




















中国农业科技导报 ›› 2025, Vol. 27 ›› Issue (6): 28-38.DOI: 10.13304/j.nykjdb.2024.0409
收稿日期:2024-05-22
接受日期:2024-07-13
出版日期:2025-06-15
发布日期:2025-06-23
通讯作者:
孙思龙
作者简介:孙吉林 E-mail:3236291157@qq.com;
基金资助:
Jilin SUN( ), Jiaqi ZHANG, Fansen MENG, Silong SUN(
), Jiaqi ZHANG, Fansen MENG, Silong SUN( )
)
Received:2024-05-22
Accepted:2024-07-13
Online:2025-06-15
Published:2025-06-23
Contact:
Silong SUN
摘要:
热激蛋白HSP70(heat shock protein 70)家族是一类结构高度保守和功能强大的蛋白家族,然而有关其在二倍体长穗偃麦草(Thinopyrum elongatum)中的研究尚未报道。为研究二倍体长穗偃麦草HSP70基因家族(TeHSP70),基于生物信息学方法对二倍体长穗偃麦草TeHSP70基因进行鉴定,并系统分析其在盐胁迫和多组织中的表达模式。结果表明,在二倍体长穗偃麦草中共鉴定到29个TeHSP70基因,可分为3个亚族,均匀地分布在7条染色体上;其中有26个TeHSP70基因定位于细胞质,且都包含α-螺旋、延伸链、β-折叠和无规则卷曲结构。29个TeHSP70基因在基序上高度保守,但基因结构存在差异,此外在各基因上游均包含多种激素和胁迫响应的顺式作用元件。共线性分析显示,29个TeHSP70基因间存在3对串联重复和片段重复事件;同时与水稻和小麦有着多对同源基因,而与拟南芥同源基因较少。蛋白质互作网络分析显示,有11个TeHSP70蛋白间存在复杂且密切的蛋白质互作网络。表达分析显示,随着盐胁迫的增加(0~300 mmol·L-1)有7个TeHSP70基因持续上调,有6个TeHSP70基因持续下调,其余16个TeHSP70基因未表达或变化特征不连续;有25个TeHSP70基因在10个不同组织中均有表达,有4个TeHSP70s基因仅在部分组织中表达。以上研究结果为深入解析二倍体长穗偃麦草HSP70功能提供了有效的借鉴和参考。
中图分类号:
孙吉林, 张家琦, 孟凡森, 孙思龙. 二倍体长穗偃麦草HSP70基因家族鉴定及表达模式分析[J]. 中国农业科技导报, 2025, 27(6): 28-38.
Jilin SUN, Jiaqi ZHANG, Fansen MENG, Silong SUN. Genome-wide Identification and Tissue Expression Pattern Analysis of HSP70 Gene Family in Thinopyrum elongatum[J]. Journal of Agricultural Science and Technology, 2025, 27(6): 28-38.
基因名称 Gene name | 基因ID Gene ID | 蛋白质长度Protein length/aa | 分子质量Molecular weight/kD | 等电点pI | 不稳定系数 Instability index | 脂肪系数Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| TeHSP70-01 | GWHGABKY001532 | 887 | 97.70 | 5.45 | 44.50 | 82.33 | -0.458 |
| TeHSP70-02 | GWHGABKY001533 | 835 | 92.56 | 5.07 | 45.59 | 75.13 | -0.513 |
| TeHSP70-03 | GWHGABKY001646 | 740 | 82.25 | 5.28 | 42.34 | 79.97 | -0.380 |
| TeHSP70-04 | GWHGABKY001898 | 734 | 81.38 | 5.43 | 50.39 | 88.26 | -0.333 |
| TeHSP70-05 | GWHGABKY003413 | 686 | 75.38 | 5.23 | 38.34 | 77.07 | -0.503 |
| TeHSP70-06 | GWHGABKY011778 | 683 | 74.88 | 5.31 | 36.86 | 80.85 | -0.427 |
| TeHSP70-07 | GWHGABKY012581 | 680 | 74.83 | 5.18 | 33.92 | 85.50 | -0.381 |
| TeHSP70-08 | GWHGABKY012586 | 690 | 73.56 | 5.01 | 29.29 | 86.19 | -0.270 |
| TeHSP70-09 | GWHGABKY016707 | 665 | 73.27 | 5.06 | 25.54 | 86.50 | -0.479 |
| TeHSP70-10 | GWHGABKY017407 | 683 | 73.26 | 5.60 | 41.92 | 84.17 | -0.325 |
| TeHSP70-11 | GWHGABKY022426 | 684 | 73.14 | 5.04 | 30.07 | 87.09 | -0.310 |
| TeHSP70-12 | GWHGABKY022433 | 665 | 72.93 | 5.04 | 23.51 | 86.78 | -0.371 |
| TeHSP70-13 | GWHGABKY022437 | 678 | 72.86 | 5.29 | 36.04 | 84.04 | -0.310 |
| TeHSP70-14 | GWHGABKY022852 | 678 | 72.84 | 5.23 | 35.63 | 84.04 | -0.303 |
| TeHSP70-15 | GWHGABKY024486 | 684 | 72.77 | 5.78 | 35.28 | 87.49 | -0.268 |
| TeHSP70-16 | GWHGABKY024546 | 677 | 72.55 | 5.36 | 38.85 | 85.48 | -0.292 |
| TeHSP70-17 | GWHGABKY024596 | 657 | 71.80 | 5.17 | 36.09 | 79.15 | -0.446 |
| TeHSP70-18 | GWHGABKY025756 | 653 | 71.65 | 5.21 | 34.54 | 79.62 | -0.458 |
| TeHSP70-19 | GWHGABKY026068 | 667 | 71.63 | 5.06 | 27.89 | 88.22 | -0.296 |
| TeHSP70-20 | GWHGABKY027854 | 640 | 71.35 | 5.44 | 31.47 | 99.83 | -0.088 |
| TeHSP70-21 | GWHGABKY030282 | 660 | 71.31 | 5.07 | 27.51 | 88.53 | -0.319 |
| TeHSP70-22 | GWHGABKY032027 | 648 | 71.19 | 5.08 | 34.79 | 83.4 | -0.394 |
| TeHSP70-23 | GWHGABKY034860 | 648 | 71.02 | 5.13 | 33.73 | 81.16 | -0.415 |
| TeHSP70-24 | GWHGABKY035578 | 651 | 70.85 | 7.82 | 47.67 | 92.78 | -0.118 |
| TeHSP70-25 | GWHGABKY035973 | 620 | 69.08 | 6.25 | 41.66 | 81.1 | -0.402 |
| TeHSP70-26 | GWHGABKY042123 | 617 | 67.68 | 8.84 | 33.40 | 87.07 | -0.173 |
| TeHSP70-27 | GWHGABKY042494 | 582 | 64.25 | 5.08 | 34.38 | 83.14 | -0.417 |
| TeHSP70-28 | GWHGABKY043121 | 583 | 63.91 | 5.10 | 35.91 | 84.19 | -0.390 |
| TeHSP70-29 | GWHGABKY043154 | 544 | 60.80 | 7.58 | 39.85 | 83.46 | -0.388 |
表1 TeHSP70s蛋白的理化性质
Table 1 Physicochemical properties of TeHSP70s protein
基因名称 Gene name | 基因ID Gene ID | 蛋白质长度Protein length/aa | 分子质量Molecular weight/kD | 等电点pI | 不稳定系数 Instability index | 脂肪系数Aliphatic index | 总平均亲水性 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|
| TeHSP70-01 | GWHGABKY001532 | 887 | 97.70 | 5.45 | 44.50 | 82.33 | -0.458 |
| TeHSP70-02 | GWHGABKY001533 | 835 | 92.56 | 5.07 | 45.59 | 75.13 | -0.513 |
| TeHSP70-03 | GWHGABKY001646 | 740 | 82.25 | 5.28 | 42.34 | 79.97 | -0.380 |
| TeHSP70-04 | GWHGABKY001898 | 734 | 81.38 | 5.43 | 50.39 | 88.26 | -0.333 |
| TeHSP70-05 | GWHGABKY003413 | 686 | 75.38 | 5.23 | 38.34 | 77.07 | -0.503 |
| TeHSP70-06 | GWHGABKY011778 | 683 | 74.88 | 5.31 | 36.86 | 80.85 | -0.427 |
| TeHSP70-07 | GWHGABKY012581 | 680 | 74.83 | 5.18 | 33.92 | 85.50 | -0.381 |
| TeHSP70-08 | GWHGABKY012586 | 690 | 73.56 | 5.01 | 29.29 | 86.19 | -0.270 |
| TeHSP70-09 | GWHGABKY016707 | 665 | 73.27 | 5.06 | 25.54 | 86.50 | -0.479 |
| TeHSP70-10 | GWHGABKY017407 | 683 | 73.26 | 5.60 | 41.92 | 84.17 | -0.325 |
| TeHSP70-11 | GWHGABKY022426 | 684 | 73.14 | 5.04 | 30.07 | 87.09 | -0.310 |
| TeHSP70-12 | GWHGABKY022433 | 665 | 72.93 | 5.04 | 23.51 | 86.78 | -0.371 |
| TeHSP70-13 | GWHGABKY022437 | 678 | 72.86 | 5.29 | 36.04 | 84.04 | -0.310 |
| TeHSP70-14 | GWHGABKY022852 | 678 | 72.84 | 5.23 | 35.63 | 84.04 | -0.303 |
| TeHSP70-15 | GWHGABKY024486 | 684 | 72.77 | 5.78 | 35.28 | 87.49 | -0.268 |
| TeHSP70-16 | GWHGABKY024546 | 677 | 72.55 | 5.36 | 38.85 | 85.48 | -0.292 |
| TeHSP70-17 | GWHGABKY024596 | 657 | 71.80 | 5.17 | 36.09 | 79.15 | -0.446 |
| TeHSP70-18 | GWHGABKY025756 | 653 | 71.65 | 5.21 | 34.54 | 79.62 | -0.458 |
| TeHSP70-19 | GWHGABKY026068 | 667 | 71.63 | 5.06 | 27.89 | 88.22 | -0.296 |
| TeHSP70-20 | GWHGABKY027854 | 640 | 71.35 | 5.44 | 31.47 | 99.83 | -0.088 |
| TeHSP70-21 | GWHGABKY030282 | 660 | 71.31 | 5.07 | 27.51 | 88.53 | -0.319 |
| TeHSP70-22 | GWHGABKY032027 | 648 | 71.19 | 5.08 | 34.79 | 83.4 | -0.394 |
| TeHSP70-23 | GWHGABKY034860 | 648 | 71.02 | 5.13 | 33.73 | 81.16 | -0.415 |
| TeHSP70-24 | GWHGABKY035578 | 651 | 70.85 | 7.82 | 47.67 | 92.78 | -0.118 |
| TeHSP70-25 | GWHGABKY035973 | 620 | 69.08 | 6.25 | 41.66 | 81.1 | -0.402 |
| TeHSP70-26 | GWHGABKY042123 | 617 | 67.68 | 8.84 | 33.40 | 87.07 | -0.173 |
| TeHSP70-27 | GWHGABKY042494 | 582 | 64.25 | 5.08 | 34.38 | 83.14 | -0.417 |
| TeHSP70-28 | GWHGABKY043121 | 583 | 63.91 | 5.10 | 35.91 | 84.19 | -0.390 |
| TeHSP70-29 | GWHGABKY043154 | 544 | 60.80 | 7.58 | 39.85 | 83.46 | -0.388 |
蛋白名称 Protein name | 亚细胞定位 Subcelluar localization | α-螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β-折叠 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|---|
| TeHSP70-01 | 周质Periplasmic | 43.29 | 13.19 | 2.59 | 40.92 |
| TeHSP70-02 | 细胞质Cytoplasmic | 43.11 | 14.37 | 2.75 | 39.76 |
| TeHSP70-03 | 细胞质Cytoplasmic | 45.41 | 14.46 | 2.70 | 37.43 |
| TeHSP70-04 | 细胞质Cytoplasmic | 48.91 | 15.12 | 3.13 | 32.83 |
| TeHSP70-05 | 细胞质Cytoplasmic | 42.71 | 17.20 | 6.27 | 33.82 |
| TeHSP70-06 | 细胞质Cytoplasmic | 42.90 | 17.72 | 6.59 | 32.80 |
| TeHSP70-07 | 细胞质Cytoplasmic | 40.74 | 18.53 | 6.62 | 34.12 |
| TeHSP70-08 | 外膜Outermembrane | 41.30 | 19.13 | 6.23 | 33.33 |
| TeHSP70-09 | 细胞质Cytoplasmic | 45.11 | 18.05 | 6.62 | 30.23 |
| TeHSP70-10 | 细胞质Cytoplasmic | 43.48 | 19.62 | 6.73 | 30.16 |
| TeHSP70-11 | 细胞质Cytoplasmic | 38.45 | 20.03 | 7.75 | 33.77 |
| TeHSP70-12 | 细胞质Cytoplasmic | 43.46 | 18.50 | 6.92 | 31.13 |
| TeHSP70-13 | 细胞质Cytoplasmic | 38.20 | 23.16 | 8.11 | 30.53 |
| TeHSP70-14 | 细胞质Cytoplasmic | 44.10 | 19.76 | 7.82 | 28.32 |
| TeHSP70-15 | 外膜Outermembrane | 44.88 | 18.86 | 6.58 | 29.68 |
| TeHSP70-16 | 细胞质Cytoplasmic | 44.90 | 19.65 | 7.98 | 27.47 |
| TeHSP70-17 | 细胞质Cytoplasmic | 41.55 | 18.57 | 6.39 | 33.49 |
| TeHSP70-18 | 细胞质Cytoplasmic | 42.57 | 18.68 | 6.28 | 32.47 |
| TeHSP70-19 | 细胞质Cytoplasmic | 44.08 | 18.59 | 8.40 | 28.94 |
| TeHSP70-20 | 细胞质Cytoplasmic | 42.34 | 22.66 | 6.56 | 28.44 |
| TeHSP70-21 | 细胞质Cytoplasmic | 43.03 | 18.64 | 8.03 | 30.30 |
| TeHSP70-22 | 细胞质Cytoplasmic | 41.67 | 19.44 | 6.79 | 32.10 |
| TeHSP70-23 | 细胞质Cytoplasmic | 41.20 | 16.98 | 6.48 | 35.34 |
| TeHSP70-24 | 细胞质Cytoplasmic | 31.18 | 21.35 | 6.14 | 41.32 |
| TeHSP70-25 | 细胞质Cytoplasmic | 43.39 | 19.03 | 7.10 | 30.48 |
| TeHSP70-26 | 细胞质Cytoplasmic | 40.36 | 19.77 | 6.97 | 32.90 |
| TeHSP70-27 | 细胞质Cytoplasmic | 45.70 | 16.84 | 6.87 | 30.58 |
| TeHSP70-28 | 细胞质Cytoplasmic | 44.08 | 16.12 | 6.69 | 33.10 |
| TeHSP70-29 | 细胞质Cytoplasmic | 46.32 | 18.20 | 6.99 | 28.49 |
表2 TeHSP70s蛋白亚细胞定位和二级结构预测
Table 2 Subcellular localization and secondary structure of TeHSP70s protein
蛋白名称 Protein name | 亚细胞定位 Subcelluar localization | α-螺旋 Alpha-helix/% | 延伸链 Extended strand/% | β-折叠 Beta-turn/% | 无规则卷曲 Random coil/% |
|---|---|---|---|---|---|
| TeHSP70-01 | 周质Periplasmic | 43.29 | 13.19 | 2.59 | 40.92 |
| TeHSP70-02 | 细胞质Cytoplasmic | 43.11 | 14.37 | 2.75 | 39.76 |
| TeHSP70-03 | 细胞质Cytoplasmic | 45.41 | 14.46 | 2.70 | 37.43 |
| TeHSP70-04 | 细胞质Cytoplasmic | 48.91 | 15.12 | 3.13 | 32.83 |
| TeHSP70-05 | 细胞质Cytoplasmic | 42.71 | 17.20 | 6.27 | 33.82 |
| TeHSP70-06 | 细胞质Cytoplasmic | 42.90 | 17.72 | 6.59 | 32.80 |
| TeHSP70-07 | 细胞质Cytoplasmic | 40.74 | 18.53 | 6.62 | 34.12 |
| TeHSP70-08 | 外膜Outermembrane | 41.30 | 19.13 | 6.23 | 33.33 |
| TeHSP70-09 | 细胞质Cytoplasmic | 45.11 | 18.05 | 6.62 | 30.23 |
| TeHSP70-10 | 细胞质Cytoplasmic | 43.48 | 19.62 | 6.73 | 30.16 |
| TeHSP70-11 | 细胞质Cytoplasmic | 38.45 | 20.03 | 7.75 | 33.77 |
| TeHSP70-12 | 细胞质Cytoplasmic | 43.46 | 18.50 | 6.92 | 31.13 |
| TeHSP70-13 | 细胞质Cytoplasmic | 38.20 | 23.16 | 8.11 | 30.53 |
| TeHSP70-14 | 细胞质Cytoplasmic | 44.10 | 19.76 | 7.82 | 28.32 |
| TeHSP70-15 | 外膜Outermembrane | 44.88 | 18.86 | 6.58 | 29.68 |
| TeHSP70-16 | 细胞质Cytoplasmic | 44.90 | 19.65 | 7.98 | 27.47 |
| TeHSP70-17 | 细胞质Cytoplasmic | 41.55 | 18.57 | 6.39 | 33.49 |
| TeHSP70-18 | 细胞质Cytoplasmic | 42.57 | 18.68 | 6.28 | 32.47 |
| TeHSP70-19 | 细胞质Cytoplasmic | 44.08 | 18.59 | 8.40 | 28.94 |
| TeHSP70-20 | 细胞质Cytoplasmic | 42.34 | 22.66 | 6.56 | 28.44 |
| TeHSP70-21 | 细胞质Cytoplasmic | 43.03 | 18.64 | 8.03 | 30.30 |
| TeHSP70-22 | 细胞质Cytoplasmic | 41.67 | 19.44 | 6.79 | 32.10 |
| TeHSP70-23 | 细胞质Cytoplasmic | 41.20 | 16.98 | 6.48 | 35.34 |
| TeHSP70-24 | 细胞质Cytoplasmic | 31.18 | 21.35 | 6.14 | 41.32 |
| TeHSP70-25 | 细胞质Cytoplasmic | 43.39 | 19.03 | 7.10 | 30.48 |
| TeHSP70-26 | 细胞质Cytoplasmic | 40.36 | 19.77 | 6.97 | 32.90 |
| TeHSP70-27 | 细胞质Cytoplasmic | 45.70 | 16.84 | 6.87 | 30.58 |
| TeHSP70-28 | 细胞质Cytoplasmic | 44.08 | 16.12 | 6.69 | 33.10 |
| TeHSP70-29 | 细胞质Cytoplasmic | 46.32 | 18.20 | 6.99 | 28.49 |
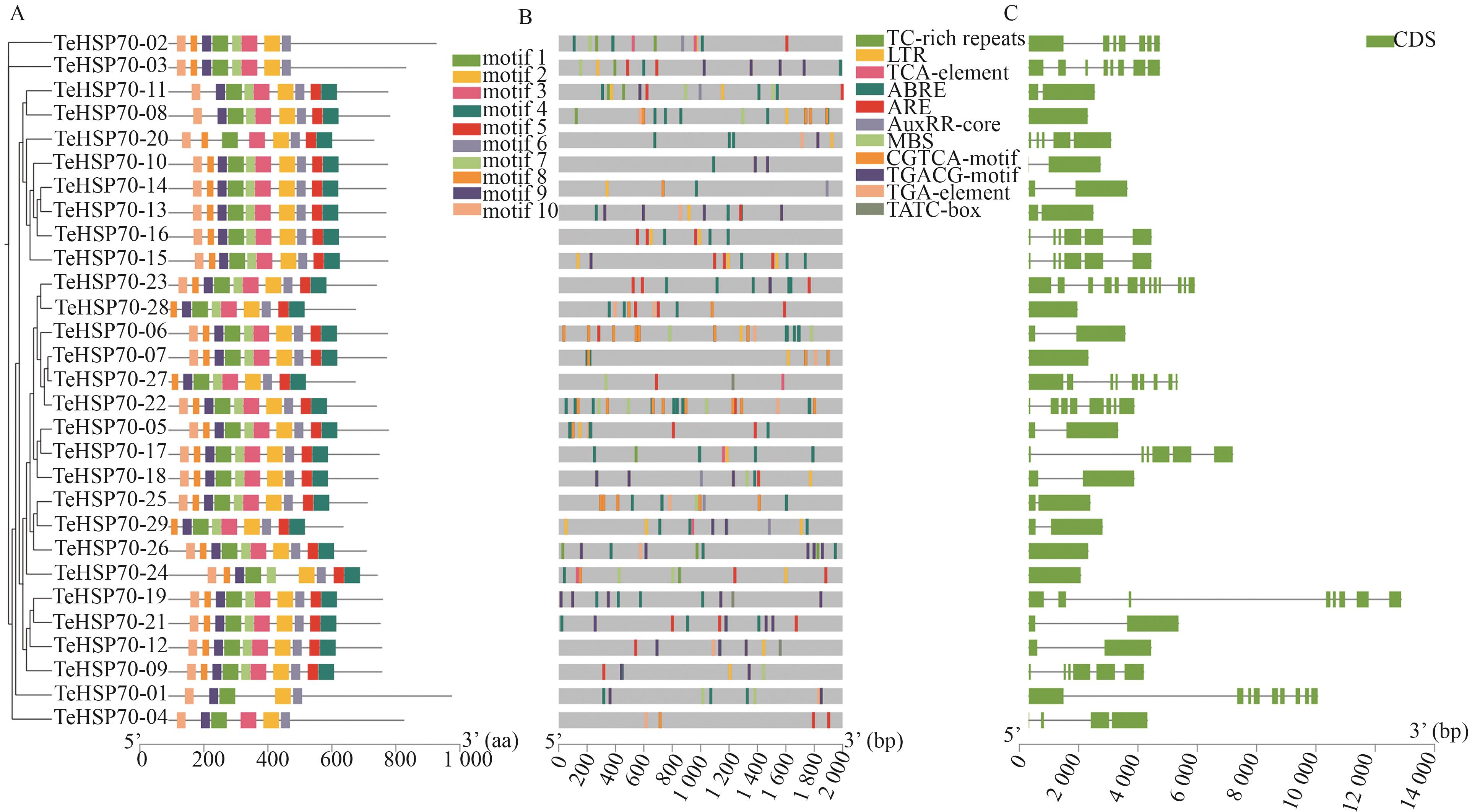
图1 TeHSP70s的保守基序、顺势作用元件及基因结构A:保守基序;B:顺式作用元件;C:基因结构
Fig. 1 Conserved motifs, cis-acting elements and gene structure of TeHSP70sA: Conserved motifs; B: Cis-acting elements; C: Gene structure

图4 TeHSP70s与拟南芥、水稻和小麦基因间的共线性分析
Fig. 4 Collinearity analysis of TeHSP70s with genes from Arabidopsis thaliana, Oryza sativa L. and Triticum aestivum L.
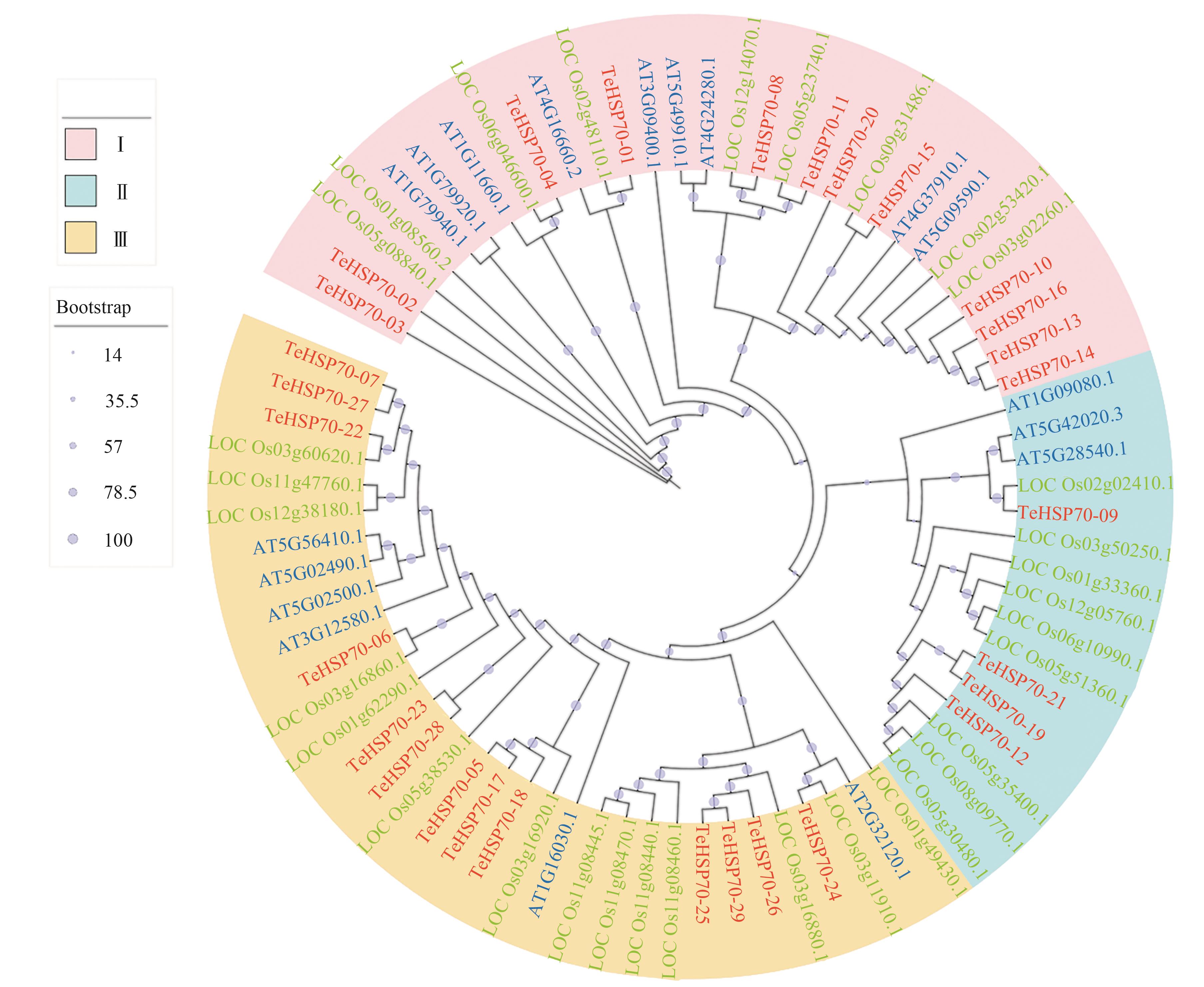
图5 TeHSP70蛋白与水稻和拟南芥HSP70蛋白系统发育进化树注:红、绿、蓝色字分别代表二倍体长穗偃麦草、水稻、拟南芥。
Fig. 5 Phylogenetic tree of TeHSP70 proteins with HSP70 proteins in Oryza sativa L. and Arabidopsis thalianaNote:Red, green and blue letters indicate Thinopyrum elongatum, Oryza sativa L. and Arabidopsis thaliana, respectively.
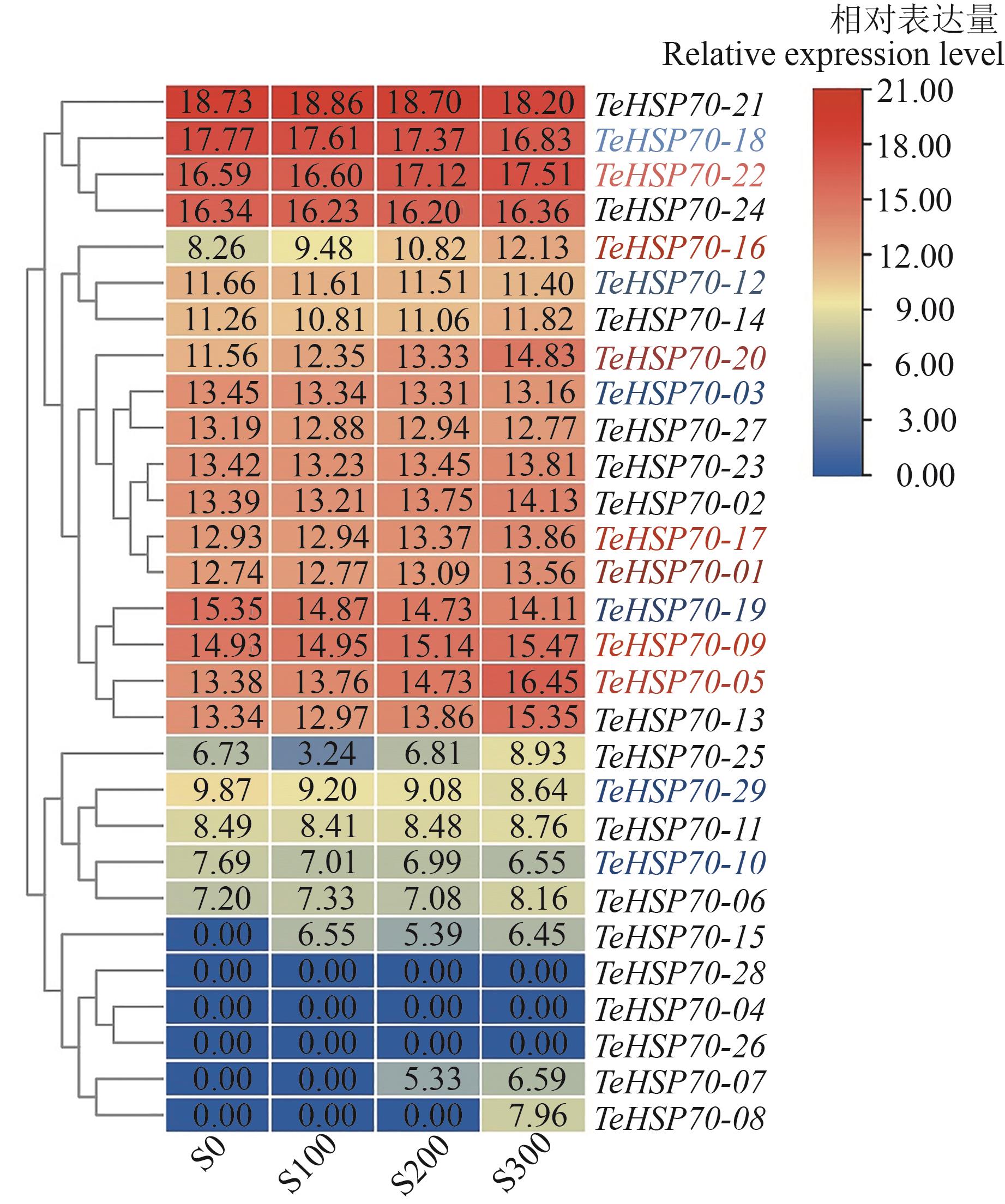
图7 TeHSP70基因在盐胁迫中的表达注:S0、S100、S200和S300分别表示0、100、200和300 mmol·L-1 NaCl处理。
Fig. 7 Expression of TeHSP70 genes in salt stressNote: S0, S100, S200 and S300 indicate 0, 100, 200 and 300 mmol·L-1 NaCl treatments, repectively.
| 1 | HENDRICK J P, HARTL F U. Molecular chaperone functions of heat-shock proteins [J]. Annu. Rev. Biochem., 1993, 62(1):349-384. |
| 2 | SHAN Q, MA F T, WEI J Y, et al.. Physiological functions of heat shock proteins [J]. Curr. Protein Pept. Sci., 2020, 21(8):751-760. |
| 3 | WANG W, VINOCUR B, SHOSEYOV O, et al.. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response [J]. Trends Plant Sci., 2004, 9(5):244-252. |
| 4 | KIANG J G, TSOKOS G C. Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology [J]. Pharmacol. Ther., 1998, 80(2):183-201. |
| 5 | DAVOUDI M, CHEN J, LOU Q. Genome-wide identification and expression analysis of heat shock protein 70 (HSP70) gene family in pumpkin (Cucurbita moschata) rootstock under drought stress suggested the potential role of these chaperones in stress tolerance [J/OL]. Int. J. Mol. Sci., 2022, 23(3):1918 [2024-06-13]. . |
| 6 | DRAGOVIC Z, BROADLEY S A, SHOMURA Y, et al.. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s [J]. EMBO J., 2006, 25(11):2519-2528. |
| 7 | KARLIN S, BROCCHIERI L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution [J]. J. Mol. Evol., 1998, 47(5): 565-577. |
| 8 | HARTL F U, BRACHER A, HAYER-HARTL M. Molecular chaperones in protein folding and proteostasis [J]. Nature, 2011, 475(7356):324-332. |
| 9 | JACOB P, HIRT H, BENDAHMANE A. The heat-shock protein/chaperone network and multiple stress resistance [J]. Plant Biotechnol. J., 2017, 15(4): 405-414. |
| 10 | KAMPINGA H H, CRAIG E A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity [J]. Nat. Rev. Mol. Cell Biol., 2010, 11(8): 579-592. |
| 11 | CHEN B, FEDER M E, KANG L. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress [J]. Mol. Ecol., 2018, 27(15): 3040-3054. |
| 12 | MADAEVA I M, KURASHOVA N A, SEMENOVA N V, et al.. Heat shock protein HSP70 in oxidative stress in apnea patients [J]. Bull. Exp. Biol. Med., 2020, 169(5):695-697. |
| 13 | PAN X, ZHENG Y, LEI K, et al.. Systematic analysis of Heat Shock Protein 70 (HSP70) gene family in radish and potential roles in stress tolerance [J/OL]. BMC Plant Biol., 2024,24(1):2 [2024-06-13]. . |
| 14 | 吴毅,冯雅岚,王添宁,等.小麦及其祖先物种HSP70基因家族鉴定与表达分析 [J].草业学报,2024,33(7):53-67. |
| WU Y, FENG Y l, WANG T N, et al.. Genome-wide identification and expression analysis of the HSP70 gene family in wheat and its ancestral species [J]. Acta Pratac. Sin., 2024, 33(7):53-67. | |
| 15 | MAO H, JIANG C, TANG C, et al.. Wheat adaptation to environmental stresses under climate change: molecular basis and genetic improvement [J]. Mol. Plant., 2023, 16(10):1564-1589. |
| 16 | ZENG J, ZHOU C, HE Z, et al.. Disomic Substitution of 3D chromosome with its homoeologue 3e in tetraploid thinopyrum elongatum enhances wheat seedlings tolerance to salt stress [J/OL]. Int. J. Mol. Sci., 2023, 24(2):1609 [2024-06-13]. . |
| 17 | BAKER L, GREWAL S, YANG C Y, et al.. Exploiting the genome of Thinopyrum elongatum to expand the gene pool of hexaploid wheat [J]. Theor. Appl. Genet., 2020, 133(7):2213-2226. |
| 18 | WANG H, SUN S, GE W, et al.. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat [J/OL]. Science, 2020, 368(6493):5435 [2024-06-13]. . |
| 19 | UL H S, KHAN A, ALI M, et al.. Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses [J/OL]. Int. J. Mol. Sci., 2019, 20(21):5321 [2024-06-13]. . |
| 20 | BERARDINI T Z, REISER L, LI D, et al.. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome [J]. Genesis, 2015, 53(8):474-485. |
| 21 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al.. Pfam: the protein families database in 2021 [J]. Nucl. Acids Res., 2021, 49(D1): D412-D419. |
| 22 | POTTER S C, LUCIANI A, EDDY S R, et al.. HMMER web server: 2018 update [J]. Nucleic Acids Res., 2018, 46(W1):200-204. |
| 23 | LETUNIC I, KHEDKAR S, BORK P. SMART: recent updates, new developments and status in 2020 [J]. Nucleic Acids Res., 2021, 49(D1):458-460. |
| 24 | PAYSAN-LAFOSSE T, BLUM M, CHUGURANSKY S, et al.. InterPro in 2022 [J]. Nucleic Acids Res., 2023, 51(D1): 418-427. |
| 25 | DUVAUD S, GABELLA C, LISACEK F, et al.. Expasy, the Swiss bioinformatics resource portal, as designed by its users [J]. Nucleic Acids Res., 2021, 49(W1):216-227. |
| 26 | YU C S, LIN C J, HWANG J K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions [J]. Protein Sci., 2004, 13(5):1402-1406. |
| 27 | DELEAGE G. ALIGNSEC: viewing protein secondary structure predictions within large multiple sequence alignments [J]. Bioinformatics, 2017, 33(24):3991-3992. |
| 28 | BAILEY T L, JOHNSON J, GRANT C E, et al.. The MEME suite [J]. Nucleic Acids Res., 2015, 43(W1):39-49. |
| 29 | LESCOT M, DEHAIS P, THIJS G, et al.. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res., 2002, 30(1):325-327. |
| 30 | CHEN C, WU Y, LI J, et al.. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol. Plant., 2023, 16(11):1733-1742. |
| 31 | KAWAHARA Y, DE LA BASTIDE M, HAMILTON J P, et al.. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data [J/OL]. Rice (N Y)., 2013, 6(1):4 [2024-06-13]. . |
| 32 | ALAUX M, ROGERS J, LETELLIER T, et al.. Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data [J/OL]. Genome Biol., 2018,19(1):111 [2024-06-13]. . |
| 33 | ROZEWICKI J, LI S, AMADA K M, et al.. MAFFT-DASH: integrated protein sequence and structural alignment [J]. Nucleic Acids Res., 2019, 47(W1):5-10. |
| 34 | MINH B Q, SCHMIDT H A, CHERNOMOR O, et al.. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era [J]. Mol. Biol. Evol., 2020, 37(5):1530-1534. |
| 35 | LETUNIC I, BORK P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation [J]. Nucleic Acids Res., 2021, 49(W1):293-296. |
| 36 | SZKLARCZYK D, KIRSCH R, KOUTROULI M, et al.. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest [J]. Nucleic Acids Res., 2023, 51(D1):638-646. |
| 37 | SHANNON P, MARKIEL A, OZIER O, et al.. Cytoscape: a software environment for integrated models of biomolecular interaction networks [J]. Genome Res., 2003, 13(11):2498-2504. |
| 38 | KIM D, PAGGI J M, PARK C, et al.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype [J]. Nat. Biotechnol., 2019, 37(8):907-915. |
| 39 | ANDERS S, PYL P T, HUBER W. HTSeq—a python framework to work with high-throughput sequencing data [J]. Bioinformatics, 2015, 31(2):166-169. |
| 40 | USMAN M G, RAFII M Y, MARTINI M Y, et al.. Molecular analysis of HSP70 mechanisms in plants and their function in response to stress [J]. Biotechnol. Genet. Eng. Rev., 2017, 33(1):26-39. |
| 41 | LEISTER D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene [J]. Trends Genet., 2004,20(3):116-122. |
| 42 | SHANER L, MORANO K A. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity [J]. Cell Stress Chaperones, 2007,12(1):1-8. |
| 43 | ROSENZWEIG R, NILLEGODA N B, MAYER M P, et al.. The Hsp70 chaperone network [J]. Nat. Rev. Mol. Cell Biol., 2019, 20(11):665-680. |
| 44 | AHMED M, TOTH Z, DECSI K. The impact of salinity on crop yields and the confrontational behavior of transcriptional regulators, nanoparticles, and antioxidant defensive mechanisms under stressful conditions: a review [J/OL]. Int. J. Mol. Sci., 2024, 25(5):2654 [2024-06-13]. . |
| 45 | PENG Z, WANG Y, GENG G, et al.. Comparative analysis of physiological, enzymatic, and transcriptomic responses revealed mechanisms of salt tolerance and recovery in Tritipyrum [J/OL]. Front. Plant Sci., 2021, 12:800081 [2024-06-13]. . |
| [1] | 马蓓, 公杰, 杜银柯, 甘雨薇, 程蓉, 朱波, 易丽霞, 马锦绣, 高世庆. 小麦花粉孔发育相关TaINP1基因鉴定及表达分析[J]. 中国农业科技导报, 2025, 27(4): 22-35. |
| [2] | 董志多, 付秋萍, 黄建, 祁通, 付彦博, 开赛尔·库尔班. 新疆棉花萌发期的耐盐能力分析[J]. 中国农业科技导报, 2025, 27(4): 57-67. |
| [3] | 屈施旭, 孙宇, 所怡祯, 苑海鹏, 张玉红. 外源钙对盐胁迫下火麻生理特性及次生代谢产物的影响[J]. 中国农业科技导报, 2025, 27(2): 80-88. |
| [4] | 徐雪雯, 王兴鹏, 王洪博, 曹振玺. 鼠李糖脂对盐胁迫下棉花幼苗根系生长的调控作用[J]. 中国农业科技导报, 2025, 27(1): 72-79. |
| [5] | 张福林, 奚瑞, 刘宇翔, 陈兆龙, 余庆辉, 李宁. 番茄BURP结构域基因家族全基因组鉴定及表达分析[J]. 中国农业科技导报, 2024, 26(8): 51-62. |
| [6] | 鲍新跃, 陈红敏, 王伟伟, 唐益苗, 房兆峰, 马锦绣, 汪德州, 左静红, 姚占军. 小麦TaCOBL-5基因克隆及表达分析[J]. 中国农业科技导报, 2024, 26(6): 11-21. |
| [7] | 陈明迪, 胡桂花, 张海文, 王旺田. 水稻RR基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(5): 20-29. |
| [8] | 李双, 王爱英, 焦浈, 池青, 孙昊, 焦涛. 盐胁迫下不同抗性小麦幼苗生理生化特性及转录组分析[J]. 中国农业科技导报, 2024, 26(2): 20-32. |
| [9] | 尼格拉·阿布都外力, 魏修振, 门婧婕, 陈凌娜, 陈永坤, 黄耿青. 薰衣草 JAZ和MYC2-like基因家族生物信息学及表达模式分析[J]. 中国农业科技导报, 2024, 26(11): 79-90. |
| [10] | 姜雪敏, 陈向前, 李红燕, 姜奇彦. 小麦盐胁迫响应的代谢组学分析[J]. 中国农业科技导报, 2023, 25(9): 43-56. |
| [11] | 吴雨露, 扈嘉鑫, 陈宇熙, 郑炳松, 闫道良. 外施α-酮戊二酸对盐胁迫下海滨锦葵生长、碳氮磷养分积累及其计量关系的影响[J]. 中国农业科技导报, 2023, 25(7): 170-177. |
| [12] | 麻仲花, 陈娟, 吴娜, 满本菊, 王晓港, 者永清, 刘吉利. 盐胁迫与供磷水平对柳枝稷苗期光合特性与总生物量的影响[J]. 中国农业科技导报, 2023, 25(6): 190-200. |
| [13] | 张帆, 王鸿, 张雪冰, 陈建军. 桃自根砧对NaCl生理生化响应及耐盐阈值分析[J]. 中国农业科技导报, 2023, 25(11): 70-79. |
| [14] | 李峰, 殷丛培, 殷冉, 王凡, 韩永亮, 杨志敏, 刘建成. 燕麦根际土壤细菌多样性对盐胁迫的响应[J]. 中国农业科技导报, 2023, 25(1): 153-165. |
| [15] | 武博琼, 崔东遥, 焦仁和, 宋坚, 湛垚垚, 常亚青. 中间球海胆己糖激酶基因克隆及高温-酸化胁迫对其表达影响的初步研究[J]. 中国农业科技导报, 2022, 24(7): 205-217. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 京公网安备11010802021197号
京公网安备11010802021197号