供磷水平对柳枝稷苗期光合特性与总生物量的影响[J]. 中国农业科技导报, 2023, 25(6): 190-200. |
| [10] | 陈春林, 王琳洋, 单梦伟, 裴甜甜, 王吉庆, 肖怀娟, 李娟起, 李猛, 杜清洁. 发酵花生壳和牛粪替代草炭基质的番茄育苗效果分析[J]. 中国农业科技导报, 2023, 25(4): 205-214. |
| [11] | 张曼, 王志城, 刘正文, 王国宁, 王省芬, 张艳. 陆地棉BGLU基因家族成员的全基因组鉴定与表达分析[J]. 中国农业科技导报, 2023, 25(2): 48-59. |
| [12] | 张帆, 王鸿, 张雪冰, 陈建军. 桃自根砧对NaCl生理生化响应及耐盐阈值分析[J]. 中国农业科技导报, 2023, 25(11): 70-79. |
| [13] | 李峰, 殷丛培, 殷冉, 王凡, 韩永亮, 杨志敏, 刘建成. 燕麦根际土壤细菌多样性对盐胁迫的响应[J]. 中国农业科技导报, 2023, 25(1): 153-165. |
| [14] | 李世民, 董琼, 金友帆, 李树萍, 李猛, 刘廷彪, 赵兴杰, 陈静, 叶平, 吕梦. 树番茄幼苗叶片性状和生理参数对遮阴的响应及评价[J]. 中国农业科技导报, 2023, 25(1): 72-82. |
| [15] | 夏秀波, 李涛, 曹守军, 姚建刚, 王虹云, 张丽莉. 液态有机肥部分替代化肥对设施番茄根区细菌群落的影响[J]. 中国农业科技导报, 2022, 24(7): 187-196. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号