




















Journal of Agricultural Science and Technology ›› 2025, Vol. 27 ›› Issue (3): 49-59.DOI: 10.13304/j.nykjdb.2024.0111
• BIOTECHNOLOGY & LIFE SCIENCE • Previous Articles Next Articles
Na LI( ), Hua ZHANG, Xinzhu XING, Zhenqi SHAO, Zhanwu YANG, Xihuan LI(
), Hua ZHANG, Xinzhu XING, Zhenqi SHAO, Zhanwu YANG, Xihuan LI( ), Caiying ZHANG
), Caiying ZHANG
Received:2024-02-12
Accepted:2024-03-28
Online:2025-03-15
Published:2025-03-14
Contact:
Xihuan LI
李娜( ), 张华, 邢馨竹, 邵振启, 杨占武, 李喜焕(
), 张华, 邢馨竹, 邵振启, 杨占武, 李喜焕( ), 张彩英
), 张彩英
通讯作者:
李喜焕
作者简介:李娜 E-mail: 1784437078@qq.com;
基金资助:CLC Number:
Na LI, Hua ZHANG, Xinzhu XING, Zhenqi SHAO, Zhanwu YANG, Xihuan LI, Caiying ZHANG. Function Analysis of Soybean Expansin Gene GmEXLA1 inPlantPod and Seed Development[J]. Journal of Agricultural Science and Technology, 2025, 27(3): 49-59.
李娜, 张华, 邢馨竹, 邵振启, 杨占武, 李喜焕, 张彩英. 大豆扩展蛋白基因GmEXLA1在荚果发育中的功能鉴定[J]. 中国农业科技导报, 2025, 27(3): 49-59.
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) | 引物用途 Primer usage |
|---|---|---|
| GmEXLA1-F | GCAGTCGACATGGCTTTCTTTATTCTCTCC | 扩增开放阅读框;检测转基因拟南芥;检测大豆突变体 Open reading frame amplification; detection of transgenic Arabidopsis and soybean mutant |
| GmEXLA1-R | TATGGATCCTGTCCCATCATCGCAGGG | |
| GmEXLA1-RT-F1 | TGGCCGTTCGAGTTGAAGAA | 转基因拟南芥实时定量分析 Real-time quantitative analysis of transgenic Arabidopsis |
| GmEXLA1-RT-R1 | ATACTGCCCCGTGGTTTCTG | |
| GmEXLA1-Mu-F | GCTGGATCCATGTGTGATCGCTGCT | gmexla1突变体目的基因原核表达 Prokaryotic expression of target gene in gmexla1 mutant |
| GmEXLA1-Mu-R | TTAGAGCTCTCATGTCCCATCATCGCAG | |
| GmEXLA1-RT-F2 | ACTGAAGCTTGGCATTGCTG | gmexla1突变体实时定量分析 Real-time quantitative analysis of gmexla1 mutant |
| GmEXLA1-RT-R2 | CATACTGCCCCGTGGTTTCT | |
| GmActin11-F | ATCTTGACTGAGCGTGGTTATTCC | |
| GmActin11-R | GCTGGTCCTGGCTGTCTCC |
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5’-3’) | 引物用途 Primer usage |
|---|---|---|
| GmEXLA1-F | GCAGTCGACATGGCTTTCTTTATTCTCTCC | 扩增开放阅读框;检测转基因拟南芥;检测大豆突变体 Open reading frame amplification; detection of transgenic Arabidopsis and soybean mutant |
| GmEXLA1-R | TATGGATCCTGTCCCATCATCGCAGGG | |
| GmEXLA1-RT-F1 | TGGCCGTTCGAGTTGAAGAA | 转基因拟南芥实时定量分析 Real-time quantitative analysis of transgenic Arabidopsis |
| GmEXLA1-RT-R1 | ATACTGCCCCGTGGTTTCTG | |
| GmEXLA1-Mu-F | GCTGGATCCATGTGTGATCGCTGCT | gmexla1突变体目的基因原核表达 Prokaryotic expression of target gene in gmexla1 mutant |
| GmEXLA1-Mu-R | TTAGAGCTCTCATGTCCCATCATCGCAG | |
| GmEXLA1-RT-F2 | ACTGAAGCTTGGCATTGCTG | gmexla1突变体实时定量分析 Real-time quantitative analysis of gmexla1 mutant |
| GmEXLA1-RT-R2 | CATACTGCCCCGTGGTTTCT | |
| GmActin11-F | ATCTTGACTGAGCGTGGTTATTCC | |
| GmActin11-R | GCTGGTCCTGGCTGTCTCC |
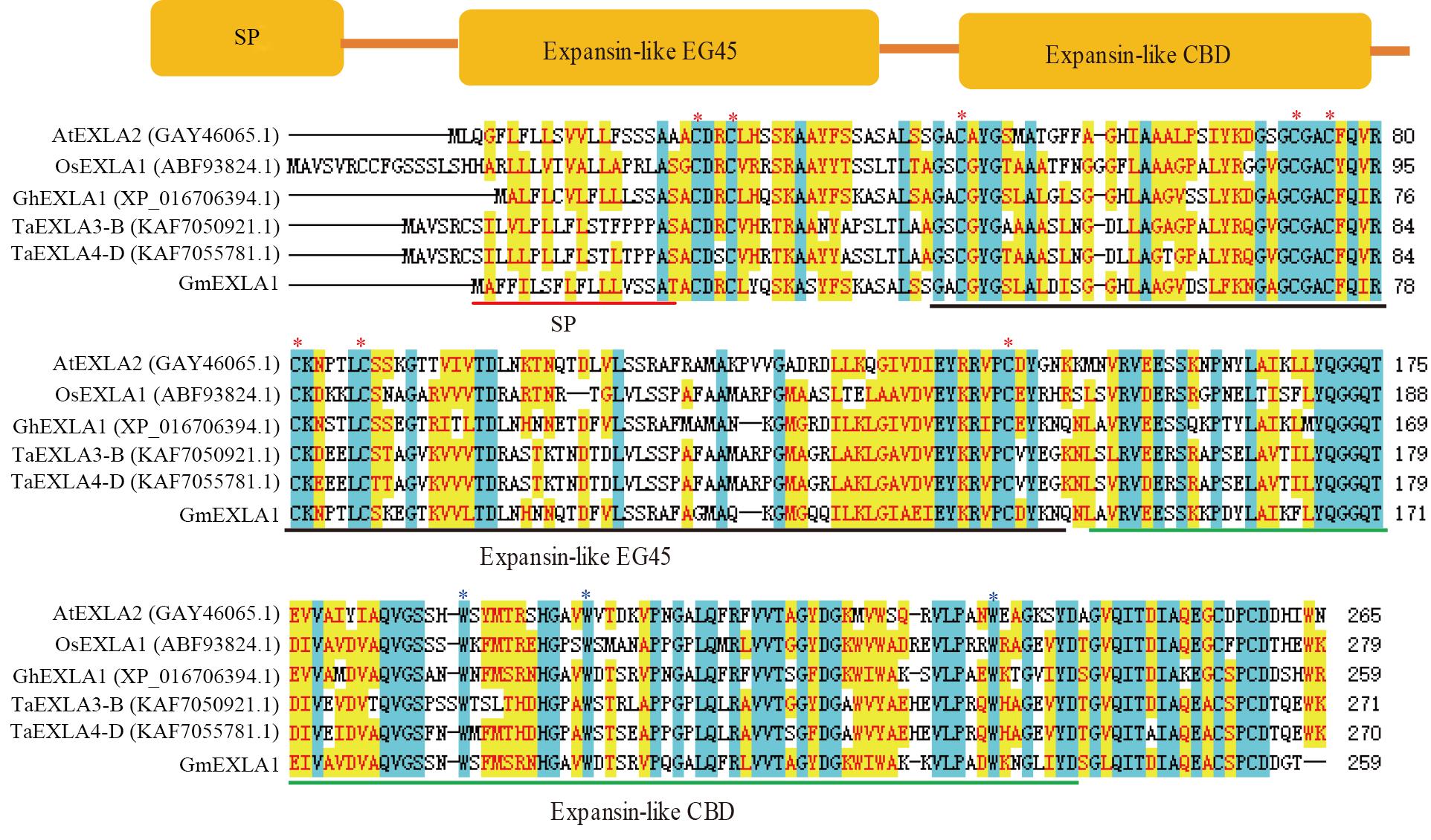
Fig. 1 Conserved domain analysis of GmEXLA1Note: SP—Signal peptide; Expansin-like EG45—Catalytic domain; Expansin-like CBD—Binding domain;* indicates conserved cysteine and tryptophan.
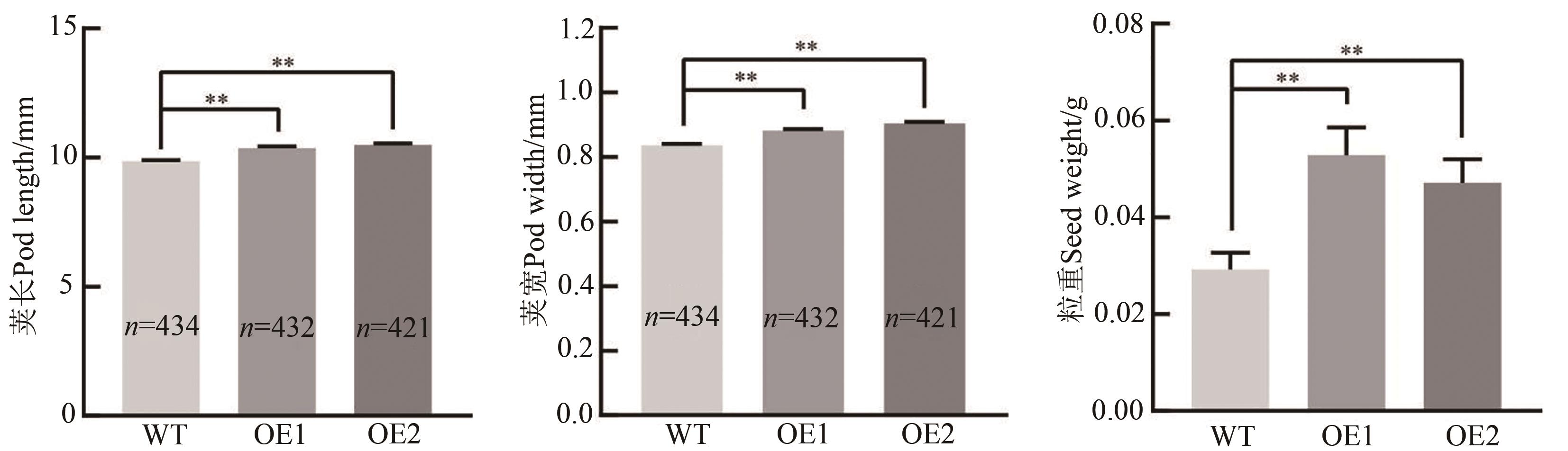
Fig. 4 Pod length, pod width and seed weight per plant of Arabidopsis over-expressed GmEXLA1Note: WT—Wild-type; OE1~OE2—Over-expression lines with GmEXLA1; ** indicates significant difference at P<0.01 level.

Fig. 5 Molecular identification of soybean gmexla1 mutant and its mutation positionA: PCR identification of soybean gmexla1 mutant, M—DNA marker DL2000, 1—Williams 82, 2—gmexla1 mutant, 3—Blank control; B: DNA sequencing result of PCR amplification, the mutant C/T was marked in yellow background; C: qRT-PCR identification of soybean gmexla1 mutant; D: Detection of induced protein in soybean gmexla1 mutant, M—Protein marker, 1—GmEXLA1 in Williams 82 before induced, 2—GmEXLA1 in Williams 82 after induced, 3—gmexla1 in mutant before induced, 4—gmexla1 in mutant after induced; E: gmexla1 mutation position
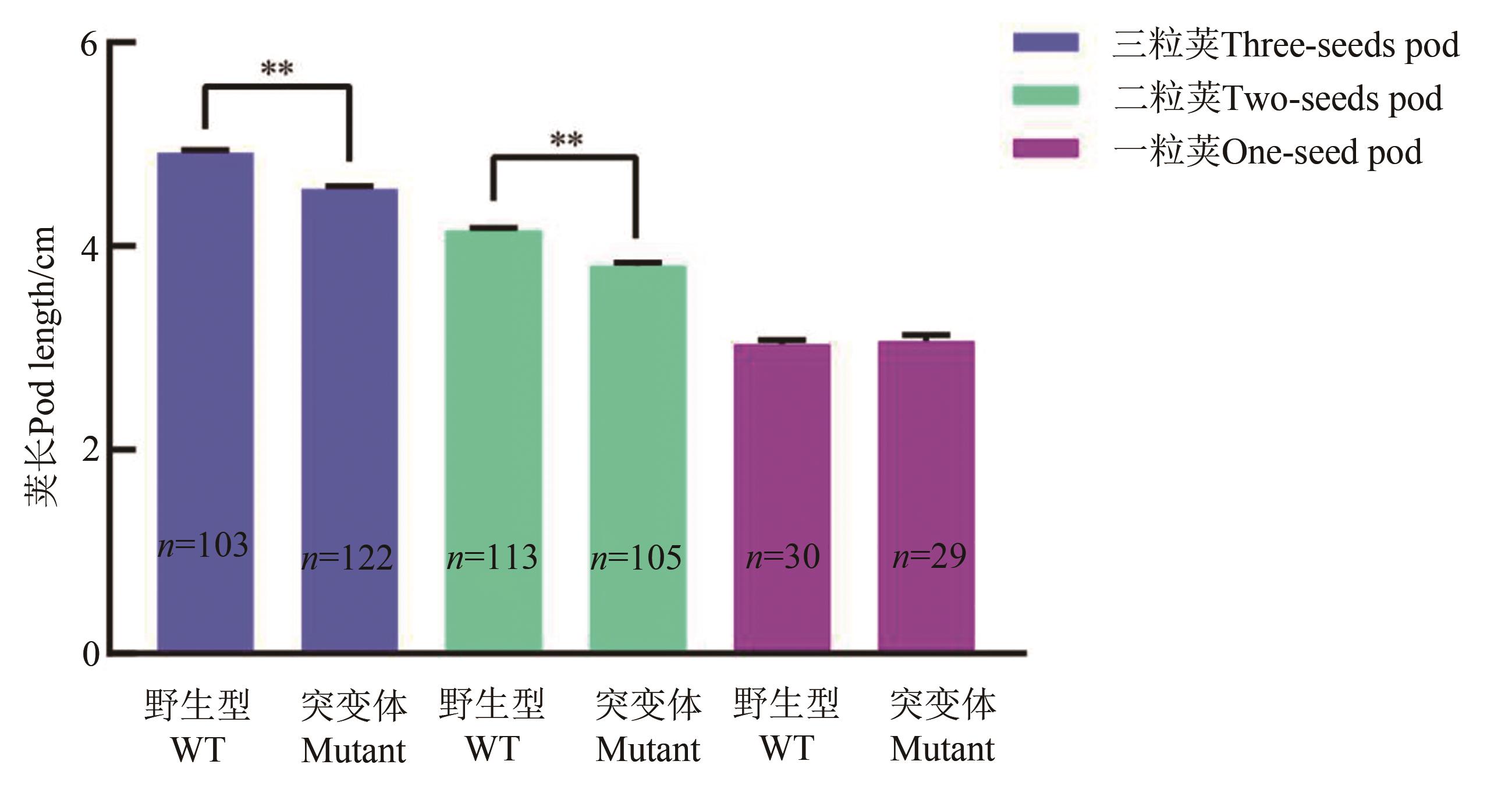
Fig. 6 Comparison of pod length between Williams82 and its gmexla1 mutantNote: WT—Williams 82 wild-type control; Mutant— gmexla1 mutant; ** indicates significant difference at P<0.01 level.
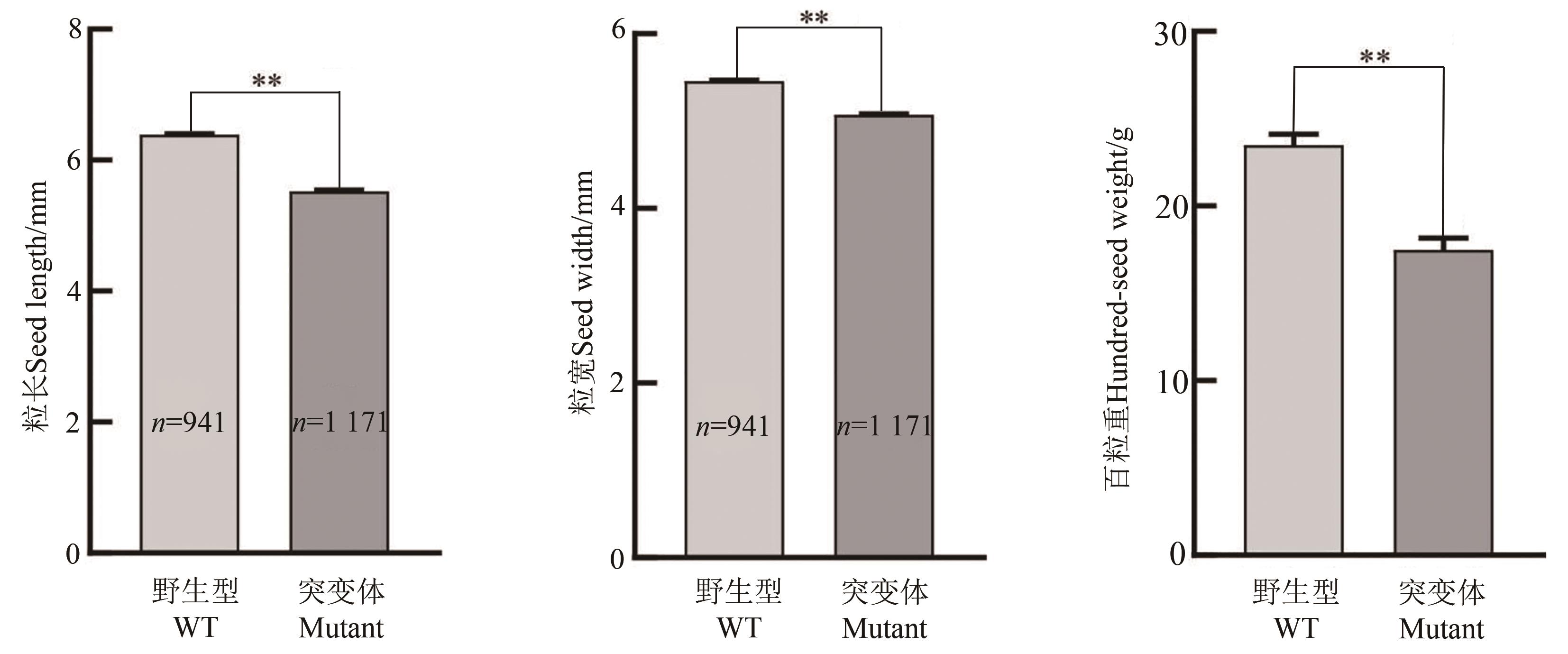
Fig. 7 Comparison of seed-length, -width and hundred-seed weight between Williams82 and its gmexla1 mutantNote: ** indicates significant difference at P<0.01 level.
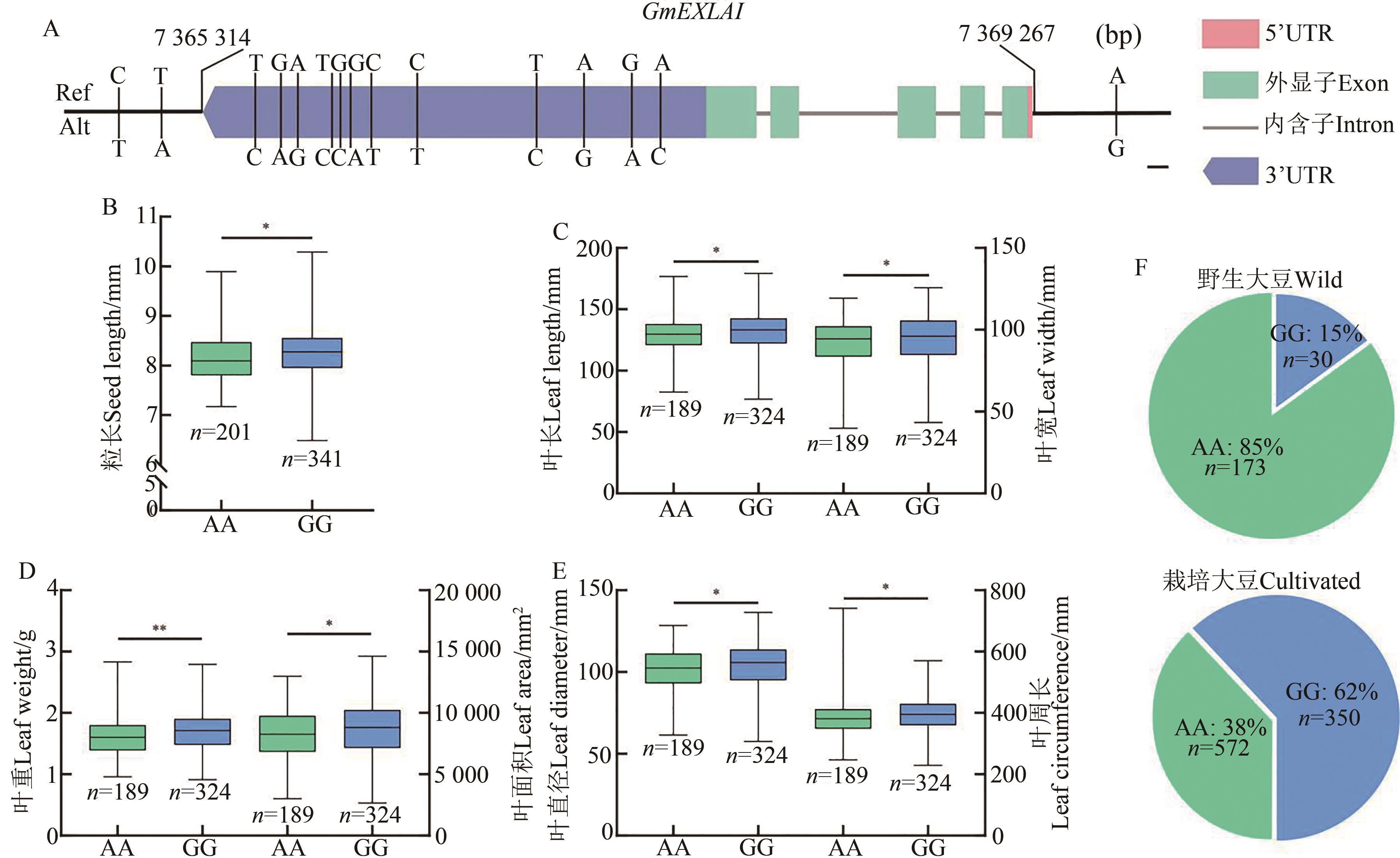
Fig. 8 SNPs and distribution frequency of GmEXLA1 in wild and cultivated soybeansA: SNPs in GmEXLA1, the SNP in promoter region is A/G variation; B~E: AA- and GG-type related traits, based of on (upstream SNP (A/G)), * and ** indicate significant difference at P<0.05 and P<0.01 levels, respectively; F: Distribution frequency of upstream SNP (A/G) in wild and cultivated soybeans;
| 1 | SHEN Y T, LIU J, GENG H Y, et al.. De novo assembly of a Chinese soybean genome [J]. Sci. China Life Sci., 2018, 61: 871-884. |
| 2 | MCQUEEN-MASON S, DURACHKO D M, COSGROVE D J. Two endogenous proteins that induce cell wall extension in plants [J]. Plant Cell, 1992, 4(11): 1425-1433. |
| 3 | KOK B Ö, ALTUNOGLU Y C, ÖNCÜL A B, et al.. Expansin gene family database: a comprehensive bioinformatics resource for plant expansin multigene family [J/OL]. J. Bioinf. Comput. Biol., 2023, 21(3): 2350015 [2024-03-13].. |
| 4 | HAN Z S, LIU Y L, DENG X, et al.. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.) [J/OL]. BMC Genomics, 2019, 20: 101 [2024-03-13]. . |
| 5 | PARK S, LI F F, RENAUD J, et al.. NbEXPA1, an α-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection [J]. Plant J., 2017, 92: 846-861. |
| 6 | HEPLER N K, BOWMAN A, CAREY R E, et al.. Expansin gene loss is a common occurrence during adaptation to an aquatic environment [J]. Plant J., 2020, 101: 666-680. |
| 7 | 蔡兆琴,龙婷晞,肖冬,等.甘薯扩展蛋白IbEXPA2基因克隆和表达分析[J].分子植物育种,2023, 21(5): 1401-1407. |
| CAI Z Q, LONG T X, XIAO D, et al.. Cloning and expression analysis of IbEXPA2 from sweet potato [J]. Mol. Plant Breeding, 2023, 21(5): 1401-1407. | |
| 8 | LIU X P, DONG S Y, MIAO H, et al.. Genome-wide analysis of expansins and their role in fruit spine development in cucumber (Cucumis sativus L.) [J]. Hortic. Plant J., 2022, 8(6): 757-768. |
| 9 | BEREZHNEVA Z A, MUSIN K G, KULUEV B R. Root growth of transgenic tobacco plants with overexpression of expansin and xyloglucan endotransglycosylase genes under cadmium stress [J/OL]. Russ. J. Plant Physiol., 2022, 69: 96 [2024-03-10]. . |
| 10 | 朱益民.桂花OfSVP和扩张蛋白基因在开花过程中的作用[D].杭州:浙江农林大学,2019. |
| ZHU Y M. The role of OfSVP transcription factors and dilated proteins in Osmanthus fragrans flowering [D]. Hangzhou: Zhejiang A&F University, 2019. | |
| 11 | 邹寒艳.水稻OsLIR1维持叶绿体功能的机制及OsEXPB2参与水稻根系发育的功能研究[D].重庆:重庆大学,2015. |
| ZOU H Y. Study on mechanisms of OsLIR1 maintaining chloroplast function and roles of OsEXPB2 in root development in rice (Oryza sativa) [D]. Chongqing: Chongqing University, 2015. | |
| 12 | YAQOOB A, BASHIR S, RAO A Q, et al.. Transformation of α-EXPA1 gene leads to an improved fibre quality in Gossypium hirsutum [J]. Plant Breeding, 2020, 139: 1213-1220. |
| 13 | 赵湾湾,冯力,胡绍彬,等.番木瓜果实软化相关CpEXPA2基因的克隆与表达分析[J].果树学报,2018, 35(7):785-793. |
| ZHAO W W, FENG L, HU S B, et al.. Cloning and expression analysis of CpEXPA2 gene related to softening of papaya fruit [J]. J. Fruit Sci., 2018, 35(7): 785-793. | |
| 14 | 廖嘉明,包钰韬,陈媛,等.黄梁木NcEXPA8基因提高拟南芥种子萌发速度的研究[J].广西植物, 2021, 41(4): 654-661. |
| LIAO J M, BAO Y T, CHEN Y, et al.. Enhancement of Arabidopsis thaliana seed germination speed by NcEXPA8 gene of Neolamarckia cadamba [J]. Guihaia, 2021, 41(4): 654-661. | |
| 15 | LI X X, ZHAO J, TAN Z Y, et al.. GmEXPB2, a cell wall β-expansin gene, affects soybean nodulation through modifying root architecture and promoting nodule formation and development [J]. Plant Physiol., 2015, 169(4): 2640-2653. |
| 16 | SUN Q, LI Y F, GONG D M, et al.. A NAC-EXPANSIN module enhances maize kernel size by controlling nucellus elimination [J/OL]. Nat. Commun., 2022, 13: 5708 [2024-03-13]. . |
| 17 | CALDERINI D F, CASTILLO F M, ARENAS-M A, et al.. Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential [J]. New Phytol., 2021, 230: 629-640. |
| 18 | MAROWA P, DING A M, KONG Y Z. Expansins: roles in plant growth and potential applications in crop improvement [J]. Plant Cell Rep., 2016, 35: 949-965. |
| 19 | GUO W B, ZHAO J, LI X X, et al.. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses [J]. Plant J., 2011, 66: 541-552. |
| 20 | YANG Z J, ZHENG J K, ZHOU H W, et al.. The soybean β-expansin gene GmINS1 contributes to nodule development in response to phosphate starvation [J]. Physiol. Plant, 2021, 172(4): 2034-2047. |
| 21 | 孙丽.大豆膨胀素GmEXPA4基因转化拟南芥及功能验证[D].哈尔滨:东北农业大学,2013. |
| SUN L. The soybean expansion GmEXPA4 gene transformed Arabidopsis thaliana and functional verification [D]. Harbin: Northeast Agricultural University, 2013. | |
| 22 | LEE D, AHN J H, SONG S, et al.. Expression of an expansin gene is correlated with root elongation in soybean [J]. Plant Physiol., 2003, 131: 985-997. |
| 23 | KONG Y B, WANG B, DU H, et al.. GmEXLB1, a soybean expansin-like B gene, alters root architecture to improve phosphorus acquisition in Arabidopsis [J/OL]. Front. Plant Sci., 2019, 10: 808 [2024-03-13]. . |
| 24 | ZHANG M Z, ZHANG X Y, JIANG X Y, et al.. iSoybean: a database for the mutational fingerprints of soybean [J]. Plant Biotechnol. J., 2022, 20: 1435-1437. |
| 25 | SHAO Z Q, SHAO J B, HUO X B, et al.. Identifcation of closely associated SNPs and candidate genes with seed size and shape via deep re-sequencing GWAS in soybean [J]. Theor. Appl. Genet., 2022, 135: 2341-2351. |
| 26 | ZHANG H Y, JIANG H, HU Z B, et al.. Development of a versatile resource for post-genomic research through consolidating and characterizing 1500 diverse wild and cultivated soybean genomes [J/OL]. BMC Genomics, 2022, 23(1): 250 [2024-03-13]. . |
| 27 | CHEN Y H, REN Y Q, ZHANG G Q, et al.. Overexpression of the wheat expansin gene TaEXPA2 improves oxidative stress tolerance in transgenic Arabidopsis plants [J]. Plant Physiol. Biochem., 2018, 124: 190-198. |
| 28 | REN Y Q, CHEN Y H, AN J, et al.. Wheat expansin gene TaEXPA2 is involved in conferring plant tolerance to Cd toxicity [J]. Plant Sci., 2018, 270: 245-256. |
| 29 | 丁安明,陈志华,杨懿德,等. NtabEXPA12基因过表达对烟草叶片发育及抗逆性的影响[J].中国烟草科学,2021,42(4):58-66. |
| DING A M, CHEN Z H, YANG Y D, et al.. Overexpression of NtabEXPA12 affects leaf development and abiotic stress tolerance in tobacco [J]. Chin. Tobacco Sci., 2021,42(4): 58-66. | |
| 30 | KRISHNAMURTHY P, MUTHUSAMY M, KIM J A, et al.. Brassica rapa expansin-like B1 gene (BrEXLB1) regulate growth and development in transgenic Arabidopsis and elicits response to abiotic stresses [J]. J. Plant Biochem. Biot., 2019, 28(4): 437-446. |
| 31 | WANG M, LI W Z, FANG C, et al.. Parallel selection on a dormancy gene during domestication of crops from multiple families [J]. Nat. Genet., 2018, 50: 1435-1441. |
| 32 | YONG B, ZHU W W, WEI S M, et al.. Parallel selection of loss-of-function alleles of Pdh1 orthologous genes in warm-season legumes for pod indehiscence and plasticity is related to precipitation [J]. New Phytol., 2023, 240(2): 863-879. |
| [1] | Bing YANG. Research Progress on Pathogenic Mechanism, Host Molecule Response, and Prevention and Control Methods of Soybean Phytophthora sojae [J]. Journal of Agricultural Science and Technology, 2025, 27(3): 133-142. |
| [2] | Guoqiang DUN, Xingpeng WU, Xinxin JI, Fuli ZHANG, Wenyi JI, Yongzhen YANG. Simulation and Optimization of Soybean Plot Metering Device with Double Swing Plate [J]. Journal of Agricultural Science and Technology, 2024, 26(6): 82-90. |
| [3] | Wenyue WANG, Zhipeng YAO, Yang YU, Yiqiang GE. Scientific and Technological Innovation of Soybean Seed Industry in China:Current Situation and Strategy [J]. Journal of Agricultural Science and Technology, 2024, 26(3): 1-6. |
| [4] | Haosen YANG. Impact of Industrialization of Soybean Bio-breeding in China on Trade Dependency [J]. Journal of Agricultural Science and Technology, 2024, 26(11): 15-22. |
| [5] | Jianmin YAO, Junkui MA, Zhongxiang WANG, Xinyuan BI, Ruizhen LI, Ruiping YANG, Zhao LIU, Fenghui GUO. Application Effect of Full Biodegradable Water Permeable Plastic Film in Soybean-Maize Belt Composite Planting [J]. Journal of Agricultural Science and Technology, 2023, 25(9): 178-185. |
| [6] | Rui TIAN, Hua ZHANG, Meihong HUANG, Zhenqi SHAO, Xihuan LI, Caiying ZHANG. Mining of Candidate Genes and Genetic Loci Conferring Drought Tolerance in Soybean [J]. Journal of Agricultural Science and Technology, 2023, 25(9): 69-82. |
| [7] | Chenyang ZHANG, Minggang XU, Fei WANG, Ran LI, Nan SUN. Effects of Manure Application on Soybean Yield and Soil Nutrients in China [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 148-156. |
| [8] | Boyang KE, Wenlong LI, Caiying ZHANG. Expressions of SWEET Genes During Pod and Seed Developments and Under Different Stress Conditions in Soybean [J]. Journal of Agricultural Science and Technology, 2023, 25(8): 33-52. |
| [9] | Yaqian SUN, Shiliang CHEN, Jiahao CHU, Xihuan LI, Caiying ZHANG. Mining of QTLs and Candidate Genes for Pod and Seed Traits via Combining BSA-seq and Linkage Mapping in Soybean [J]. Journal of Agricultural Science and Technology, 2023, 25(7): 29-42. |
| [10] | Jian HU, Gang CHE, Lin WAN, Huiru ZHOU, Guang LI. Study of Soybean Row Line Extraction Method Under High Light Conditions [J]. Journal of Agricultural Science and Technology, 2023, 25(5): 106-111. |
| [11] | Zhenqi SHAO, Wenlong LI, Youbin KONG, Hui DU, Zhanjun LI, Xihuan LI, Caiying ZHANG. Identification of Soybean Quality Traits and Screening of Elite Germplasms in Huang-Huai-Hai Ecoregion [J]. Journal of Agricultural Science and Technology, 2023, 25(11): 58-69. |
| [12] | Zhanwu YANG, Hui DU, Xinzhu XING, Wenlong LI, Youbin KONG, Xihuan LI, Caiying ZHANG. Functional Analysis of Cytochrome P450 Family GmCYP78A71 in Soybean Nodulation [J]. Journal of Agricultural Science and Technology, 2023, 25(1): 50-57. |
| [13] | Zhenxiang TIAN, Wei DING, Zhuo CHENG, Hangyu DAI. Isolation of Endophytic Bacteria in Soybean and Its Action Effect [J]. Journal of Agricultural Science and Technology, 2022, 24(6): 47-57. |
| [14] | Kuiyuan CHEN, Hui LIU, Wei DING. Effect of Glyphosate on Soil Nutrient and the Functional Enzyme Activities in Soybean Fields [J]. Journal of Agricultural Science and Technology, 2022, 24(5): 180-188. |
| [15] | Ming CHENG, Ying ZHU, Xiaonan WANG, Ping LUO, Yong CHEN, Zhuanfang HAO, Zhangying XI. Drought Resistance Regulated by Allelic Variations of ZmSNAC13 in Maize [J]. Journal of Agricultural Science and Technology, 2022, 24(5): 24-31. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号